REVIEW: Role of MicroRNAs in the Regulation of Redox-Dependent Processes
E. V. Kalinina1,a*, V. I. Ivanova-Radkevich1, and N. N. Chernov1
1Peoples’ Friendship University of Russia (RUDN University), 117198 Moscow, Russia* To whom correspondence should be addressed.
Received May 31, 2019; Revised July 4, 2019; Accepted July 4, 2019
Cellular redox homeostasis involves a combination of redox processes and corresponding regulatory systems and represents an important factor ensuring cell viability. Redox-dependent regulation of cellular processes is a multi-level system including not only proteins and enzyme complexes, but also non-coding RNAs, among which an important role belongs to microRNAs. The review focuses on the involvement of miRNAs in the redox-dependent regulation of both ROS (reactive oxygen species)-generating enzymes and antioxidant enzymes with special emphasis on the effects of miRNAs on redox-dependent processes in tumor cells. The impact of ROS on the miRNA expression and the role of the ROS/miRNA feedback regulation in the cell redox state are discussed.
KEY WORDS: reactive oxygen species, cellular redox homeostasis, microRNADOI: 10.1134/S0006297919110026
Abbreviations: ARE, antioxidant-responsive element; GSH, glutathione; HIF, hypoxia-induced factor; miRNA, microRNA; NF-κB, nuclear factor κB; Nrf2, NF-E2-dependent factor 2; OTA, ochratoxin A; Pdcd4, programmed cell death protein 4; Prx, peroxiredoxin; ROS, reactive oxygen species; SOD, superoxide dismutase; Trx, thioredoxin; TrxR, thioredoxin reductase; UTR, untranslated region.
Cellular redox homeostasis determined by a combination of redox
processes and activity of systems controlling their balance is
considered as an important factor ensuring cell viability. Many
biochemical processes in aerobic organisms are associated with the
generation of active oxygen intermediates, such as superoxide
anion-radical
(O·2–), hydroxyl
radical (·OH), hydrogen peroxide
(H2O2), singlet oxygen
(1O2), which have been termed reactive oxygen
species (ROS) [1-4].
Modulation of intracellular ROS levels is essential in maintaining cellular homeostasis. At low and intermediate concentrations, ROS can act as signaling molecules to maintain cell proliferation and differentiation, bioenergetic processes, and apoptosis, as well as participate in the cell stress response via regulation of redox-dependent signaling [4]. Changes in the intracellular ROS levels underlie redox-dependent regulation of gene expression by controlling the activity of transcription factors, such as Nrf2, AP-1, NF-κB, HIF-1, and p53 [3, 4].
On the other hand, ROS exhibit deleterious genotoxic and cytotoxic effects related to their high activity. To prevent the damaging effect of ROS and to maintain cellular homeostasis, aerobic organisms have developed in the evolution a system for the antioxidant defense. The key antioxidant enzymes of this system are catalase, superoxide dismutase (SOD), and glutathione peroxidase (GPx) [3]. The antioxidant system neutralizes ROS and maintains cell homeostasis. It was also demonstrated that its components cooperatively interact with the enzymes involved in the metabolism and detoxification of xenobiotics [5].
However, various endogenous and exogenous factors can shift the cell redox balance via either suppression of the antioxidant system activity or hyperproduction of ROS, which might lead to the emergence of the oxidative/nitrosative stress and subsequent development of various pathologies including atherosclerosis, ischemic heart disease, diabetes, chronic obstructive pulmonary diseases (COPD), Alzheimer’s and Parkinson diseases, and malignant tumors [1, 3].
An important role in the maintenance of redox homeostasis belongs to the redox-dependent proteins of the thioredoxin, glutaredoxin, and peroxiredoxin enzyme systems that control the redox state of the cell through the -SН ↔ -S–S- exchange [6-8].
The dynamic balance between protein glutathionylation and deglutathionylation (protein-SSG ↔ protein-SH) is an important marker of the oxidative stress, because it characterizes the cell protection against oxidation of SH-groups of Cys residues. In this case, the tissue specificity of expression of genes encoding antioxidant enzymes is very important [8].
At present, redox-dependent regulation of cellular homeostasis is considered as a multi-level process that involves not only proteins and enzyme complexes, but also non-coding RNAs [3, 9], i.e., a large group of untranslated RNAs participating in the transcriptional and post-transcriptional mechanisms of RNA-directed gene silencing [9, 10]. This group, which includes transport RNAs (tRNAs), ribosomal RNAs (rRNA), small nuclear RNAs (snRNAs), small nucleolar RNAs (snoRNAs), antisense RNAs (aRNAs), small interfering RNAs (siRNAs), Piwi-interacting RNA (piRNAs), long non-coding RNAs (lncRNA), and microRNAs (miRNA), plays a significant role in the regulation of cell redox status [9, 11, 12].
This review summarizes recent data on the role of miRNAs in the regulation of cell redox-dependent processes.
BIOGENESIS AND FUNCTIONS OF miRNAs
The discovery of RNA-directed suppression of gene expression (gene silencing, RNA interference) with the involvement of miRNAs allowed researchers to suggest a new role of RNA in cell metabolism and regulation of genome function. By October 2018, the number of miRNAs with different functions in the specialized database miRBase (http://mirbase.org) supported by the Manchester University reached 38,589.
miRNAs are non-coding single-stranded RNA molecules 16 to 27 nucleotides (nt) in length (mostly, 22-24 nt). They can be present in the blood serum as extracellular miRNAs from damaged tissues and as circulating miRNAs in the content of microvesicles and in complexes with RNA-binding protein (high-density lipoproteins) [13]. miRNAs cause transcriptional and post-transcriptional gene silencing. Transcriptional gene silencing is usually mediated through methylation of the silenced gene (according to some data, methylation of cytosines in promoter regions) and changes in histone structure (usually, caused by methylation and deacetylation). In both post-transcriptional and transcriptional gene silencing, miRNA and the bound Argonaute protein (endonuclease) form the polyprotein RISC (RNA-induced silencing complex) and RITS (RNA-induced transcriptional silencing complex) [14].
As a rule, miRNAs are not completely homologous to their targets (mRNA or DNA); the same miRNA can act on several targets, which differentiates it from siRNAs that are strictly complementary to their targets. This is related to the fact that siRNAs target heterologous genes from which they are transcribed. According to some data, partial or complete homology to the target mRNA determines either termination of mRNA translation or mRNA degradation, respectively [14, 15].
miRNAs can be encoded by specific genes, gene fragments (both exons and introns), and genome intergenic regions [14].
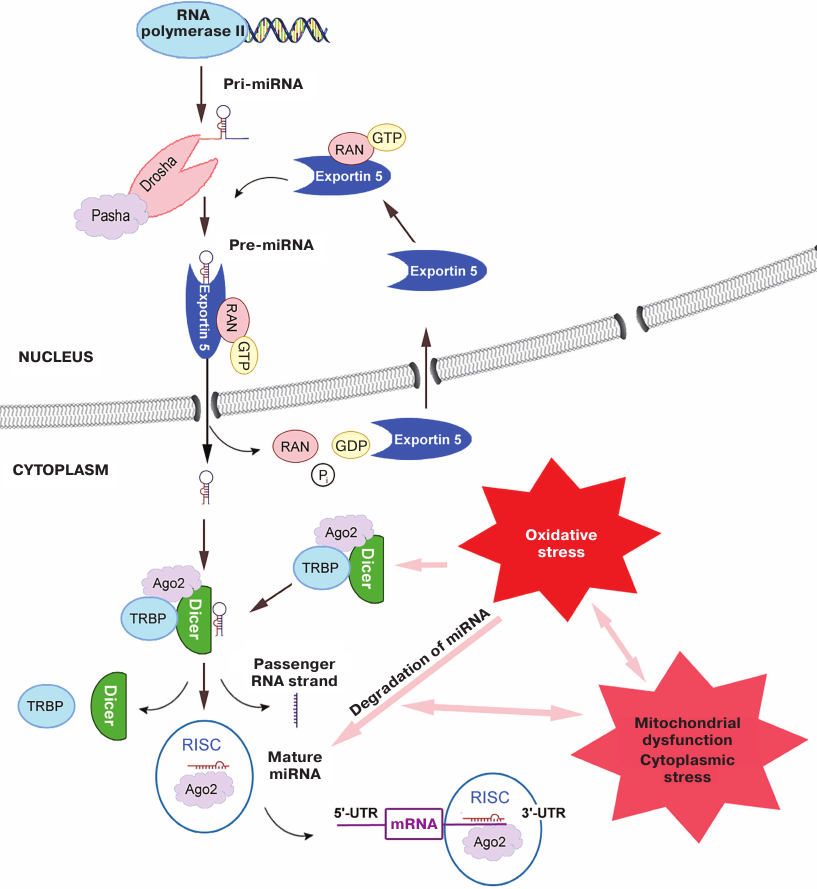
The canonical pathway of miRNA biogenesis starts with the synthesis of pri-miRNA (several thousand nucleotides in size) transcribed from DNA, usually by RNA polymerase II (Fig. 1). pri-miRNA is capped and polyadenylated similarly to mRNA. Due to the presence of inverted repeats (self-complementary regions), pri-miRNAs form a stem-loop structure. In the nucleus, this structure is cleaved by the proteins Drosha (RNase III) and Pasha (or its mammalian analog DGCR8) with the production of pre-miRNA (~70 nt). Pre-miRNA is transported to the cytoplasm with the help of exportin-5 protein. Exportin-5 binds to pre-miRNA in the presence of high levels of RAN-GTP in the nucleus and transfers it through the nuclear pore complex to the cytoplasm, where pre-miRNA is released from the RAN/GTP/exportin-5 complex after GTP hydrolysis. Free exportin-5 reenters the nucleus [15, 16].
Fig. 1. Biogenesis of miRNAs and influence of ROS. RAN is a small GTPase from the family of RAS-like GTPases that provides coupling of the transfer to the GTP hydrolysis; TRBP is an RNA-binding protein; Ago2 is a endonuclease from the Argonaute family; RISC (miRNA-induced silencing complex) is a complex of Ago2 with a mature miRNA single chain and other proteins; 5′- and 3′-UTR are mRNA 5′- and 3′-untranslated regions, respectively.
Due to the presence of sticky 3′-end, pre-miRNA binds in the cytoplasm to the triple protein complex formed by Dicer (ribonuclease III), TRBP (RNA-binding protein), and Ago2 (Argonaute family protein with the endonuclease activity) [17]. During biogenesis of some miRNAs, Ago2 participates in the formation of specific mature miRNA precursor consisting of a shortened hairpin bound to a fragment of ~11-12 nt by cleaving a bond in the 3′-arm. The precursor is transformed into the single-stranded miRNA of ~20 nt by Dicer [15, 17]. The other complementary (passenger) strand of pre-miRNA dissociates and gets degraded. Experiments with mice lacking the DICER gene indicated an extremely important role of this protein in the miRNA maturation, although in the case of RNA interference, the effect of Dicer was less pronounced [18, 19].
After formation of the single-stranded miRNA, Dicer and TRBP are separated, while Ago2 complex with the mature miRNA forms RISC or RITS complex by recruiting other proteins [17].
miRNA in the RISC complex binds to its the target. In the majority of cases, miRNAs suppress translation by binding to the 3′-UTR of the mRNA target [14]. However, they can also interact with other mRNA regions, including 5′-UTRs and exons [20].
Further stages depend on the complementation between miRNA in the content of RISC and target mRNA. In the case of incomplete complementation, translation of the mRNA target is repressed, while full complementation results in mRNA hydrolysis by Ago2 [15].
The above-described pathway of miRNA biogenesis with participation of Drosha, Pasha, exportin-5, and Dicer is considered to be canonical. However, numerous studies have demonstrated the existence of alternative pathways of the miRNA synthesis and maturation. Thus, it is found that activities of some miRNAs were not affected by the knockdown of the DICER, XPO5, or AGO2 genes [21].
miRNAs AND INTRACELLULAR ROS. CONTROL OF REDOX HOMEOSTASIS AND
DEVELOPMENT OF OXIDATIVE STRESS
It has been commonly established that oxidative stress alters expression of many miRNAs (Fig. 1). However, many researchers believe that changes in the redox-dependent processes and their relations with the mechanisms of oxidative stress development have been studied insufficiently [11].
Redox-dependent regulation of miRNA biogenesis. One of the possible redox-dependent mechanisms of miRNA regulation is downregulation of the endonuclease activity of Dicer (ribonuclease III) responsible for the processing of pre-miRNAs. In chronic hypoxia, the level of the DICER gene transcription, stability of the corresponding mRNA, and the content of the Dicer protein itself are reduced, leading to the suppression of expression of certain miRNAs [22]. Thus, a considerable decrease in the Dicer level was observed under hypoxic conditions in the breast cancer cell line [18]; similar results have been obtained in other cell lines, i.e., the effect of hypoxia was not limited to tumor cells. Treatment of cells with inhibitors of HIF (hypoxia-induced factors) hydroxylases also led to the decrease in the Dicer content. At the same time, a decrease in the content of HIF-1α and HIF-2α virtually did not influence the amount of Dicer in the cell. Supposedly, the influence of hypoxia on Dicer (and on the levels of miRNAs through it) is not mediated by HIF but rather characterizes the development of adaptive response to stress. The same conclusion was made by Ho et al. [22] and Wiesen et al. [23], who demonstrated that the activity of Dicer decreased under the action of various stress factors, including ROS.
It was shown that the activity of Dicer in rat microvascular epithelium cells was suppressed by the hydrogen peroxide-induced oxidative stress, leading to the dysregulation of miRNA maturation [24]. Sulforaphane and resveratrol esters (Nrf2 inducers of plant origin) upregulated DICER expression in the same cells. Identification of the ARE (antioxidant-responsive element)-binding sequence in the 5′-flanking region of human and mouse DICER genes led to the conclusion that redox-dependent signaling (including Nrf2/ARE-dependent pathway) plays an important role in the regulation of miRNA biosynthesis [25].
miRNA biogenesis can be also redox-regulated via changes in the activity of the endonuclease Drosha, in particular, through the action of the redox-dependent enzyme glycogen synthase kinase that phosphorylates Drosha at Ser300 and Ser302, thus promoting its translocation to the nucleus [26].
Cleavage of miRNAs precursors can also be regulated in a redox-dependent manner. Thus, activation of the transmembrane serine/threonine protein kinase and endoribonuclease IRE1α that hydrolyzes miRNA precursors at the sites different from the sites of hydrolysis by Dicer by the endoplasmic reticulum (ER) stress caused by accumulation of unfolded proteins, leads to the decrease in the levels of miR-17, miR-34a, miR-96, and miR-125b. This induces activation of caspase-2 mRNA translation with subsequent apoptosis development [27]. Moreover, miRNAs themselves can be directly oxidized by ROS, which alters their stability, structure, and ability to bind to target mRNAs [28].
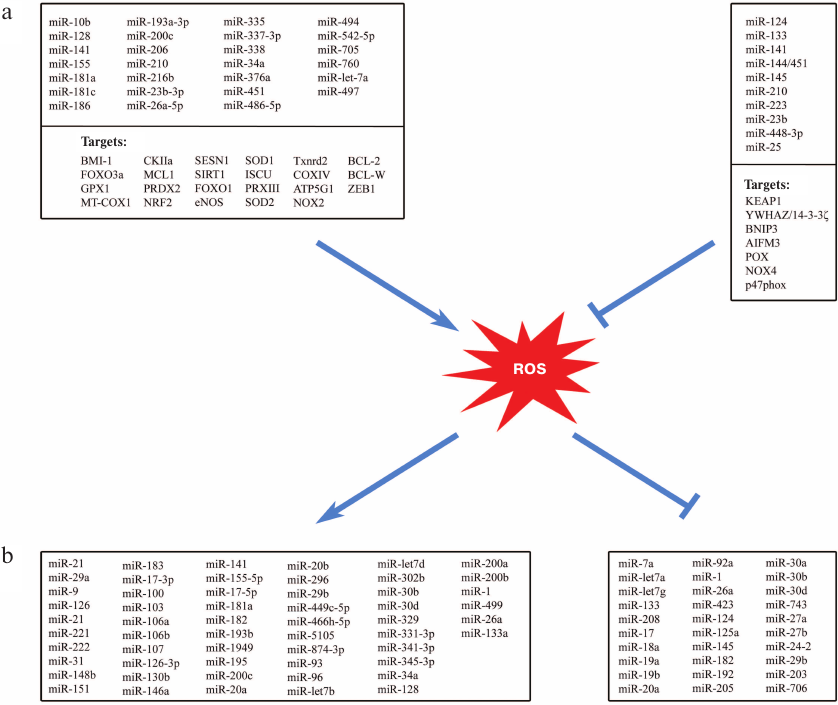
miRNA and redox regulation of antioxidant and ROS-producing systems. Multiple studies demonstrate not only effects of ROS and oxidative stress on miRNAs and their expression but also the influence of miRNA on redox-dependent signaling and enzymes of ROS generation and of antioxidant defense (Fig. 2 and table). Some miRNAs (so-called redox miRs) participate in cell response to ROS and oxidative stress, in particular, via changing the expression of genes encoding antioxidant enzymes (SOD, catalase, peroxiredoxin, glutathione transferase) [29].
Fig. 2. Relationship between miRNAs and ROS. a) Influence of miRNAs on the enzyme systems controlling ROS intracellular levels. b) Influence of oxidative stress on the miRNA expression (→ activation, ⊥ repression).
Regulators of cell redox status as redox-dependent targets of miRNAs
The impact of ROS on the miRNA expression can be different depending on the cell type and experimental conditions. Thus, both increase and decrease in the expression caused by the oxidative stress have been described for miR-1, miR-133, miR-182, miR-20a, miR-26a, miR-29b, miR-30b, and miR-30d, depending on the type of human or rat tissue [9].
Using the TargetScanB database and special software, Engedal et al. predicted that some miRNAs participate in the oxidative stress development and described ROS influence on the miRNA synthesis [11]. This allowed to suggest that miR-15a/b, miR-106a/b, miR-20a, and miR-195 might be involved in the oxidative stress development. Later, the levels of these miRNAs were shown to change in the cells subjected to stress-induced aging associated with the development of oxidative stress [30]. Using this method, the authors classified more than 20 miRNAs as potential participants of the cell response to oxidative stress. Experimental data obtained for six of these miRNAs (miR-9, miR-16, miR-29b, miR-128, miR-144, and miR-200c) confirmed their participation in the cell response to oxidative stress [11].
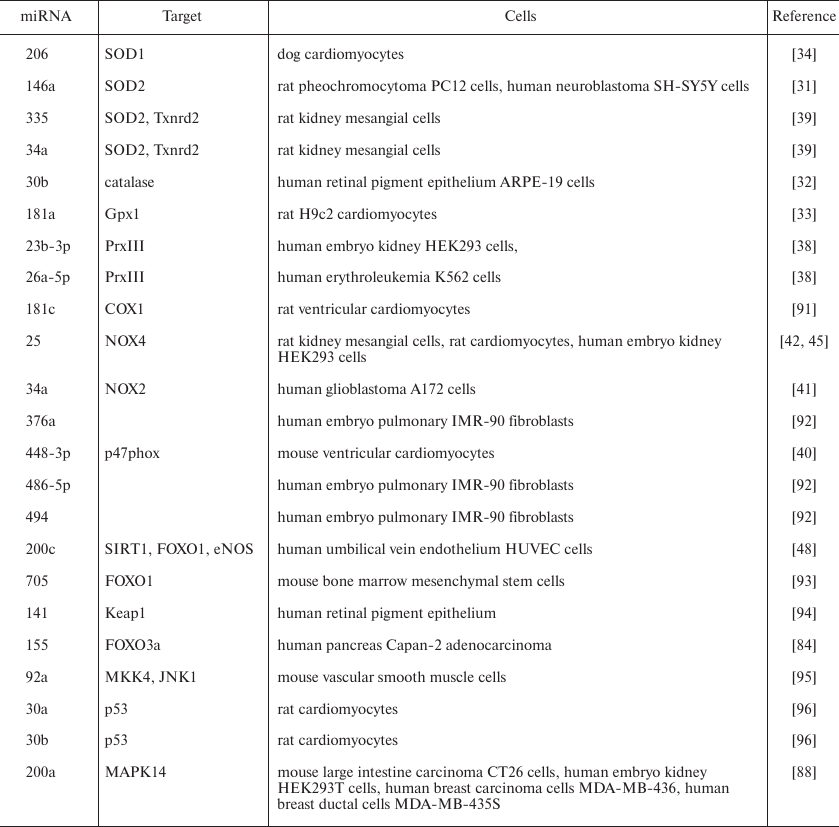
miRNAs that affect expression of the antioxidant enzymes, leading to the oxidative stress aggravation, have been identified. At the same time, ROS induce miRNAs that control expression of genes encoding key antioxidant enzymes, e.g., miR-146a and miR-335 (target – Mn-superoxide dismutase 2, SOD2), miR-206 (target – Cu,Zn-superoxide dismutase 1, SOD1), miR-30b (target – catalase), and miR-181a (target – glutathione peroxidase, isoform GPx1) [31-33]. Upregulation of miR-146a, miR-30b, and miR-181a promotes oxidative stress [9].
SOD catalyzes dismutation of superoxide (O2–) with the formation of less reactive H2O2 that is then reduced to H2O by catalase and glutathione peroxidase [3]. SOD isoforms can be targets for miRNAs. Thus, miR-206 binding to the SOD1 mRNA induces ROS accumulation both in vitro and in vivo, which might be a cause of cardiovascular diseases, in particular, atrial fibrillation [34]. SOD2 mRNA is a target for miR-21-5p, miR-23a-3p, and miR-222-3p, which can be potentially used as biomarkers for predicting heart failure [35].
Some miRNAs (e.g., miR-27a-5p, miR-575, miR-24-3p, miR-143-3p, miR-9, and miR-204-5p) play an important role in the regulation of expression of glutathione peroxidase family members [36]. Application of bioinformatics approaches allowed to predict over 100 miRNAs capable of interacting with the 3′-UTRs in mRNAs of glutathione peroxidases. Note that some miRNAs can bind to mRNAs of several glutathione peroxidase isoforms. Thus, miR-146b-5p can regulate translation of Gpx3 and Gpx6 mRNA [36].
The influence of miRNAs on mRNAs of the thioredoxin family redox-dependent enzymes promotes the antioxidant protective effect of the latter (Fig. 2a). Thus, the action of miR-200c targeting peroxiredoxin 2 (Prx2) and miR-26a-5p and miR-23b-3p, both targeting peroxiredoxin 3 (Prx3) leads to the reduction in the control of the H2O2 content in the cytoplasm and mitochondria by peroxiredoxins, which aggravates oxidative stress [37, 38]. Expression of miR-335 and miR-34a leads to the suppression of TrxR2 activity in the redox-dependent reduction of oxidized mitochondrial Trx2, thus inhibiting reduction of disulfides and promoting deleterious action of ROS [39].
At the same time, some miRNAs reduce oxidative stress via suppressing ROS-generating enzyme systems (Fig. 2b). Thus, miR-448-3p, miR-34a, and miR-25, whose targets are p47ph°x subunit, NOX2, and NOX4 (the latter two proteins are isoforms of ROS-producing NADPH oxidase) and miR-23b, whose target is proline oxidase (POX), suppress ROS generation [40-42]. miR-1 causes decrease in the intracellular level of NOX5 and inhibits production of superoxide anion, whereas miR-1283 expression leads to the decrease in the NOX1-dependent generation of ROS [43, 44]. Suppression of miR-25 expression in rats receiving cholesterol with food was accompanied by the increase in the NOX4 protein level and oxidative stress development in the myocardium cells, resulting in the cardiac muscle dysfunction [45]. Moreover, NOX4 is also a target of miR-182 and miR-9-5p. Impairments in the expression of these miRNAs cause the damage of neurons and development of fibroses, respectively [46, 47]. Downregulation of miR-448-3p, whose target is NOX2, is associated with cardiomyopathy mediated by the NOX2 activation and ROS accumulation in the myocardium cells [40].
Some miRNAs control expression of nitric oxide synthase (NOS) isoforms that produce NO. In the presence of elevated amounts of superoxide anion, NO can form highly reactive peroxynitrite. miR-155 and miR-200c suppress NO generation by decreasing the intracellular levels of eNOS (endothelial NOS) [48, 49], whereas miR-Let7a, miR-294, and miR-721 suppress expression of iNOS (inducible NOS) [50, 51].
Changes in the cell redox status under the influence of miRNAs can be promoted by miRNA-mediated suppression of pentose phosphate pathway of glucose oxidation. Overexpression of miR-1 and miR-206 (whose targets are glucose-6-phosphate dehydrogenase, 6-phosphogluconate dehydrogenase, and transketolase) is accompanied by a dramatic decrease in the intracellular level of NADPH [52], which causes a decrease in the activity of NADPH-dependent oxidoreductases (including thioredoxin reductase, glutathione reductase, cytochrome P-450 reductase, and NADPH oxidase), leading to further imbalance of the ROS/antioxidant ratio and disturbance of cell redox homeostasis.
Mitochondria are major providers of ROS in the cell. miRNAs can regulate the metabolic activity of mitochondria [53]. Under hypoxic conditions, some types of cell upregulate expression of miR-210, resulting in the activation of ROS generation by the mitochondria. A decrease in the production of carnitine O-acetyltransferase, a mitochondrial enzyme involved in the fatty acid metabolism, by miR-378 proves the ability of miRNAs to control the energy state of the cell.
Many studies have indicated that miRNAs can regulate redox-depending signaling and its relationship with other pathways of signal transduction.
It was found that miR-205 binds to the PHD1/EGLN2 gene encoding prolyl hydroxylase 1 and suppressed transcription of EGLN2 (α-ketoglutarate-dependent hydroxylase 2), which leads to the inhibition of transcription factors HIF and ATF4 (transcription activating factor 4) involved in the expression of the SOD1, SOD2 and НO-1 genes. As a result, miR-205 decreases ROS levels in the cell and protects it against oxidative stress [54]. An increase in the content of miR-205 in the cells does not affect cell growth and morphology but increases cell resistance to the oxidative and ER stresses. Based on these data, miR-205 might be a promising candidate for a potential therapeutic agent [54].
Cell redox homeostasis can be regulated by miRNAs through modulating the intracellular content of the redox-dependent transcription factor Nrf2. Some genes encoding proteins that play an essential role in the protection against oxidative stress and xenobiotics contain antioxidant-responsive elements (AREs) in their promoter regions. These are genes for the antioxidant enzymes (Mn-SOD, catalase, heme oxygenase 1), enzymes supporting the intracellular level of GSH via de novo synthesis and GSSG reduction (H- and L-subunits of γ-glutamyl-cysteine ligase, γ-glutamyl transferase, glutathione reductase), redox-dependent proteins (thioredoxin 1, thioredoxin reductase 1, peroxiredoxin 1, peroxiredoxin 2), glutathione S-transferase isoforms GSTP1-1 and GSTA4-4, NADPH:quinone oxidoreductase 1, ferritin H- and L-subunits, etc. AREs are regulated through binding with Nrf2 that forms a heterodimer with the low-molecular-weight MafK protein. Normally, Nrf2 is present in the cytoplasm in a complex with the protein Keap1. In response to oxidative stress and some inducers, cysteine residues within Keap1 are oxidized, leading to the Nrf2 release and translocation to the nucleus, where it binds with the ARE-containing gene promoters [55].
miR-93 represses the NRF2 gene with the following reduction in the cell antioxidant defense, while miR-153, miR-27a, miR-142-5p, and miR-144 decrease the Nrf2 level post-transcriptionally, leading to a similar effect [56, 57]. In the p53-dependent tumor cells, miR-34a suppresses expression of the NRF2 gene, thus activating development of the oxidative stress [58]. Similar effect is produced by miR-93-5p, miR-27a-5p, miR-28, and miR-142-5p. miRNAs can influence NRF2 expression indirectly; however, the mechanism of such regulation is studied insufficiently [59]. Nrf2 mRNA is a target for more than 85 miRNAs that bind to this mRNA and prevent its translation [60].
A decrease in the intracellular level of Nrf2 due to the upregulation of miR-144 expression in erythroid cells led to the decrease in the intracellular level of basic low-molecular-weight antioxidant glutathione (GSH), resulting in the reduction of cell antioxidant defense [61]. The same effect was demonstrated for the neuronal SH-SY5Y cells, in which overexpression of miR-144, miR-153, miR-27a, and miR-142-5p suppressed the NRF2 expression and led to the decrease in the GSH level as a result of the downregulation of expression of the catalytic subunit of γ-glutamylcysteine ligase, an enzyme limiting the rate of GSH de novo synthesis [62]. miR-153 and miR-30a-5p regulate expression of the gene for glutathione peroxidase (Gpx1) located in both the cytoplasm and mitochondria via suppressing transcription factors Nrf2 and NeuroD1 [63].
The toxic effects of the mycotoxin ochratoxin A (OTA) in a culture of epithelial cells of kidney proximal tubules are mediated by a decrease in the antioxidant response through suppressing the expression of the NRF2 and HO-1 genes, leading to the increase in the ROS content and activation of lipid peroxidation [64]. It was established that OTA decreases the intracellular level of Nrf2 but does not influence all miRNAs participating in the Nrf2 regulation. Thus, OTA did not affect expression of miR-144 and miR-34a, but upregulated expression of miR-200c and miR-132. It was also shown that induction of miR-132 plays a crucial role in the decrease in the intracellular level of Nrf2. Downregulation of the НO-1 expression by OTA is associated with a decrease in the Nrf2 content and increase in the expression of miR-200c via independent pathways. Assessment of the cell response to the inhibition of various miRNAs demonstrated that inhibition of miR-132 prevents the development of the OTA-induced oxidative stress. Inhibition of miR-200c also resulted in the decrease in the OTA toxicity and suppression of ROS generation [64].
miRNAs can activate the Nrf2 signaling pathway by decreasing the levels of Keap1. Thus, miR-7 that binds with the Keap1 mRNA downregulates the intracellular content of Keap1 protein, resulting in the nuclear localization of Nrf2 and subsequent transcriptional activation of its target genes in SH-SY5Y cells [65]. miR-200a destabilizes Keap1 mRNA and leads to a considerable increase in the Nrf2 activity [66].
miRNAs that control expression of genes encoding isoforms of the transcription factor FOXO (fork-head box protein O1) significantly contribute to the redox regulation. FOXO isoforms play an important role in the cell cycle control, apoptosis, autophagy, stress response, and antioxidant protection [67]. Suppression of the FOXO gene by miR-182 activates the cell cycle and proliferation. On the contrary, downregulation of miR-182 leads to the suppression of proliferation, cell cycle arrest, and/or activation of apoptosis caused by the activity of FOXO isoforms [67]. It was found that the action of H2O2 on the neuroblastoma SK-N-MC cells is accompanied by inactivation of the transcription factor STAT5, resulting in the 10-fold decrease in the expression of miR-182, which in its turn, causes a 4-fold increase in the FOXO1 protein level (however, the content of FOXO1 mRNA remains virtually unchanged) with subsequent increase in the levels of proapoptotic proteins Bax and Bim and activation of caspase 3 [68].
The deficiency of miR-155 in the culture of endothelial cells from the human brain microvessels leads to the suppression of ROS generation and increase in the production of NO via the PI3K/Akt signaling pathway. The cells deficient by miR-155 were resistant to the oxidative stress associated with the increase in the content of oxidized low-density lipoproteins. Knocking down the gene encoding miR-155 lowered the cell ability to initiate apoptosis and promoted proliferation under both normal conditions and in oxidative stress. The authors believe that these changes were associated with suppression of genes coding for the epidermal growth factor (EGFR), kinase ERK1/2, and p38 MAPK [69].
It should be noted that a high degree of correlation has been found between the antioxidant protection, circadian rhythm, and miRNA functions [70]. In particular, such correlation has been demonstrated for circadian regulation of the key antioxidant enzymes (SOD1, SOD2, catalase, and glutathione peroxidase isoforms Gpx1, Gpx2, and Gpx3), thiol oxidoreductases of the thioredoxin family (Trx1, Trx2, TrxR1, TrxR2, and Txnip), and peroxiredoxins (Prx1, Prx2, Prx3, and Prx6). Therefore, it is believed that dysfunction of miRNAs caused, in particular, by the oxidative stress, can considerably contribute to the development of various pathologies. The role of miRNAs in the regulation of circadian rhythms of redox homeostasis needs further investigation for probable prediction of some diseases and their treatment.
miRNAs AND REDOX-DEPENDENT PROCESSES IN TUMOR CELLS
Metabolic reprogramming of cancer cells leading to the upregulation of intracellular ROS levels is a characteristic feature of malignant development [3]. In tumor cells, the content of ROS is maintained at the critical level, which facilitates triggering of many signaling pathways that can accelerate proliferation without inducing cell death. Such regulatory mechanisms are associated with the activities of specific ROS-sensitive proteins (redox sensors) [71]. Currently, it is commonly accepted that maintaining the redox balance is crucial for preventing oncogenesis. At the same time, promoting the death of tumor cells by increasing the levels of ROS may be a promising strategy in the antitumor therapy.
Because miRNAs are sensitive to the action of ROS, their functional activity is determined by the redox status of tumor cells [12]. Moreover, expression of miRNAs can be regulated by various transcription factors, including p53, c-Myc, and NF-κB, whose activities in tumors cells are significantly altered. Numerous studies have shown that some of these transcription factors are redox-sensitive; therefore, an abnormal expression of miRNAs in tumor tissues is likely to be at least partially associated with the ROS-induced dysregulation of transcription factors [12].
Tumor suppressor p53 causes cell cycle arrest and apoptosis activation. The manifestation of its antitumor activity involves of a number of miRNAs. Thus, p53 activates transcription of miR-34a, which triggers apoptosis of lung cancer cells. p53 participates in the activation of transcription of many tumor-associated miRNAs, including miR-124, miR-145, miR-192, miR-194, and miR-215 [72-74]. On the other hand, p53 is sensitive to ROS; its transcriptional activity changes depending on the ROS level, thus regulating the tumorigenesis. Cysteine residues in p53 can undergo oxidation/reduction with the involvement of Trx and GSH, which regulates the ability of p53 for DNA binding and subsequent activation of apoptosis. The existence of the redox-dependent control of miRNA expression through the regulation of p53 was confirmed by the observation that H2O2 causes p53 phosphorylation at Ser33 residue, leading to the activation of miR-200 transcription by p53 and subsequent death of liver cells [75]. A decrease in the miR-200 level is associated with hepatocellular carcinoma metastasis; therefore, it may be assumed that p53 functions as a tumor suppressor through the redox-dependent increase in the miR-200 level [12]. Moreover, an increase in the ROS generation activates p53, that in turn activates transcription of miR-506 and suppresses lung tumor growth both in vitro and in vivo [76]. These data indicate that ROS control the transcriptional activity of p53 and its functioning as a tumor suppressor, thus facilitating the regulation of miRNA expression.
c-Myc is an oncoprotein that can either activate transcription of oncogenic miRNAs (e.g., miR-17-92) or inhibit transcription of tumor-suppressing miRNAs, including miR-15a and let-7, which promotes oncogenesis [77]. The activity of c-Myc is also regulated by ROS. Overexpression of the MYC gene induced by ROS can activate transcription of miR-27a/b, thereby suppressing expression of PHB1 and NRF2 genes. Knocking down c-Myc and miR-27a/b promotes cell defense against the oxidative stress in liver cells. Since miR-27a/b activates proliferation of hepatocellular carcinoma cells, we can suggest that the signaling pathway underlying progression of liver cancer is the ROS/c-Myc/miR-27a/b cascade [78].
Transcription factor NF-κB controls expression of genes participating in immune response, apoptosis, and cell cycle, in particular, through their involvement in the inflammatory responses and stress monitoring. Oxidative stress induces NF-κB dissociation from its inhibitory protein IκB, leading to the NF-κB activation, translocation into the nucleus, binding to DNA, and activation of gene transcription [79]. Some of the genes controlled by NF-κB encode miRNAs. By regulating the activity of NF-κB, ROS can indirectly influence transcription of these genes. Thus, induction of TGFβ1 (transforming growth factor beta 1) by ROS promotes NF-κB translocation to the nucleus and its subsequent binding to the promoter of the gene encoding miR-21 in a medium with a high glucose content. At the same time, activation of miR-21 is associated with the progression of colorectal cancer [80]. ROS can inhibit the activity of NF-κB by downregulating expression of miR-19a and activation of apoptosis in pheochromocytoma PC12 cells [81]. Moreover, the TNF-α-mediated oxidative stress suppresses the activity of NF-κB and transcription of its target miR-155 (oncogenic miRNA associated with breast cancer) [82]. In general, oxidative stress can either activate or inhibit NF-κB, which makes redox-dependent regulation of miRNA transcription more flexible and dependent on the tumor cell type and environmental conditions [12].
The ability for chronic proliferation is a specific trait of tumor cells. Proliferation is associated with the activation of PI3K/Akt/mTOR, Ras/Raf/MEK/MAPK, and Wnt/β-catenin signaling pathways [83]. Both ROS and miRNAs are involved in the regulation of these signaling cascades, which indicates their important role in the malignant transformation of cells. Activation of K-Ras/MAPK signaling induces expression of miR-155, whose target is mRNA of the transcription factor FOXO3a that controls transcription of genes for some antioxidant enzymes (e.g., SOD2 and catalase). FOXO3a deficiency caused by the action of miR-155 increases the intracellular content of ROS and stimulates proliferation of pancreatic tumor cells [84]. Moreover, the redox-dependent regulation of miRNAs can facilitate proliferation of cancer cells due to the changes in metabolic processes. Thus, miR-1 and miR-206, whose targets are genes encoding enzymes of the pentose phosphate pathway, suppress the growth of tumor cells. Silencing of genes for these two miRNAs prevents their oncosuppressive function, leading to the elimination of ROS and oncogenesis acceleration [52].
Moreover, redox-dependent regulation of miRNAs is observed during activation of apoptosis of tumor cells. Thus, the oncogene PAX7 contributing to the tumor cell resistance to apoptosis is a target for miR-206. The action of the antimalaria preparation artesunate includes facilitation of ROS generation and activation of p38 MAPK, which results in the upregulation of miR-206 expression and decrease in the protein PAX7 level, leading to the activation of apoptosis in the rhabdomyosarcoma cells [85]. The target of miR-23b is the mRNA for the tumor suppressor proline oxidase that increases production of ROS and activates apoptosis. Overexpression of miR-23b correlates with the suppression of proline oxidase activity in patients with renal cancer [86]. miR-21 targets the mRNA for the programmed cell death 4 protein (Pdcd4). An increase in the ROS level caused by the epithelial growth factor (EGF) stimulates miR-21 expression, resulting in the inactivation of Pdcd4, which promotes the development of colorectal cancer [87].
Metastases are associated with a poor clinical prognosis and unfavorable prospects in the therapy of malignant tumors. Dysregulation of ROS generation and expression of miRNAs can stimulate metastasis of tumor cells. Thus, an increase in the ROS content induces expression of miR-141 and miR-200a targeting the p38α mRNA. This results in the suppression of metastasis of ovarian cancer cells by increasing their sensitivity to chemotherapy and can be considered a favorable prognosis in patients with ovarian cancer [88]. It was established that the antioxidant resveratrol lowers the expression of miR-21, causes an increase in the Pdcd4 level, and inhibits the growth and metastasis of prostate tumor cells [89]. miR-212 targets SOD2, and its action leads to the increase in the cellular level of ROS and suppression of epithelial-mesenchymal transition and metastasis of colorectal cancer cells. By contrast, the knockout of the miR-212 gene causes an increase in the intracellular SOD2 level, thus promoting the ability of the cells to withstand oxidative stress, which is associated with a poor prognosis and more aggressive tumor phenotype in patients with colorectal cancer [90].
In conclusion, the studies of the last decade have convincingly demonstrated the importance of the redox-dependent regulation for maintaining cell homeostasis and the role of miRNAs in this process. The mutual feedback regulation of ROS and miRNAs, i.e., the interplay between the ROS action on expression of miRNAs and control of the ROS level via miRNAs expression, is very important for the cell functioning, since it regulates the ratio between the activities of ROS-generating enzymatic systems and cell antioxidant defense. Further studies are required for better comprehension of the role of miRNAs in the hierarchy of redox-dependent regulatory systems with the purpose of miRNA application in the target therapy of diseases in which oxidative stress is a pathogenic factor.
Funding. The publication has been prepared with the support of the “RUDN University Program 5-100”.
Conflict of interest. The authors declare no conflict of interest. Compliance with ethical norms. This article does not contain description of studies with participation of humans and animals performed by any of the authors.
REFERENCES
1.Singh, A., Kukreti, R., Saso, L., and Kukreti, S.
(2019) Oxidative stress: a key modulator in neurodegenerative diseases,
Molecules, 24, E1583, doi: 10.3390/molecules24081583.
2.Massaro, M., Scoditti, E., Carluccio, M. A., and De
Caterina, R. (2019) Oxidative stress and vascular stiffness in
hypertension: a renewed interest for antioxidant therapies? Vasc.
Pharmacol., 116, 45-50, doi: 10.1016/j.vph.2019.03.004.
3.Sies, H., Berndt, C., and Jones, D. P. (2017)
Oxidative stress, Annu. Rev. Biochem., 86, 715-748, doi:
10.1146/annurev-biochem-061516-045037.
4.Ursini, F., Maiorino, M., and Forman, H. J. (2016)
Redox homeostasis: the golden mean of healthy living, Redox
Biol., 8, 205-215, doi: 10.1016/j.redox.2016.01.010.
5.Klotz, L. O., and Steinbrenner, H. (2017) Cellular
adaptation to xenobiotics: interplay between xenosensors, reactive
oxygen species and FOXO transcription factors, Redox Biol.,
13, 646-654, doi: 10.1016/j.redox.2017.07.015.
6.Jones, D. P. (2006) Redefining oxidative stress,
Antioxid. Redox Signal., 8, 865-1879, doi:
10.1089/ars.2006.8.1865.
7.Hopkins, B. L., and Neumann, C. A. (2019) Redoxins
as gatekeepers of the transcriptional oxidative stress response,
Redox Biol., 21, 101104, doi:
10.1016/j.redox.2019.101104.
8.Kalinina, E. V., Chernov, N. N., and Saprin, A. N.
(2008) Involvement of thio-, peroxi-, and glutaredoxins in cellular
redox-dependent processes, Biochemistry (Moscow), 73,
1493-1510, doi: 10.1134/S0006297908130099.
9.Leisegang, M. S., Schroder, K., and Brandes, R. P.
(2018) Redox regulation and noncoding RNAs, Antioxid. Redox
Signal., 29, 793-812, doi: 10.1089/ars.2017.7276.
10.Uchida, S., and Bolli, R. (2018) Short and long
noncoding RNAs regulate the epigenetic status of cells, Antioxid.
Redox Signal., 29, 832-845, doi: 10.1089/ars.2017.7262.
11.Engedal, N., Zerovnik, E., Rudov, A., Galli, F.,
Olivieri, F., Procopio, A. D., Rippo, M. R., Monsurro, V., Betti, M.,
and Albertini, M. C. (2018) From oxidative stress damage to pathways,
networks, and autophagy via microRNAs, Oxid. Med. Cell. Longev.,
2018, 4968321, doi: 10.1155/2018/4968321.
12.Lan, J., Huang, Z., Han, J., Shao, J., and Huang,
C. (2018) Redox regulation of microRNAs in cancer, Cancer Lett.,
418, 250-259, doi: 10.1016/j.canlet.2018.01.010.
13.Koroleva, I. A., Nazarenko, M. S., and Kucher, A.
N. (2018) Role of microRNA in development of instability of
atherosclerotic plaques, Biochemistry (Moscow), 82,
1380-1390, doi: 10.1134/S0006297917110165.
14.Vishnoi, A., and Rani, S. (2017) MiRNA biogenesis
and regulation of diseases: an overview, Methods Mol. Biol.,
1509, 1-10, doi: 10.1007/978-1-4939-6524-3_1.
15.Drusco, A., and Croce, C. M. (2017) MicroRNAs and
cancer: a long story for short RNAs, Adv. Cancer Res.,
135, 1-24, doi: 10.1016/bs.acr.2017.06.005.
16.Wu, K., He, J., Pu, W., and Peng, Y. (2018) The
role of exportin-5 in microRNA biogenesis and cancer, Genom.
Proteom. Bioinform., 16, 120-126, doi:
10.1016/j.gpb.2017.09.004.
17.Chendrimada, T. P., Gregory, R. I., Kumaraswamy,
E., Norman, J., Cooch, N., Nishikura, K., and Shiekhattar, R. (2005)
TRBP recruits the Dicer complex to Ago2 for microRNA processing and
gene silencing, Nature, 436, 740-744, doi:
10.1038/nature03868.
18.Bandara, V., Michael, M. Z., and Gleadle, J. M.
(2014) Hypoxia represses microRNA biogenesis proteins in breast cancer
cells, BMC Cancer, 14, 533, doi:
10.1186/1471-2407-14-533.
19.Suarez, Y., Fernandez-Hernando, C., Pober, J. S.,
and Sessa, W. C. (2007) Dicer dependent microRNAs regulate gene
expression and functions in human endothelial cells, Circ. Res.,
100, 1164-1173, doi: 10.1161/01.RES.0000265065.26744.17.
20.Orom, U. A., Nielsen, F. C., and Lund, A. H.
(2008) MicroRNA-10a binds the 5′-UTR of ribosomal protein mRNAs
and enhances their translation, Mol. Cell, 30, 460-471,
doi: 10.1016/j.molcel.2008.05.001.
21.Havens, M. A., Reich, A. A., Duelli, D. M., and
Hastings, M. L. (2012) Biogenesis of mammalian microRNAs by a
non-canonical processing pathway, Nucleic Acids Res., 40,
4626-4640, doi: 10.1093/nar/gks026.
22.Ho, J. J., Metcalf, J. L., Yan, M. S., Turgeon,
P. J., Wang, J. J., Chalsev, M., Petruzziello-Pellegrini, T. N., Tsui,
A. K., He, J. Z., Dhamko, H., Man, H. S., Robb, G. B., The, B. T., Ohh,
M., and Marsden, P. A. (2012) Functional importance of Dicer protein in
the adaptive cellular response to hypoxia, J. Biol. Chem.,
287, 29003-29020, doi: 10.1074/jbc.M112.373365.
23.Wiesen, J. L., and Tomasi, T. B. (2009) Dicer is
regulated by cellular stresses and interferons, Mol. Immunol.,
46, 1222-1228, doi: 10.1016/j.molimm.2008.11.012.
24.Ungvari, Z., Tucsek, Z., Sosnowska, D., Toth, P.,
Gautam, T., Podlutsky, A., Csiszar, A., Losonczy, G., Valcarcel-Ares,
M. N., Sonntag, W. E., and Csiszar, A. (2012) Aging-induced
dysregulation of Dicer1-dependent microRNA expression impairs
angiogenic capacity of rat cerebromicrovascular endothelial cells,
J. Gerontol. A Biol. Sci. Med. Sci., 68, 877-891, doi:
10.1093/gerona/gls242.
25.Cheng, X., Ku, C.-H., and Siow, R. C. M. (2013)
Regulation of the Nrf2 antioxidant pathway by microRNAs: new players in
micromanaging redox homeostasis, Free Radic. Biol. Med.,
64, 4-11, doi: 10.1016/j.freeradbiomed.2013.07.025.
26.Tang, X., Li, M., Tucker, L., and Ramratnam, B.
(2011) Glycogen synthase kinase 3 beta (GSK3β) phosphorylates the
RNase III enzyme Drosha at S300 and S302, PLoS One, 6,
e20391, doi: 10.1371/journal.pone.0020391.
27.Upton, J. P., Wang, L., Han, D., Wang, E. S.,
Huskey, N. E., Lim, L., Truitt, M., McManus, M. T., Ruggero, D., Goga,
A., Papa, F. R., and Oakes, S. A. (2012) IRE1α cleaves select
microRNAs during ER stress to derepress translation of proapoptotic
caspase-2, Science, 338, 818-822, doi:
10.1126/science.1226191.
28.Poulsen, H. E., Specht, E., Broedbaek, K.,
Henriksen, T., Ellervik, C., Mandrup-Poulsen, T., Tonnesen, M.,
Nielsen, P. E., Andersen, H. U., and Weimann, A. (2012) RNA
modifications by oxidation: a novel disease mechanism? Free Radic.
Biol. Med., 52, 1353-1361, doi:
10.1016/j.freeradbiomed.2012.01.009.
29.Karihtala, P., Porvari, K., Soini, Y., and
Haapasaari, K. M. (2017) Redox regulating enzymes and connected
microRNA regulators have prognostic value in classical Hodgkin
lymphomas, Oxid. Med. Cell. Longev., 2017, 2696071, doi:
10.1155/2017/2696071.
30.Li, G., Luna, C., Qiu, J., Epstein, D. L., and
Gonzalez, P. (2009) Alterations in microRNA expression in
stress-induced cellular senescence, Mech. Ageing Dev.,
130, 731-741, doi: 10.1016/j.mad.2009.09.002.
31.Ji, G., Lv, K., Chen, H., Wang, T., Wang, Y.,
Zhao, D., Qu, L., and Li, Y. (2013) MiR-146a regulates SOD2 expression
in H2O2 stimulated PC12 cells, PLoS One,
8, e69351, doi: 10.1371/journal.pone.0069351.
32.Haque, R., Chun, E., Howell, J. C., Sengupta, T.,
Chen, D., and Kim, H. (2012) MicroRNA-30b-mediated regulation of
catalase expression in human ARPE-19 cells, PLoS One, 7,
e42542, doi: 10.1371/journal.pone.0042542.
33.Wang, L., Huang, H., Fan, Y., Kong, B., Hu, H.,
Hu, K., Guo, J., Mei, Y., and Liu, W. L. (2014) Effects of
downregulation of microRNA-181a on H2O2-induced
H9c2 cell apoptosis via the mitochondrial apoptotic pathway, Oxid.
Med. Cell. Longev., 2014, 960362, doi:
10.1155/2014/960362.
34.Zhang, Y., Zheng, S., Geng, Y., Xue, J., Wang,
Z., Xie, X., Wang, J., Zhang, S., and Hou, Y. (2015) MicroRNA profiling
of atrial fibrillation in canines: miR-206 modulates intrinsic cardiac
autonomic nerve remodeling by regulating SOD1, PLoS One,
10, e0122674, doi: 10.1371/journal.pone.0122674.
35.Dubois-Deruy, E., Cuvelliez, M., Fiedler, J.,
Charrier, H., Mulder, P., Hebbar, E., Pfanne, A., Beseme, O.,
Chwastyniak, M., Amouyel, P., Richard, V., Bauters, C., Thum, T., and
Pinet, F. (2017) MicroRNAs regulating superoxide dismutase 2 are new
circulating biomarkers of heart failure, Sci. Rep., 7,
14747, doi: 10.1038/s41598-017-15011-6.
36.Matouskova, P., Hanouskova, B., and Skalova, L.
(2018) MicroRNAs as potential regulators of glutathione peroxidases
expression and their role in obesity and related pathologies, Int.
J. Mol. Sci., 19, E1199, doi: 10.3390/ijms19041199.
37.Cortez, M. A., Valdecanas, D., Zhang, X., Zhan,
Y., Bhardwaj, V., Calin, G. A., Komaki, R., Giri, D. K., Quini, C. C.,
Wolfe, T., Peltier, H. J., Bader, A. G., Heymach, J. V., Meyn, R. E.,
and Welsh, J. W. (2014) Therapeutic delivery of miR-200c enhances
radio-sensitivity in lung cancer, Mol. Ther., 22,
1494-1503, doi: 10.1038/mt.2014.79.
38.Jiang, W., Min, J., Sui, X., Qian, Y., Liu, Y.,
Liu, Z., Zhou, H., Li, X., and Gong, Y. (2015) MicroRNA-26a-5p and
microRNA-23b-3p up-regulate peroxiredoxin III in acute myeloid
leukemia, Leuk. Lymphoma, 56, 460-471, doi:
10.3109/10428194.2014.924115.
39.Bai, X. Y., Ma, Y., Ding, R., Fu, B., Shi, S.,
and Chen, X. M. (2011) MiR-335 and miR-34a promote renal senescence by
suppressing mitochondrial antioxidative enzymes, J. Am. Soc.
Nephrol., 22, 1252-1261, doi: 10.1681/ASN.2010040367.
40.Kyrychenko, S., Kyrychenko, V., Badr, M. A.,
Ikeda, Y., Sadoshima, J., and Shirokova, N. (2015) Pivotal role of
miR-448 in the development of ROS-induced cardiomyopathy,
Cardiovasc. Res., 108, 324-334, doi:
10.1093/cvr/cvv238.
41.Li, S. Z., Hu, Y. Y., Zhao, J., Zhao, Y. B., Sun,
J. D., Yang, Y. F., Ji, C. C., Liu, Z. B., Cao, W. D., Qu, Y., Liu, W.
P., Cheng, G., and Fei, Z. (2014) MicroRNA-34a induces apoptosis in the
human glioma cell line, A172, through enhanced ROS production and NOX2
expression, Biochem. Biophys. Res. Commun., 444, 6-12,
doi: 10.1016/j.bbrc.2013.12.136.
42.Fu, Y., Zhang, Y., Wang, Z., Wang, L., Wei, X.,
Zhang, B., Wen, Z., Fang, H., Pang, Q., and Yi, F. (2010) Regulation of
NADPH oxidase activity is associated with miRNA-25-mediated NOX4
expression in experimental diabetic nephropathy, Am. J.
Nephrol., 32, 581-589, doi: 10.1159/000322105.
43.Chen, F., Yin, C., Dimitropoulou, C., and Fulton,
D. J. (2016) Cloning, characteristics, and functional analysis of
rabbit NADPH oxidase 5, Front. Physiol., 7, 284, doi:
10.3389/fphys.2016.00284.
44.Yang, S., Gao, Y., Liu, G., Li, J., Shi, K., Du,
B., Si, D., and Yang, P. (2015) The human ATF1 rs11169571 polymorphism
increases essential hypertension risk through modifying miRNA binding,
FEBS Lett., 589, 2087-2093, doi:
10.1016/j.febslet.2015.06.029.
45.Varga, Z. V., Kupai, K., Szucs, G., Gaspar, R.,
Paloczi, J., Farago, N., Zvara, A., Puskas, L. G., Razga, Z.,
Tiszlavicz, L., Bencsik, P., Gorbe, A., Csonka, C., Ferdinandy, P., and
Csont, T. (2013) MicroRNA-25-dependent up-regulation of NADPH oxidase 4
(NOX4) mediates hypercholesterolemia-induced oxidative/nitrative stress
and subsequent dysfunction in the heart, J. Mol. Cell. Cardiol.,
62, 111-121, doi: 10.1016/j.yjmcc.2013.05.009.
46.Wang, Y., Zhao, X., Wu, X., Dai, Y., Chen, P.,
and Xie, L. (2016) MicroRNA-182 mediates Sirt1-induced diabetic corneal
nerve regeneration, Diabetes, 65, 2020-2031, doi:
10.2337/db15-1283.
47.Fierro-Fernandez, M., Busnadiego, O., Sandoval,
P., Espinosa-Diez, C., Blanco-Ruiz, E., Rodriguez, M., Pian, H., Ramos,
R., Lopez-Cabrera, M., Garcia-Bermejo, M. L., and Lamas, S. (2015)
MiR-9-5p suppresses pro-fibrogenic transformation of fibroblasts and
prevents organ fibrosis by targeting NOX4 and TGFBR2, EMBO
Reports, 16, 1358-1377, doi: 10.15252/embr.201540750.
48.Carlomosti, F., D’Agostino, M., Beji, S.,
Torcinaro, A., Rizzi, R., Zaccagnini, G., Maimone, B., Di Stefano, V.,
De Santa, F., Cordisco, S., Antonini, A., Ciarapica, R., Dellambra, E.,
Martelli, F., Avitabile, D., Capogrossi, M. C., and Magenta, A. (2017)
Oxidative stress-induced miR-200c disrupts the regulatory loop among
SIRT1, FOXO1, and eNOS, Antioxid. Redox Signal., 27,
328-344, doi: 10.1089/ars.2016.6643.
49.Kim, J., Lee, K. S., Kim, J. H., Lee, D. K.,
Park, M., Choi, S., Park, W., Kim, S., Choi, Y. K., Hwang, J. Y., Choe,
J., Won, M. H., Jeoung, D., Lee, H., Ryoo, S., Ha, K. S., Kwon, Y. G.,
and Kim, Y. M. (2017) Aspirin prevents TNF-α-induced endothelial
cell dysfunction by regulating the NF-κB-dependent miR-155/eNOS
pathway: role of a miR-155/eNOS axis in preeclampsia, Free Radic.
Biol. Med., 104, 185-198, doi:
10.1016/j.freeradbiomed.2017.01.010.
50.Cho, K. J., Song, J., Oh, Y., and Lee, J. E.
(2015) MicroRNA-Let-7a regulates the function of microglia in
inflammation, Mol. Cell. Neurosci., 68, 167-176, doi:
10.1016/j.mcn.2015.07.004.
51.Muxel, S. M., Laranjeira-Silva, M. F., Zampieri,
R. A., and Floeter-Winter, L. M. (2017) Leishmania (Leishmania)
amazonensis induces macrophage miR-294 and miR-721 expression and
modulates infection by targeting NOS2 and L-arginine metabolism,
Sci. Rep., 7, 44141, doi: 10.1038/srep44141.
52.Singh, A., Happel, C., Manna, S. K.,
Acquaah-Mensah, G., Carrerero, J., Kumar, S., Nasipuri, P., Krausz, K.
W., Wakabayashi, N., Dewi, R., Boros, L. G., Gonzalez, F. J.,
Gabrielson, E., Wong, K. K., Girnun, G., and Biswal, S. (2013)
Transcription factor NRF2 regulates miR-1 and miR-206 to drive
tumorigenesis, J. Clin. Invest., 123, 2921-2934, doi:
10.1172/JCI66353.
53.Sripada, L., Tomar, D., and Singh, R. (2012)
Mitochondria: one of the destinations of miRNAs, Mitochondrion,
12, 593-599, doi: 10.1016/j.mito.2012.10.009.
54.Muratsu-Ikeda, S., Nangaku, M., Ikeda, Y.,
Tanaka, T., Wada, T., and Inagi, R. (2012) Downregulation of miR-205
modulates cell susceptibility to oxidative and endoplasmic reticulum
stresses in renal tubular cells, PLoS One, 7, e41462,
doi: 10.1371/journal.pone.0041462.
55.Wu, S., Lu, H., and Bai, Y. (2019) Nrf2 in
cancers: a double-edged sword, Cancer Med., 8, 2252-2267,
doi: 10.1002/cam4.2101.
56.Singh, B., Ronghe, A. M., Chatterjee, A., Bhat,
N. K., and Bhat, H. K. (2013) MicroRNA-93 regulates NRF2 expression and
is associated with breast carcinogenesis, Carcinogenesis,
34, 1165-1172, doi: 10.1093/carcin/bgt026.
57.Narasimhan, M., Patel, D., Vedpathak, D.,
Rathinam, M., Henderson, G., and Mahimainathan, L. (2012)
Identification of novel microRNAs in posttranscriptional control of
Nrf2 expression and redox homeostasis in neuronal, SH-SY5Y cells,
PLoS One, 7, e51111, doi:
10.1371/journal.pone.0051111.
58.Do, M. T., Kim, H. G., Choi, J. H., and Jeong, H.
G. (2014) Metformin induces microRNA-34a to downregulate the
Sirt1/Pgc-1α/Nrf2 pathway, leading to increased susceptibility of
wild-type p53 cancer cells to oxidative stress and therapeutic agents,
Free Radic. Biol. Med., 74, 21-34, doi:
10.1016/j.freeradbiomed.2014.06.010.
59.Yaribeygi, H., Atkin, S. L., and Sahebkar, A.
(2018) Potential roles of microRNAs in redox state: an update, J.
Cell. Biochem., doi: 10.1002/jcb.27475 [Epub ahead of print].
60.Papp, D., Lenti, K., Modos, D., Fazekas, D., Dul,
Z., Turei, D., Foldvari-Nagy, L., Nussinov, R., Csermely, P., and
Korcsmaros, T. (2012) The NRF2-related interactome and regulome contain
multifunctional proteins and fine-tuned autoregulatory loops, FEBS
Lett., 586, 1795-1802, doi:
10.1016/j.febslet.2012.05.016.
61.Sangokoya, C., Telen, M. J., and Chi, J. T.
(2010) MicroRNA miR-144 modulates oxidative stress tolerance and
associates with anemia severity in sickle cell disease, Blood,
116, 4338-4348, doi: 10.1182/blood-2009-04-214817.
62.Franklin, C. C., Backos, D. S., Mohar, I., White,
C. C., Forman, H. J., and Kavanagh, T. J. (2009) Structure, function,
and post-translational regulation of the catalytic and modifier
subunits of glutamate cysteine ligase, Mol. Aspects Med.,
30, 86-98, doi: 10.1016/j.mam.2008.08.009.
63.Zhou, C., Zhao, L., Zheng, J., Wang, K., Deng,
H., Liu, P., Chen, L., and Mu, H. (2017) MicroRNA-144 modulates
oxidative stress tolerance in SH-SY5Y cells by regulating nuclear
factor erythroid 2-related factor 2-glutathione axis, Neurosci.
Lett., 655, 21-27, doi: 10.1016/j.neulet.2017.06.045.
64.Stachurska, A., Ciesla, M., Kozakowska, M.,
Wolffram, S., Boesch-Saadatmandi, C., Rimbach, G., Jozkowicz, A.,
Dulak, J., and Loboda, A. (2013) Cross-talk between microRNAs, nuclear
factor E2-related factor 2, and heme oxygenase-1 in ochratoxin
A-induced toxic effects in renal proximal tubular epithelial cells,
Mol. Nutr. Food Res., 57, 504-515, doi:
10.1002/mnfr.201200456.
65.Kabaria, S., Choi, D. C., Chaudhuri, A. D., Jain,
M. R., Li, H., and Junn, E. (2015) MicroRNA-7 activates Nrf2 pathway by
targeting Keap1 expression, Free Radic. Biol. Med., 89,
548-556, doi: 10.1016/j.freeradbiomed.2015.09.010.
66.Eades, G., Yang, M., Yao, Y., Zhang, Y., and
Zhou, Q. (2011) MiR-200a regulates Nrf2 activation by targeting Keap1
mRNA in breast cancer cells, J. Biol. Chem., 286,
40725-40733, doi: 10.1074/jbc.M111.275495.
67.Urbanek, P., and Klotz, L. O. (2017)
Posttranscriptional regulation of FOXO expression: microRNAs and
beyond, Br. J. Pharmacol., 174, 1514-1532, doi:
10.1111/bph.13471.
68.Gheysarzadeh, A., and Yazdanparast, R. (2015)
STAT5 reactivation by catechin modulates
H2O2-induced apoptosis through miR-182/FOXO1
pathway in SK-N-MC cells, Cell Biochem. Biophys., 71,
649-656, doi: 10.1007/s12013-014-0244-6.
69.Liu, Y., Pan, Q., Zhao, Y., He, C., Bi, K., Chen,
Y., Zhao, B., Chen, Y., and Ma, X. (2015) MicroRNA-155 regulates ROS
production, NO generation, apoptosis and multiple functions of human
brain microvessel endothelial cells under physiological and
pathological conditions, J. Cell. Biochem., 116,
2870-2881, doi: 10.1002/jcb.25234.
70.Kinoshita, C., Aoyama, K., and Nakaki, T. (2018)
Neuroprotection afforded by circadian regulation of intracellular
glutathione levels: a key role for miRNAs, Free Radic. Biol.
Med., 119, 17-33, doi:
10.1016/j.freeradbiomed.2017.11.023.
71.Helfinger, V., and Schroder, K. (2018) Redox
control in cancer development and progression, Mol. Aspects
Med., 63, 88-98, doi: 10.1016/j.mam.2018.02.003.
72.Jeong, D., Kim, J., Nam, J., Sun, H., Lee, Y. H.,
Lee, T. J., Aguiar, R. C., and Kim, S. W. (2015) MicroRNA-124 links p53
to the NF-κB pathway in B-cell lymphomas, Leukemia,
29, 1868-1874, doi: 10.1038/leu.2015.101.
73.Sachdeva, M., Zhu, S., Wu, F., Wu, H., Walia, V.,
Kumar, S., Elble, R., Watabe, K., and Mo, Y. Y. (2009) p53 represses
c-Myc through induction of the tumor suppressor miR-145, Proc. Natl.
Acad. Sci. USA, 106, 3207-3212, doi:
10.1073/pnas.0808042106.
74.Pichiorri, F., Suh, S. S., Rocci, A., De Luca,
L., Taccioli, C., Santhanam, R., Zhou, W., Benson, D. M., Jr.,
Hofmainster, C., Alder, H., Garofalo, M., Di Leva, G., Volinia, S.,
Lin, H. J., Perrotti, D., Kuehl, M., Aqeilan, R. I., Palumbo, A., and
Croce, C. M. (2010) Downregulation of p53-inducible microRNAs 192, 194,
and 215 impairs the p53/MDM2 autoregulatory loop in multiple myeloma
development, Cancer Cell, 18, 367-381, doi:
10.1016/j.ccr.2010.09.005.
75.Xiao, Y., Yan, W., Lu, L., Wang, Y., Lu, W., Cao,
Y., and Cai, W. (2015) p38/p53/miR-200a-3p feedback loop promotes
oxidative stress-mediated liver cell death, Cell Cycle,
14, 1548-1558, doi: 10.1080/15384101.2015.1026491.
76.Yin, M., Ren, X., Zhang, X., Luo, Y., Wang, G.,
Huang, K., Feng, S., Bao, X., Huang, K., He, X., Liang, P., Wang, Z.,
Tang, H., He, J., and Zhang, B. (2015) Selective killing of lung cancer
cells by miRNA-506 molecule through inhibiting NF-κB p65 to evoke
reactive oxygen species generation and p53 activation, Oncogene,
34, 691-703, doi: 10.1038/onc.2013.597.
77.Chang, T. C., Yu, D., Lee, Y. S., Wentzel, E. A.,
Arking, D. E., West, K. M., Dang, C. V., Thomas-Tikhonenko, A., and
Mendell, J. T. (2008) Widespread microRNA repression by Myc contributes
to tumorigenesis, Nat. Genet., 40, 43-50, doi:
10.1038/ng.2007.30.
78.Yang, H., Li, T. W., Zhou, Y., Peng, H., Liu, T.,
Zandi, E., Martinez-Chantar, M. L., Mato, J. M., and Lu, S. C. (2015)
Activation of a novel c-Myc-miR27-prohibitin 1 circuitry in cholestatic
liver injury inhibits glutathione synthesis in mice, Antioxid. Redox
Signal., 22, 259-274, doi: 10.1089/ars.2014.6027.
79.Lingappan, K. (2018) NF-κB in oxidative
stress, Curr. Opin. Toxicol., 7, 81-86, doi:
10.1016/j.cotox.2017.11.002.
80.Toiyama, Y., Takahashi, M., Hur, K., Nagasaka,
T., Tanaka, K., Inoue, Y., Kusunoki, M., Boland, C. R., and Goel, A.
(2013) Serum miR-21 as a diagnostic and prognostic biomarker in
colorectal cancer, J. Natl. Cancer Inst., 105, 849-859,
doi: 10.1093/jnci/djt101.
81.Hong, J., Wang, Y., Hu, B. C., Xu, L., Liu, J.
Q., Chen, M. H., Wang, J. Z., Han, F., Zheng, Y., Chen, X., Li, Q.,
Yang, X. H., Sun, R. H., and Mo, S. J. (2017) Transcriptional
downregulation of microRNA-19a by ROS production and NF-κB
deactivation governs resistance to oxidative stress-initiated
apoptosis, Oncotarget, 8, 70967-70981, doi:
10.18632/oncotarget.20235.
82.Jiang, S., Zhang, H. W., Lu, M. H., He, X. H.,
Li, Y., Gu, H., Liu, M. F., and Wang, E. D. (2010) MicroRNA-155
functions as an OncomiR in breast cancer by targeting the suppressor of
cytokine signaling 1 gene, Cancer Res., 70, 3119-3127,
doi: 10.1158/0008-5472.CAN-09-4250.
83.Nusse, R., and Clevers, H. (2017)
Wnt/β-catenin signaling, disease, and emerging therapeutic
modalities, Cell, 169, 985-999, doi:
10.1016/j.cell.2017.05.016.
84.Wang, P., Zhu, C. F., Ma, M. Z., Chen, G., Song,
M., Zeng, Z. L., Lu, W. H., Yang, J., Wen, S., Chiao, P. J., Hu, Y.,
and Huang, P. (2015) Micro-RNA-155 is induced by K-Ras oncogenic signal
and promotes ROS stress in pancreatic cancer, Oncotarget,
6, 21148-21158, doi: 10.18632/oncotarget.4125.
85.Beccafico, S., Morozzi, G., Marchetti, M. C.,
Riccardi, C., Sidoni, A., Donato, R., and Sorci, G. (2015) Artesunate
induces ROS- and p38 MAPK-mediated apoptosis and counteracts tumor
growth in vivo in embryonal rhabdomyosarcoma cells,
Carcinogenesis, 36, 1071-1083, doi:
10.1093/carcin/bgv098.
86.Liu, W., Zabirnyk, O., Wang, H., Shiao, Y. H.,
Nickerson, M. L., Khalil, S., Anderson, L. M., Perantoni, A. O., and
Phang, J. M. (2010) MicroRNA-23b targets proline oxidase, a novel tumor
suppressor protein in renal cancer, Oncogene, 29,
4914-4924, doi: 10.1038/onc.2010.237.
87.Saxena, A., Shoeb, M., Ramana, K. V., and
Srivastava, S. K. (2013) Aldose reductase inhibition suppresses colon
cancer cell viability by modulating microRNA-21 mediated programmed
cell death 4 (PDCD4) expression, Eur. J. Cancer, 49,
3311-3319, doi: 10.1016/j.ejca.2013.05.031.
88.Mateescu, B., Batista, L., Cardon, M., Gruosso,
T., de Feraudy, Y., Mariani, O., Nicolas, A., Meyniel, J. P., Cottu,
P., Sastre-Garau, X., and Mechta-Grigoriou, F. (2011) MiR-141 and
miR-200a act on ovarian tumorigenesis by controlling oxidative stress
response, Nat. Med., 17, 1627-1635, doi:
10.1038/nm.2512.
89.Sheth, S., Jajoo, S., Kaur, T., Mukherjea, D.,
Sheehan, K., Rybak, L. P., and Ramkumar, V. (2012) Resveratrol reduces
prostate cancer growth and metastasis by inhibiting the Akt/microRNA-21
pathway, PLoS One, 7, e51655, doi:
10.1371/journal.pone.0051655.
90.Meng, X., Wu, J., Pan, C., Wang, H., Ying, X.,
Zhou, Y., Yu, H., Zuo, Y., Pan, Z., Liu, R. Y., and Huang, W. (2013)
Genetic and epigenetic down-regulation of microRNA-212 promotes
colorectal tumor metastasis via dysregulation of MnSOD,
Gastroenterology, 145, 426-436, doi:
10.1053/j.gastro.2013.04.004.
91.Das, S., Ferlito, M., Kent, O. A., Fox-Talbot,
K., Wang, R., Liu, D., Raghavachari, N., Yang, Y., Wheelan, S. J.,
Murphy, E., and Steenbergen, C. (2012) Nuclear miRNA regulates the
mitochondrial genome in the heart, Circ. Res., 110,
1596-1603, doi: 10.1161/CIRCRESAHA.112.267732.
92.Faraonio, R., Salerno, P., Passaro, F., Sedia,
C., Iaccio, A., Bellelli, R., Nappi, T. C., Comegna, M., Romano, S.,
Salvatore, G., Santoro, M., and Cimino, F. (2012) A set of miRNAs
participates in the cellular senescence program in human diploid
fibroblasts, Cell Death Differ., 19, 713-721, doi:
10.1038/cdd.2011.143.
93.Liao, L., Su, X., Yang, X., Hu, C., Li, B., Lv,
Y., Shuai, Y., Jing, H., Deng, Z., and Jin, Y. (2016) TNF-α
inhibits FoxO1 by upregulating miR-705 to aggravate oxidative damage in
bone marrow-derived mesenchymal stem cells during osteoporosis, Stem
Cells, 34, 1054-1067, doi: 10.1002/stem.2274.
94.Cheng, L. B., Li, K. R., Yi, N., Li, X. M., Wang,
F., Xue, B., Pan, Y. S., Yao, J., Jiang, Q., and Wu, Z. F. (2017)
miRNA-141 attenuates UV-induced oxidative stress via activating
Keap1-Nrf2 signaling in human retinal pigment epithelium cells and
retinal ganglion cells, Oncotarget, 8, 13186-13194, doi:
10.18632/oncotarget.14489.
95.Zhang, L., Zhou, M., Wang, Y., Huang, W., Qin,
G., Weintraub, N. L., and Tang, Y. (2014) MiR-92a inhibits vascular
smooth muscle cell apoptosis: role of the MKK4-JNK pathway,
Apoptosis, 19, 975-983, doi:
10.1007/s10495-014-0987-y.
96.Li, J., Donath, S., Li, Y., Qin, D., Prabhakar,
B. S., and Li, P. (2010) MiR-30 regulates mitochondrial fission through
targeting p53 and the dynamin-related protein-1 pathway, PLoS
Genet., 6, e1000795, doi: 10.1371/journal.pgen.1000795.