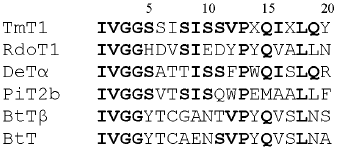Digestive Proteinases of Yellow Mealworm (Tenebrio molitor) Larvae: Purification and Characterization of a Trypsin-Like Proteinase
T. A. Tsybina1,2, Y. E. Dunaevsky2, M. A. Belozersky2, D. P. Zhuzhikov3, B. Oppert4, and E. N. Elpidina2*
1Bach Institute of Biochemistry, Russian Academy of Sciences, Leninsky pr. 33, 119071 Moscow, Russia2Belozersky Institute of Physico-Chemical Biology, Lomonosov Moscow State University, 119992 Moscow, Russia; fax: (7-095) 939-3181; E-mail: elp@belozersky.msu.ru
3Department of Entomology, Faculty of Biology, Lomonosov Moscow State University, 119992 Moscow, Russia
4Grain Marketing and Production Research Center, USDA Agricultural Research Service, 1515 College Ave, Manhattan KS, 66502, USA
* To whom correspondence should be addressed.
Received October 14, 2004; Revision received November 1, 2004
A new trypsin-like proteinase was purified to homogeneity from the posterior midgut of Tenebrio molitor larvae by ion-exchange chromatography on DEAE-Sephadex A-50 and gel filtration on Superdex-75. The isolated enzyme had molecular mass of 25.5 kD and pI 7.4. The enzyme was also characterized by temperature optimum at 55°C, pH optimum at 8.5, and Km value of 0.04 mM (for hydrolysis of Bz-Arg-pNA). According to inhibitor analysis the enzyme is a trypsin-like serine proteinase stable within the pH range of 5.0-9.5. The enzyme hydrolyzes peptide bonds formed by Arg or Lys residues in the P1 position with a preference for relatively long peptide substrates. The N-terminal amino acid sequence, IVGGSSISISSVPXQIXLQY, shares 50-72% identity with other insect trypsin-like proteinases, and 44-50% identity to mammalian trypsins. The isolated enzyme is sensitive to inhibition by plant proteinase inhibitors and it can serve as a suitable target for control of digestion in this stored product pest.
KEY WORDS: digestive proteinase, Tenebrio molitor, trypsin
Abbreviations: Bz) benzoyl; FPLC) fast performance liquid chromatography; For) formyl; PMSF) phenylmethylsulfonyl fluoride; pNA) p-nitroanilide; TLCK) Nalpha-p-tosyl-L-lysine chloromethyl ketone; TmT1) trypsin-like proteinase from T. molitor larvae; E-64) L-trans-epoxysuccinyl-L-leucine amido(4-guanidine)butane; Z) N-benzoyloxycarbonyl.
Trypsin-like proteinases play an important role in digestion in most
insects studied [1-4]. They are
also involved in processing of insecticide toxins of Bacillus
thuringiensis, and this underlines protease-dependent resistance of
these species to the toxins [5]. Although a
significant number of sequences typical for trypsins have been found
during analysis of cDNA libraries [6-11], only a small number of trypsin-like insect
proteinases have been purified to homogeneity and characterized [12-14].
In this paper, we describe a relatively simple method for purification of the main trypsin-like enzyme of the posterior part of the midgut of yellow mealworm (Tenebrio molitor) larvae (TmT1) and characterize the proteinase of this stored product pest.
MATERIALS AND METHODS
Yellow mealworm (Tenebrio molitor) larvae were reared on a mixture of wheat flour, bran, and brewer's yeast and transferred to milled oat flakes (Raisio, Finland) 1-1.5 weeks prior to dissection.
Proteinase purification. Isolated larvae midguts were separated into anterior and posterior parts. The collected posterior parts were homogenized in 0.14 M NaCl and centrifuged at 15,000g for 10 min. The supernatant dialyzed against 20 mM K,Na-phosphate buffer, pH 6.9, containing 0.02% NaN3, was centrifuged again and subjected to batch-chromatography on DEAE-Sephadex A-50 using the same buffer. The filtrate containing the major proportion of trypsin-like activity was concentrated and subjected to FPLC gel filtration using a Superdex-75 column (Pharmacia-LKB, Sweden) equilibrated with 20 mM K,Na-phosphate buffer, pH 6.9. Fractions exhibiting the highest activity with Bz-Arg-pNA as a substrate were concentrated and used in subsequent work.
Enzyme activity was assayed by the rate of hydrolysis of synthetic p-nitroanilide substrates by the method of Erlanger et al. [15]. The following compounds were used as substrates: Bz-DL-Arg-pNA (Fluka, Switzerland), Bz-DL-Lys-pNA (Merck, Germany), Z-Gly-Pro-Arg-pNA, D-Pro-Phe-Arg-pNA, For-Ala-Phe-Lys-pNA, D-Val-Leu-Lys-pNA, Z-Ala-Phe-Arg-pNA, and Z-Ala-Ala-Trp-Arg-pNA (synthesized by T. L. Voyushina at the Scientific Research Institute of Genetics and Selection of Industrial Microorganisms) [16]. The purification procedure was monitored by hydrolysis of 0.5 mM Bz-Arg-pNA. The mixture containing enzyme and substrate was incubated at 37°C for 15-40 min and then absorbance of p-nitroaniline formed was measured at 410 nm. Substrates were initially dissolved in dimethylformamide to 10 mM concentration; subsequent dilutions were made using 0.02 M Tris-HCl buffer, pH 8.0. The reaction was terminated by adding acetic acid (final concentration 5%). One unit of enzyme activity was defined as the amount of the enzyme required for formation of 1 µmol of product during 1 min at 37°C.
Kinetic studies were carried out using a Hitachi 557 spectrophotometer (Japan) at 25°C. Substrate concentrations were varied within 0.005-0.25 mM range using 0.02 M Tris-HCl buffer, pH 8.0, containing 2.5% dimethylformamide. The dependence of the reaction rate on substrate concentration was analyzed using Lineweaver-Burk plots by least-squares method using MS Excel 97 software.
Protein concentration was determined by the method of Lowry et al. [17] or spectrophotometrically at 280 nm. Protein solutions were concentrated by ultrafiltration in Amicon cells (The Netherlands) using YM-5 membranes (Amicon) or by evaporation in a vacuum Speedvac concentrator (Savant, USA).
Chromatofocusing was carried out using a PBI-94 column (Pharmacia-LKB). A pH gradient of 6.0-8.7 was formed with Polybuffer-96 (Pharmacia-LKB). The pH value corresponding to the peak of catalytic activity with Bz-Arg-pNA as substrate was defined as the isoelectric point.
For determination of functional groups in the active site, we used specific inhibitors of proteases from different classes. These included serine protease inhibitors: phenylmethylsulfonyl fluoride (PMSF; Serva, Germany), tosyl-lysine chloromethyl ketone (TLCK, trypsin inhibitor; Fluka), and tosyl-phenylalanine chloromethyl ketone (TPCK, chymotrypsin inhibitor; Fluka); cysteine protease inhibitor, L-trans-epoxysuccinyl-L-leucine amido(4-guanidine)butane (E-64; Sigma, USA), and metalloprotease inhibitor EDTA (Reanal, Hungary). Enzyme solution was incubated in the presence of an inhibitor for 30 min, the substrate was added, and residual activity was assayed using the method of Erlanger et al. [15].
pH-stability. For determination of enzyme stability at various pH values, 0.1 M universal buffer [18] was used in the pH range 3.0-11.5 (with an interval of 0.5 pH unit). The enzyme was incubated at various pH values for 2 h at 25°C. After that pH was adjusted to 8.0 (using 0.5 M Tris-HCl buffer, pH 8.0), and enzyme activity was assayed as above. Maximal enzyme activity was defined as 100%.
Thermal stability. For determination of thermal stability the enzyme solution in 0.02 M Tris-HCl buffer, pH 8.0, was incubated in water bath thermostat for 30 min at 25-60°C and after rapid cooling in an ice bath the enzyme activity was assayed at 37°C using Bz-Arg-pNA as substrate.
Temperature optimum was determined after preincubation at 30-60°C in a water bath thermostat for 5 min using Bz-Arg-pNA as substrate.
Molecular mass of the isolated enzyme was determined by electrophoresis in 14% polyacrylamide gel (14% T, 0.5% C) in the presence of SDS (SDS-PAGE). The following proteins were used as molecular mass markers: cellulase (94.6 kD), BSA (66.2 kD), ovalbumin (45.4 kD), carbonic anhydrase (31 kD), trypsin inhibitor (21.5 kD), and lysozyme (14.4 kD).
For post-electrophoretic determination of enzymatic activity, polyacrylamide gel was co-polymerized with 0.026% gelatin. The protein was loaded onto the gel without heating. The electrophoretic procedure was carried out at 4°C and constant current of 25 mA. After termination of electrophoretic separation the slab was washed twice in 2.5% Triton X-100 and twice in 20 mM Tris-HCl buffer, pH 8.0, and then it was incubated in the same buffer for 2 h at 37°C. After incubation the slab was stained with 0.1% Coomassie R-250 in the mixture of methanol-acetic acid-water (4 : 1 : 5 v/v), and washed in the same solution but without the dye. The proteinase was detected as a clear band on a dark background.
Amino acid composition of the proteinase was analyzed by a standard procedure with amino acid analyzer Hitachi 835 (Japan). Cysteines were determined as cysteic acid after protein oxidation with a mixture of H2O2 and 88% performic acid (1 : 9 v/v) followed by hydrolysis with 5.7 M HCl (110°C, 22 h). Tryptophans were determined after protein hydrolysis with 4 M methanesulfonic acid containing 0.2% tryptamine.
For determination of N-terminal amino acid sequence, the protein obtained after SDS-PAGE was electroblotted to Immobilon P membrane (Millipore, USA) during 1.5 h at 20°C and constant current of 250 mA, using Tris-glycine electrode buffer, pH 8.6, with 20% methanol. Amino acid sequence was determined by the Edman automated degradation procedure using an Applied Biosystems model 492 sequencer (USA).
Purification of trypsin inhibitors from plant seeds. Trypsin inhibitors from buckwheat and triticale seeds were purified by affinity chromatography (using trypsin-Sepharose) and ion-exchange FPLC (using Mono Q) [19].
RESULTS AND DISCUSSION
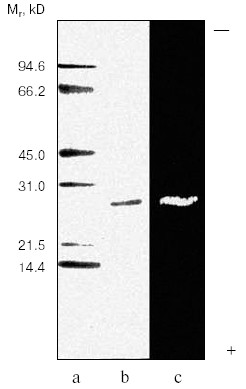
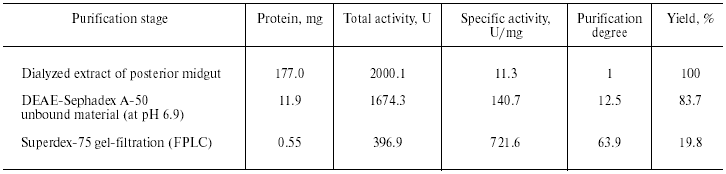
Purification of TmT1 proteinase included two main stages: ion-exchange chromatography on DEAE-Sephadex A-50 and gel filtration on Superdex-75 by means of FPLC. The first step resulted in separation of not more than 16% of the trypsin-like activity adsorbed on this anion exchanger. The remainder of the trypsin-like activity was separated as a single peak from ballast proteins during the second purification step. Results of the purification procedure are summarized in Table 1. Thus, use of two chromatographic steps resulted in 64-fold purification of TmT1 responsible for 84% of the total trypsin-like activity in the posterior midgut of T. molitor larvae; the yield of the resultant enzyme preparation was 20%. TmT1 was homogenous according to SDS-PAGE (Fig. 1b). The purified enzyme was detected as one clear band of gelatinase activity during polyacrylamide gel electrophoresis with co-polymerized gelatin (Fig. 1c). During purification, there was a problem in separation of TmT1 from a chymotrypsin-like proteinase similar to the former in charge and molecular mass. This problem was overcome using gel filtration on a column with Superdex-75 equilibrated with 20 mM K,Na-phosphate buffer without addition of NaCl. Due to nonspecific sorption, the chymotrypsin-like enzyme was retained on this column and eluted later than TmT1.
Table 1. Purification of TmT1 proteinase
from the posterior midgut of T. molitor larvae
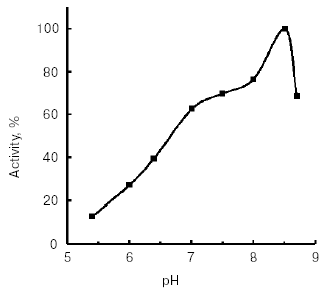
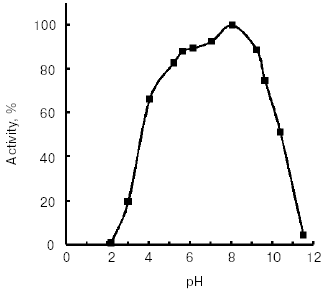
Molecular mass of the purified enzyme evaluated by SDS-PAGE was 25.5 kD. The isoelectric point detected by chromatofocusing was 7.4. The maximal activity of the TmT1 assayed with a specific substrate of trypsin-like proteinases, Bz-Arg-pNA, was observed at pH 8.5 (Fig. 2). The alkaline pH optimum determined for TmT1 from T. molitor larvae is typical for most insect and mammalian trypsins and its value is also close to the pH of the posterior part of the midgut (7.9) where this enzyme is localized. The enzyme is highly stable (>80% activity) over a wide range of pH values (from 5.0 to 9.5) (Fig. 3). Sharp decrease in catalytic activity (by more than 95%) was observed at pH < 4.0 or pH > 11.5. In contrast to mammalian trypsins, TmT1 (as well as most insect trypsin-like enzymes [2, 3]) was unstable at highly acidic or alkaline pH values. These differences may be attributed to existence of special digestive compartment in mammals (stomach) with highly acidic pH, whereas most insects lack such a compartment (with pH < 4.0) [20].Fig. 1. SDS-Electrophoresis of purified TmT1 proteinase from posterior midgut of T. molitor larvae: a) protein markers; b) purified proteinase; c) proteinase zymogram in the same gel co-polymerized with 0.026% gelatin.
Fig. 2. Effect of pH on activity of TmT1 proteinase from the midgut of T. molitor larvae. Maximal enzyme activity with 0.25 mM Bz-Arg-pNA was taken as 100%.
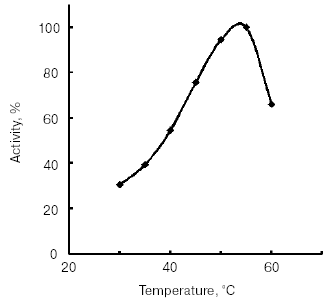
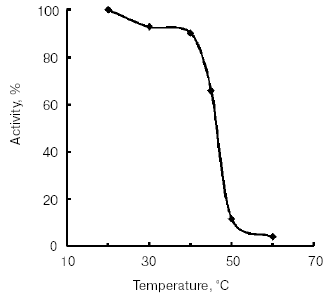
The temperature optimum for catalytic activity assayed with Bz-Arg-pNA as substrate at pH 8.0 was 55°C (Fig. 4). TmT1 was stable for 30 min at temperatures of 40°C and below (Fig. 5).Fig. 3. Effect of pH on stability of TmT1 proteinase from the midgut of T. molitor larvae (substrate 0.25 mM Bz-Arg-pNA).
Fig. 4. Effect of temperature on activity of TmT1 proteinase from the midgut of T. molitor larvae.
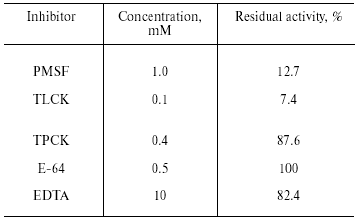
Inhibitory analysis revealed that TmT1 was almost totally inhibited by a specific inhibitor of serine proteases, PMSF, and specific trypsin inhibitor, TLCK. Specific inhibitors of cysteine proteases (E-64), metalloproteases (EDTA), and chymotrypsin-like serine proteinases (TPCK) had little to no effect on the activity of TmT1 (Table 2).Fig. 5. Temperature stability of TmT1 proteinase from the midgut of T. molitor larvae.
Table 2. Inhibitory analysis of TmT1
proteinase from the midgut of T. molitor larvae
The purified enzyme hydrolyzed substrates containing Arg and Lys residues at P1 position (Table 3). Initial rates of hydrolysis of substrates containing Arg residue in this position were higher than those for hydrolysis of substrates containing a Lys residue in P1. Initial rates for hydrolysis of longer substrates were higher than the initial rates for hydrolysis of shorter substrates. The rate of hydrolysis also depended on the nature of the amino acid residues in P2 and P3 positions, and it was significantly lower when Pro was in these positions.
Table 3. Substrate specificity of TmT1
proteinase from the midgut of T. molitor larvae
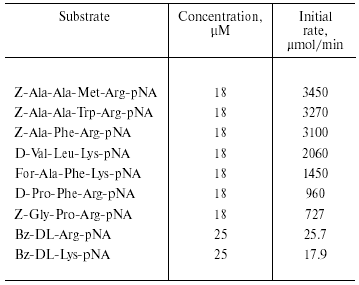
Thus, analysis of substrate specificity and inhibitor sensitivity indicates that TmT1 purified from the posterior midgut of T. molitor larvae is a trypsin-like serine proteinase.
The Km values determined for Bz-Arg-pNA and Bz-Lys-pNA substrates were 0.04 and 0.09 mM, respectively. Among insects studied, only a trypsin-like proteinase from the cricket Teleogryllus commodus was characterized by similar (low) Km value, 0.08 mM [21]. Km values for another trypsin-like enzyme from T. molitor larvae (0.93 mM) [12] and similar proteinases from other insect species (0.12-0.47 mM) [13, 14] or bovine enzyme (0.65 mM) [22] were significantly higher.
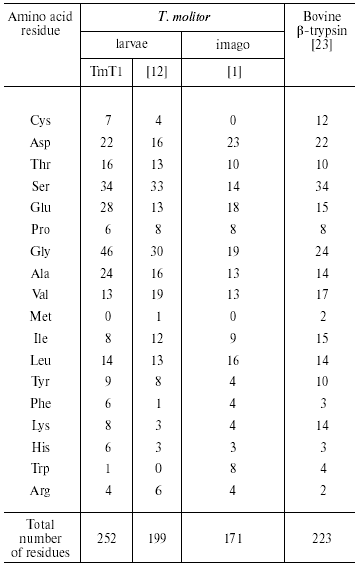
Analysis of the amino acid composition of the purified TmT1 from T. molitor larvae revealed certain differences from trypsins previously detected in this species (Table 4). These included primarily content of Cys, Glu, Gly, and Lys residues. There is an odd number of Cys residues (seven in TmT1); which was also found in some other insect trypsins [7, 9]. We cultivated the larvae on milled oat flakes lacking active proteinase inhibitors (data not shown); under these conditions TmT1 activity dominated in the T. molitor midgut. In larvae fed with 90% wheat bran containing various trypsin inhibitors another trypsin-like proteinase, TLE, dominated; this enzyme differs from TmT1 protease in amino acid composition and kinetic constants for hydrolysis of specific substrates [12]. It is possible that the genome of T. molitor contains several genes encoding trypsin-like proteinases. It is known that preferential expression of certain genes encoding certain trypsins may be related to insect adaptation to diets containing proteinaceous trypsin inhibitors [10, 24].
Table 4. Amino acid composition of
trypsin-like proteinases (number of residues per molecule)
N-Terminal sequencing of purified TmT1 revealed the following sequence--IVGGSSISISSVPXQIXLQY. Comparative analysis of N-terminal sequences of TmT1 and some trypsin-like proteinases isolated from Coleoptera, Diptera, and Lepidoptera and also mammalian trypsins revealed that TmT1 contained 50-72% amino acid residues identical with the insect trypsin-like enzymes and 44-50% - with mammalian trypsins (Fig. 6).
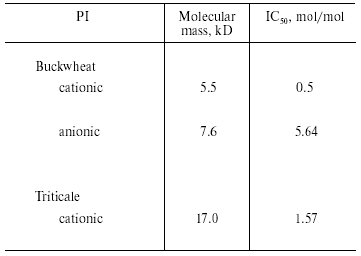
We investigated the effects of proteinaceous proteinase inhibitors isolated from buckwheat and triticale seeds on the activity of TmT1 (Table 5). Cationic inhibitors from buckwheat seeds caused the most potent enzyme inhibition. They might be good candidates for inhibition of digestion in T. molitor larvae, known as stored product pest.Fig. 6. N-Terminal amino acid sequences of some trypsin-like insect and mammalian proteinases. TmT1, the proteinase from the midgut of T. molitor larvae investigated in this study; RdoT1, Rhyzopertha dominica proteinase [8]; DeTalpha, Drosophila erecta proteinase [6]; PiT2b, Plodia interpunctella proteinase [9]; BtTbeta and BtT, bovine (Bos taurus) proteinases [23, 25].
Table 5. Effect of proteinaceous proteinase
inhibitors (PI) from buckwheat and triticale seeds on the activity of
TmT1 from the midgut of T. molitor larvae
Thus, results of the present study indicate that TmT1 purified to homogeneity from the midgut of T. molitor larvae is a trypsin-like serine proteinase, which may be used as a target for potential bio-pesticides developed on the basis of plant proteinase inhibitors.
This work was supported by the Grant Council of the President of Russian Federation (grant No. MK-1395.2004.4), Russian Foundation for Basic Research (grant No. 02-04-48808), Civil Research and Development Foundation (grant No. RB2-2396-MO-02), and USDA Agricultural Research Service.
REFERENCES
1.Applebaum, S. W. (1985) in Comprehensive
Physiology, Biochemistry and Pharmacology of Insects (Kerkut, G.
A., and Gilbert, L. I., eds.) Pergamon Press, Oxford, pp. 279-311.
2.Terra, W. R., and Ferreira, C. (1994) Comp.
Biochem. Physiol., 109B, 1-62.
3.Terra, W. R., Ferreira, C., Jordao, B. P., and
Dillon, R. J. (1996) in Biology of the Insect Midgut (Lehane, M.
J., and Billingsley, P. F., eds.) Chapman and Hall, London, pp.
153-194.
4.Reeck, G., Oppert, B., Denton, M., Kanost, M.,
Baker, J., and Kramer, K. (1999) in Proteases. New Perspective
(Turk, V., ed.) Birkhauser Verlag, Basel, pp. 125-148.
5.Oppert, B. (1999) Arch. Insect Biochem.
Physiol., 42, 1-12.
6.Wang, S., Magoulas, C., and Hickey, D. (1999)
Mol. Biol. Evol., 16, 1117-1124.
7.Bown, D. P., Wilkinson, H. S., and Gatehouse, J. A.
(1997) Insect Biochem. Mol. Biol., 27, 625-638.
8.Zhu, Y.-C., and Baker, J. E. (1999) Insect
Biochem. Mol. Biol., 29, 1053-1063.
9.Zhu, Y.-C., Kramer, K. J., Dowdy, A. K., and Baker,
J. E. (2000) Insect Biochem. Mol. Biol., 30,
1027-1035.
10.Mazumdar-Leighton, S., and Broadway, R. M. (2001)
Insect Biochem. Mol. Biol., 31, 645-657.
11.Zhu, Y.-C., Zeng, F., and Oppert, B. (2003)
Insect Biochem. Mol. Biol., 33, 889-899.
12.Levinsky, H., Birk, Y., and Applebaum, S. W.
(1977) Int. J. Peptide Protein Res., 10, 252-264.
13.Lam, W., Coast, G. M., and Rayne, R. C. (2000)
Insect Biochem. Mol. Biol., 30, 85-94.
14.Lopes, A. R., and Terra, W. R. (2003) Insect
Biochem. Mol. Biol., 33, 407-415.
15.Erlanger, B. F., Kokowsky, N., and Cohen, W.
(1961) Arch. Biochem. Biophys., 95, 271-278.
16.Stepanov, V. M., Terentyeva, E. Yu., Voyushina,
T. L., and Gololobov, M. Yu. (1995) Bioorg. Med. Chem.,
3, 479-485.
17.Lowry, O. H., Rosebrough, N. J., Farr, A. L., and
Randall, R. J. (1951) J. Biol. Chem., 193, 265-275.
18.Lurie, Yu. Yu. (1979) Hand-Book on Analytical
Chemistry [in Russian], Khimiya, Moscow.
19.Dunaevsky, Y. E., Pavlukova, E. B., Beliakova, G.
A., Tsybina, T. A., Gruban, T. N., and Belozersky, M. A. (1998) J.
Plant Physiol., 152, 696-702.
20.Terra, W. R., Ferreira, C., and Baker, J. E.
(1996) in Biology of the Insect Midgut (Lehane, M. J., and
Billingsley, P. F., eds.) Chapman and Hall, London, pp. 206-235.
21.Christeller, J. T., Laing, W. A., Shaw, B. D.,
and Burgess, E. P. J. (1990) Insect Biochem., 20,
157-164.
22.Asgeirsson, B., Fox, J. W., and Bjarnason, J. B.
(1989) Eur. J. Biochem., 180, 85-94.
23.Mikes, O., Holeysovsky, V., Tomasek, V., and
Sorm, F. (1966) Biochem. Biophys. Res. Commun., 24,
346-352.
24.Jongsma, M. A., Bakker, P. L., Peters, J., Bosch,
D., and Stiekema, W. J. (1995) Proc. Natl. Acad. Sci. USA,
92, 8041-8045.
25.Le Huerou, I., Wicker, C., Guilloteau, P.,
Toullec, R., and Puigserver, A. (1990) Eur. J. Biochem.,
193, 767-773.