REVIEW: Phenoptosis and the Various Types of Natural Selection
Giacinto Libertini1,2
1Italian Society for Evolutionary Biology (ISEB), 14100 Asti, Italy2Department of Translational Medical Sciences, Federico II University of Naples, 80131 Naples, Italy
Received July 27, 2023; Revised September 14, 2023; Accepted September 17, 2023
In the first description of evolution, the fundamental mechanism is the natural selection favoring the individuals best suited for survival and reproduction (selection at the individual level or classical Darwinian selection). However, this is a very reductive description of natural selection that does not consider or explain a long series of known phenomena, including those in which an individual sacrifices or jeopardizes his life on the basis of genetically determined mechanisms (i.e., phenoptosis). In fact, in addition to (i) selection at the individual level, it is essential to consider other types of natural selection such as those concerning: (ii) kin selection and some related forms of group selection; (iii) the interactions between the innumerable species that constitute a holobiont; (iv) the origin of the eukaryotic cell from prokaryotic organisms; (v) the origin of multicellular eukaryotic organisms from unicellular organisms; (vi) eusociality (e.g., in many species of ants, bees, termites); (vii) selection at the level of single genes, or groups of genes; (viii) the interactions between individuals (or more precisely their holobionts) of the innumerable species that make up an ecosystem. These forms of natural selection, which are all effects and not violations of the classical Darwinian selection, also show how concepts as life, species, individual, and phenoptosis are somewhat not entirely defined and somehow arbitrary. Furthermore, the idea of organisms selected on the basis of their survival and reproduction capabilities is intertwined with that of organisms also selected on the basis of their ability to cooperate and interact, even by losing their lives or their distinct identities.
KEY WORDS: natural selection, holobiont, eusociality, superorganism, phenoptosis, selfish gene, symbiont, living beingDOI: 10.1134/S0006297923120052
INTRODUCTION
Natural selection is often described or known as the preferential reproduction of the individuals best suited to survive and reproduce, a description that could be defined as the classical Darwinian selection.
However, as it is possible to document extensively on the basis of a large body of works, the Darwinian description of natural selection appears to be an oversimplified description of selective phenomena. This is not a reductive criticism of the huge contribution of Charles Darwin because a great scientific revolution initially describes the fundamental concepts and subsequently these concepts are extended into the infinite complexity of phenomena. Indeed, it is possible to describe various “types” of natural selection, much more complex than selection at the individual level and leading to well-known phenomena that would be impossible to explain by the Darwinian description of natural selection. These phenomena do not invalidate or contradict the classic Darwinian selection, while representing a more complete and necessarily more complex description of the effects of natural selection, with results that are sometimes even paradoxical.
Each type of selective phenomenon has certainly been studied better and in more depth in authoritative works, some of which are then cited in the subsequent specific sections. The fundamental idea that natural selection acts at multiple levels has already been proposed in valuable works (e.g., [1-3]). The fact that in the course of evolution there have been fundamental transitions, for example towards increasing complexity [4] and towards increasing degrees of cooperation [5] has been the subject of other valid works.
However, this review aims to comprehensively and generally examine the various forms of natural selection, including natural selection related to holobionts and ecosystems, and selective phenomena at the level of individual gene sequences. Furthermore, it is considered the concept of phenoptosis, which is unlikely in terms of selection at the individual level while well explainable in terms of supra-individual selection and fundamental for the description and understanding of a wide series of phenomena.
DISCUSSION
In fact, eight general “types” of natural selection can be described.
Individual selection. In the classical Darwinian description of evolution, individuals who are the fittest to survive reproduce more than those less fit. The survival and preferential reproduction of individuals with better morphological and functional characteristics gradually change a species, so determining its evolution [6].
With the subsequent discovery that the characteristics of a species are determined by genes, it was also possible to explain how the necessary transmission of the characteristics of a species from one generation to the next took place. Genes also made it possible to define the mechanism of natural selection and evolution as the preferential diffusion of each gene based on the survival and reproduction capabilities that it determined. This can be summed up in a single very simple formula (1):
where ΔC is frequency variation between one generation and the next of a gene C manifesting its action in the individual I; S, advantage or disadvantage for I caused by the gene C; P, residual reproductive capacity of I at the age when the gene acts.
These concepts were summarized by Spencer, a great defender and popularizer of Darwin’s ideas, with the expression “survival of the fittest” individuals [7] which was later adopted by Darwin himself [8].
Kin selection and some related form of supra-individual selection. However, the idea of natural selection, and so of evolution, limited to the survival capacity of the single individual appeared insufficient even on the basis of simple considerations.
In fact, reproduction and the parental actions allowing the survival of the offspring force us to consider not only the single individual but also other genetically related individuals. For example, a mother who breastfeeds a newborn transfers part of her energies to it, reducing her own survival capacities and increasing those of the newborn. Actions of this kind cannot be summed up only in the concept of survival of the fittest individuals.
Furthermore, according to Darwin himself, the actions of members of a tribe who sacrifice their lives or put them in grave danger to save or defend the other members of the tribe would be favored by natural selection (see [9], p. 500). These actions also fall outside the literal definition of “survival of the fittest”, but are described by Darwin as favored by natural selection.
Consequently, although not excluding the selection at individual level, it is necessary to consider the consequences of genetic effects not only on the individual in whom a gene acts but also on other individuals.
Indeed, this clear difficulty was resolved by the proposal of the concept of “inclusive fitness” [10-13].
In formal terms, in the calculation of natural selection, considering a gene C which determines or influences an action in the individual I1 where it is present, causing an advantage (S1 > 0) or a disadvantage (S1 < 0) for I1, it is indispensable also consider possible advantageous or disadvantageous effects on other genetically related individuals (Sx for Ix, with a coefficient rx of genetic relationship between Ix and I1). Furthermore, for each of the individuals, it is necessary to consider the residual value of the reproductive capacity (Px) at the moment in which the gene acts. These concepts are summarized by the formula (2):
where ΔC is frequency variation between one generation and the next of a gene C that acts in the individual I1; n is number of the individuals IX (I1, I2, ..., In) genetically related to I1 for which the actions of the gene C have any effect.
The gene C is favored by natural selection and so increases its frequency when the summation is positive, while the contrary happens when the summation is negative.
When the gene C acts only on individual I1, as r1 = 1, the formula (2) becomes (3):
which is the same of formula (1).
In particular cases, the inclusive fitness formula can easily be extended to group selection.
Case 1. Let us consider a species divided into demes each composed of identical individuals being originated from a single clone and hypothesize that:
– Following particular events, if no individual sacrifices himself, there is damage for each individual equal to –S;
– Conversely, let us hypothesize that, within a deme, among n individuals with a gene C, nd individuals sacrifice themselves and die (therefore with a damage Sd equal to –1), and ns individuals survive and enjoy an advantage Ss. So, since in a monoclonal deme the kinship coefficient rx is always equal to 1, neglecting for the sake of simplicity the reproductive value which is assumed to be the same for all, the diffusion or elimination of gene C by natural selection will depend on the formula (4):
This means that C will be favored if (5):
Case 2. Let us now consider the case in which the deme is composed of individuals deriving from several clones (1, 2 ... z). If gene C exists in clone 1, the probability that it exists in clone x will be equal to the kinship coefficient between clones 1 and x (rx). Gene C will be favored by natural selection if (6):
where, nx,d are the individuals in a clone X that sacrifice themselves and nx,s the survivors in the same clone in the case of an event similar to that of case 1 [14].
It should be noted that formulas (4), (5) and (6) are all developments of formula (2) that describes inclusive fitness. These formulas demonstrate that, at least in particular cases, inclusive fitness describes forms of group selection.
The importance of inclusive fitness is that, among other things, it can very well explain actions that damage the individual in which a specific gene acts, a fact that is inexplicable if only selection at the individual level is considered.
More generally, forms of supra-individual selection, of which inclusive fitness is a part, can very well explain the innumerable phenomena defined by the general term phenoptosis proposed by Vladimir Skulachev [15, 16], in which an individual sacrifices his own life or exposes it to a serious danger due to actions that are genetically determined or influenced [17].
Phenomena of this type are widespread in nature and have been well known for some time [18] but only after the definition proposed by Skulachev they have been highlighted as phenoptotic phenomena [17].
Even in recent times, such actions are considered unlikely as they are certainly disadvantaged by selection at the individual level. E.g., for the case of possible aging-causing genes: “… any hypothetical ‘accelerated ageing gene’ would be disadvantageous to the individual. It is therefore difficult to see how genes for accelerated ageing could be maintained in stable equilibrium, as individuals in whom the genes were inactivated by mutation would enjoy a selection advantage.” [19]; “The anomalous nature of ageing as a putative adaptation is that it is bad for the individual in which the process is exhibited. An animal that grows to maturity and thereafter reproduces indefinitely has, other things being equal, a greater Darwinian fitness than one that grows to maturity and then survives and reproduces for only a fixed period of time.” [20]
However, overcoming these objections that are invalidated by the consideration of supra-individual selection mechanisms, aging, which may be defined as “increasing mortality with increasing chronological age in populations in the wild” [21], has been described as a sort of “slow phenoptosis” [22] and classified as “obligatory and slow phenoptosis” [17]. The theoretical considerations and empirical data supporting this thesis as an alternative to aging as a phenomenon determined by insufficient selection against harmful agents are widely discussed elsewhere [14].
Holobiontic selection. By limiting the discussion to multicellular species like ours, with regard to natural selection it is misleading to consider individuals as isolated organisms continually undermined and exploited by countless other species, which can be microbial (viruses, bacteria – including mycoplasmas – fungi, protozoa, etc.) or even multicellular (mites, intestinal worms, etc.). These other species are considered as foreign organisms that are generally harmful to survival, with particular rare exceptions, for example bacteria that are useful because capable of synthesizing vitamins [23, 24]. Even when these species behave as harmless commensals, they are considered a potential danger which is held back by the body’s defenses but manifests itself when the body is weakened and is no longer able to fight them. According to this conception, these organisms are important for the individual selection mainly because they represent a threat to survival.
For our species, this conception is strongly contradicted by the empirical evidence which leads to a radical rethinking.
First of all, it is necessary to consider the variety and number of species that are present on the external and internal body surfaces and also in more internal parts of the human organism:
– The number of bacteria living in the gut, and on skin and mucous surfaces has been estimated at 3.8 × 1013, i.e., a higher number than that of the cells in the human body estimated at 3 × 1013 [25];
– The number of viruses present in each part of the human body [26, 27] has been estimated at around 1013 [28], i.e., a number comparable to that of bacteria. Viruses are also present in areas of the body that are typically considered sterile. For example: bloodstream [29]; cerebrospinal fluid [28, 30] and breast milk [31];
– In every part of the internal and external surfaces of our organism, there are complex ecosystems composed of numerous species of microorganisms. For example, the mucous membranes of the oral cavity host “fungi, viruses and protozoa” and “over 700 species of bacteria” [32]. The microorganisms present in the gut ecosystem “have a combined metabolic capacity equivalent to that of the liver, justifying their description as an additional human organ ...” [33]. The ecosystems hosted by the respiratory tract and the skin have a similar complexity and it has been estimated that the species belonging to all ecosystems present in the human body have genetic information which is over 150 times that present in the human genome [34];
– Archaea, a vast group of prokaryote microorganisms metabolically very different from bacteria, and capable of living even in extreme environments where bacteria could not resist, were discovered over 40 years ago [35]. For a long time, also due to the difficulties for their isolation and in vitro cultivation, archaea were not considered relevant for human physiology and pathology [36]. However, archaeal species are normal components of human body ecosystems such as those of intestine, oral cavity, vagina, and skin [37]. For the archaea present in the human intestine, a recent study has highlighted the presence of over 1000 distinct genomes [38]. The importance of archaea in both physiological and pathological conditions is the subject of growing interest [39, 40];
– Two species of mites (Demodex folliculorum and Demodex brevis), small arthropods belonging to the subclass Acari, are present in the human hair bulbs, in an estimated number of about 1000-2000 mites for each individual [41].
Now consider the large number of species that coexist with our body, in every part of it, the huge number of genes present in these species, most of which are microbial [42], and the large number of relationships between all these species, and between them and our species. This extraordinary set of species and their mutual relationships, has been defined as a holobiont, a concept proposed in 1943 – see [43] – and later reproposed in 1991 [44], or also as supraorganism/superorganism [33], where there is a host organism (host, or main biont) and the other organisms that constitute it (other bionts).
As examples of the interrelationships:
– There are about 200 special oligosaccharides (HMOs) in human breast milk [45], which in the newborn are useful for the growth of particular intestinal bacteria and perform other functions related to gut microbiome [46], so demonstrating close interrelationships between the host organism and the other components of the human holobiont;
– In populations that do not live according to modern lifestyles and where, among other things, the eradication of intestinal worms is not practiced, the presence of helminths and other parasites is the rule [47, 48]. These infestations are the subject of great attention due to the pathologies they can cause, but it is also known that the presence of these parasites is important for the regulation of the immune system [49, 50]. Furthermore, the eradication of helminths and alterations of the intestinal microbiome cause autoimmune diseases [51, 52] and allergic diseases [53].
However, considering the close relationships among all the component species of a holobiont, in particular the dependence of the host for many functions on the other bionts and the relationship between the survival of the host and the survival of the other bionts, it is unlikely that natural selection would not exercise actions on the bionts that constitute the holobiont and on the relationships among the bionts, and so, indirectly, on the entire holobiont. It is likely that this type of selection, which we can briefly define as holobiontic, cannot be described with the same conciseness as individual selection, and indeed there are no formulas available or which can be easily developed.
The concept of selection at the holobiont level does not mean that for natural selection the holobiont is a distinct unit (i.e., a unit of selection), or something that evolves as a species for the effects of natural selection. However, certainly natural selection acts on the countless species that constitute the holobiont, also as a function of the numberless interrelationships among these species, and therefore indirectly models the holobiont. In short, holobiontic selection is not a different form of selection but the overall effects of the usual selective mechanisms on a complex set of interacting species.
Origin of eukaryote cell. The hypothesis that the eukaryotic cell has its origin in the symbiosis of several microbial species was formulated for the first time by Mereschkowsky, in 1905, with regard to chloroplasts. These organelles were explained as individuals of one microbial species being phagocytosed and becoming endosymbionts of another microbial species. The different origin was mainly motivated on the basis of their autonomous duplication [54]. This thesis was neglected or considered unsustainable for a long time until 1967, when it was re-proposed for plastids (of which chloroplasts are an important type), and, for the first time, for mitochondria, also with autonomous duplication and endowed of DNA distinct from that of the cell nucleus [55].
The re-proposed idea initially met with considerable resistance (Sagan’s article was proposed and rejected several times before publication), but subsequently became widely shared following many observations that supported it [56-58].
The first is that prokaryotes are divided into two distinct domains on the basis of multiple biochemical differences: archaea (formerly archaebacteria) and bacteria (formerly eubacteria) [35, 59]. Indeed, the nucleic DNA and ribosomes of eukaryotes (eucarya) appear related to those of archaea while the DNA of mitochondria (mtDNA) and plastids (plDNA), and their ribosomes, appear phylogenetically related to that of bacteria. In particular, the correlation with chloroplasts appears to be greater, which is a sign of a less ancient symbiosis.
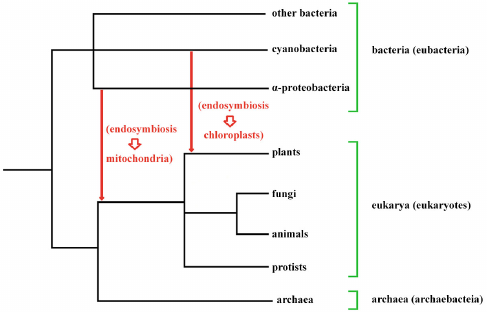
From this and other more specific observations, it has been deduced that in a first phase an archaea prokaryote (likely an Asgard archaea [60]) developed a symbiosis with a eubacterium, an α-proteobacteria, giving rise to the mitochondria and so constituting the progenitor of all eukaryotic cells. Subsequently, about a billion years ago, a eukaryotic cell developed a symbiosis with another eubacterium, a cyanobacterium, giving rise to chloroplasts [61] and thus constituting the progenitor of all plants (Fig. 1). Moreover, “Plastids have on multiple occasions also moved horizontally from eukaryote to eukaryote by secondary and tertiary endosymbiotic events.” [61].
Fig. 1. Eukaryotes probably originated from the endosymbiosis of a bacterium (an α-proteobacterium) in an archaebacterium, with the bacterium constituting the progenitors of the mitochondria. In one eukaryote branch, the subsequent endosymbiosis of another bacterium (a cyanobacterium) gave rise to chloroplasts and so to plants. This second endosymbiosis did not affect the ancestors of animals, fungi and protists.
After each endosymbiosis was stabilized, it is likely that there was a progressive reduction in the number of endosymbiont genes, either because they were no longer needed or because they existed in DNA host, and that there have also been transfers of genes from the endosymbiont to the host, or even replacements of host genes with genes from the endosymbiont [62].
Moreover, phylogenomic analysis of many archaeal and bacterial genomes has shown that the incorporation in archaeal genomes of many bacterial genes is characteristic of the major groups of archaea and that probably at the origin of each archaeal branch there is “massive acquisition” of bacterial genes [63]: “These findings have obvious and striking implications for the origin of eukaryotes. Acquisitions of numerous bacterial genes that amount to genomic chimaerism and lead to substantial remolding of cell physiology and emergence of groups with new lifestyles appears to be a recurrent rather than unique event in evolution, at least in archaea. Could it be that most if not all major groups of archaea emerged from botched endosymbiotic events? Should that be the case, eukaryogenesis only differs in that the endosymbiont survived, retaining part of its physical and genetic identity.” [63]
The symbiosis that gave rise to the eukaryotes are very ancient and very intimate, so much so that (i) the independent life of the single symbionts is impossible; and (ii) the organelle genome has been largely replaced by analogous host genes or transferred into the host nuclear genome [64]. Nonetheless, possible situations of conflict between the symbionts can arise, in particular because the organelles are endowed with their own genome with selective factors that can be in conflict with those of the host. For example, possible mutants of the mitochondria, with “selfish” actions, could favor their own reproduction and at the same time damage the entire symbiotic organism. Perhaps, precisely to limit the spread of selfish mitochondrial genes, the transmission of mitochondria from one generation to the next is limited to a single sex. In mammals, the paternal mitochondria are marked during spermatogenesis with a particular small protein, the ubiquitin, which then after fertilization activates the destruction of the mitochondria [65]. In this way the mitochondria of the progeny are only of maternal origin and the variability of the mitochondria is curbed, hindering the possible transmission of selfish mitochondria.
However, these brief notes are by no means intended to be a precise and complete history of the fascinating events which are at the origin and development of the eukaryotic organisms. They just want to indicate that in such a story there are events of symbiosis and gene transfers between very different species and that these events go beyond the simple logic of the evolution of a single species determined by individuals more suited to survive.
Selection in multicellular organisms. Multicellular eukaryotic organisms are in fact a clone of genetically uniform cells, in which there are different types of cells with differentiated roles and, in particular, with the role of the reproduction of the whole organism entrusted to specialized cell types. Multicellularity in eukaryotic organisms has developed distinctly multiple times. Restricting the definition of multicellularity to the intrinsically multicellular organisms, i.e., those with a heritable phenotype and cellular sustained cell-to-cell interconnection, communication, and cooperation, it has been estimated that multicellularity developed at least 11-fold among eukaryotes. [66]. Namely: once in the origin of animalia (metazoa) and amoebozoa, three times in the fungi (for chytrids, ascomycetes, and basidiomycetes), and twice in each of the photosynthetic eukaryotic clades (rhodophytes, stramenopiles, and chlorobionta) [67, 68].
Even among prokaryotes, multicellularity has evolved several times (e.g., for actinobacteria, myxobacteria, and cyanobacteria [69]). Among cyanobacteria it has been demonstrated that multicellularity developed several times and then often there was a return to the single-celled condition [70].
However, here we do not want to investigate or illustrate the conditions and mechanisms that led to the development of multicellularity and which then conditioned its subsequent developments (or sometimes the return to unicellularity), very interesting topics widely debated in other works (e.g., [66, 71, 72]), but only to focus on some essential aspects of eukaryote multicellularity useful for the discussion of the present work.
In a eukaryote multicellular organism:
– the reproduction of the organism is entrusted exclusively to specialized cell types;
– the other cells can reproduce, inside the organism and when this is useful or necessary for the whole organism, but this does not mean reproduction of the organism as a whole;
– each cell, when this is useful or necessary for the organism, sacrifices itself by mechanisms that can be defined as programmed cell death (PCD).
In animals, the various types of PCD derive from the needs for morphologic development or physiological functions or even to allow continuous cell renewal [73].
With regard to the last necessity, in tissues not subject to particular physiological needs or conditions of wear, there is a type of genetically regulated spontaneous cell death, defined as apoptosis, which was discovered quite late [74] because it is present in apparently unchanging tissues. In reality, apoptosis is present in most cell types [14, Chapt. 6], and even, improperly, the term is used as a synonym for PCD [75], when in fact it is only one of PCD forms.
Apoptosis and other kinds of PCD are types of cellular sacrifice of some of the cells of a multicellular organism, which is useful for the multicellular organism and does not mean the death of the whole organism. The term inspired the birth of the word phenoptosis [15, 16], which includes numberless types of sacrifice of an individual in favor of other genetically related individuals, a typology of widespread phenomena favored in the context of supra-individual selection [17]. Thus, apoptosis and other forms of PCD are not a particular type of phenoptosis, although somehow the selective logic that leads to PCD is analogous to that at the origin of phenoptotic phenomena.
In fact, the birth of multicellular organisms has been defined as the result of a kind of “phenoptotic pact” [14, Chapt. 2] in which the cells of a clone are united in a multicellular organism with the differentiation of their functions and simultaneously with the availability to sacrifice where this is useful for the new multicellular organism.
For the new organism, natural selection follows the same rules valid for unicellular organisms, while for the single cells of the new organism the rules change according to the adaptive needs of the new organism.
Selection and eusociality. The term eusociality defines the particular organization of numerous species in groups where, in general, there is: reproduction reserved to few individuals, division of roles among various types of individuals (defined as castes) with physiological and morphological differentiation according to the role, and joint rearing of the young [13].
Species that fall under the definition of eusociality are the ants, many species of bees, wasps, and termites, some species of thrips, aphids, snapping shrimps, a beetle species [76], and two mammal species, the naked mole rat (Heterocephalus glaber) [77] and Damaraland mole-rat (Fukomys damarensis) [78].
For a time eusociality, starting from the fact that ants, bees, and wasps have a particular sex-determination system defined as haplodiploidy, was explained as a direct consequence of kin selection mechanisms [13]. Subsequently, this explanation proved to be insufficient (non-significant relationship between haplodiploidy and eusociality; presence of many species that were both eusocial and non-haplodiploid) and was overcome by means of precise models of population structure. These turned out to be a simpler and better approach which allowed the comparison between alternative conditions and provided a precise context for interpreting the natural observations [79].
A key element to explain the origin and maintenance of eusociality has been considered the ancestral condition of monogamy, which allows the formation of a genetically homogeneous group: “We found that mating with a single male, which maximizes relatedness, is ancestral for all eight independent eusocial lineages that we investigated.” [80] However, this condition is necessary but insufficient and certainly other particular conditions are necessary [79] as, for example, for the only two species of eusocial mammals [77, 78].
The organization of eusocial species, particularly that of ants and eusocial termite and bee species is extraordinary [13] and a eusocial colony has been defined as a “superorganism, essentially a new kind of organism built up of organisms of the old kind.” [76]
However, this definition does not highlight a further degree of complexity since such a “superorganism” is more precisely composed of holobionts.
Let us consider the case of termites, animals essential for the metabolization of plant cellulose:
– “Culture-independent studies indicate that a single termite species harbors several hundred species of gut microbes unique to termites, and that the microbiota is consistent within a host termite species.” [81];
– “Termite-gut microbiota form a complex, highly structured, but stable microbial community comprising largely yet-uncultivated, novel and diverse species” [81-84];
– Among other things, termites need other species present in their intestines to metabolize cellulose, their main food [85, 86].
Furthermore, adding a further degree of complexity: (i) in the gut of many termites there is the prevalence of “cellulolytic flagellates of the genus Trichonympha, which are consistently associated with bacterial symbionts” necessary for cellulose lysis [87]; and (ii) for the recycling of uric acid nitrogen, a critical function for animals that are predominantly wood-feeding, termites need particular bacteria present in their intestines [84, 88, 89].
Selection at the level of single genes, or groups of genes. Natural selection, both for unicellular and multicellular organisms, also acts within single cells at the level of single genes, or groupings of genes, which are somehow favored in their diffusion by actions defined as “selfish” [90]. This adjective, excluding any impossible ethical meaning, simply indicates that particular effects of a genetic sequence favor an increase in the frequency of the sequence even if this involves damage to the organism in which the sequence expresses its action. A detailed discussion of this huge topic is impossible and incongruous here and we refer for a fuller exposition to the work cited above [90].
Here only some general concepts and facts will be set forth which may be useful for the discussion of this paper.
An increased frequency of a genetic sequence can be achieved by many types of mechanisms. The most widespread and best studied belong to two main categories [90]:
– Actions of duplication of the sequence of a gene, or of a group of genes, with the insertion of copies in other points of the genome;
– Actions defined as “drive” by which a gene, or a group of genes, increases its frequency, favoring its diffusion and hindering that of alleles.
(1) Genes increasing their frequencies by duplication. Transposable elements. Among the “selfish” genes, the so-called transposable elements constitute an important group. While other types of genes “compete” with their alleles to be present at a given locus, transposable elements try to place copies of their respective sequences elsewhere in cell’s genome. Because they try to duplicate themselves, i.e., move, to other parts of the DNA they have also been known as “mobile DNA” [91]. Their discovery dates back to 1951 by Barbara McClintock [92], and for many years the acceptance of their existence met with strong resistance, but their discoverer was awarded the Nobel prize in 1983 [93]. They have been and are the subject of a large number of studies. An authoritative 2002 book dedicated to this topic offered 50 contributions based on thousands of references [94] and there are many more recent works (e.g., [95, 96]).
It should be emphasized that transposable elements are not only present in eukaryotic cells but have also been described for prokaryotes and viruses (e.g., for DNA Transposons see [97]).
Among the transposable elements, there are three main types that are quite different in terms of structure and duplication mechanisms:
* DNA Transposons [98]
The sequence of DNA Transposons mostly encodes a single protein whose action is the excision of the sequence from where it is present, and its insertion in another point of the DNA. It is useful to add that DNA transposons can be inactivated by mutations and that they are also capable of horizontal transmission with the passage from one species to another [99, 100].
* LINEs (long interspersed nuclear elements) and SINEs (short interspersed nuclear elements) [101, 102]
In general, a LINE codes for one or two proteins whose actions are as follows [103, 104]: (i) using an internal promoter the sequence is transcribed into RNA; (ii) this RNA goes to the cytoplasm where it is translated forming one or two proteins; (iii) one of these proteins binds to the aforementioned RNA; (iv) the RNA–protein complex then moves into the nucleus and is inserted into a point of the DNA different from that of origin (depending on the type of LINE, it can be random or limited to some particular sites).
The SINE sequences do not code for any protein but have particular short sequences which allow the exploitation of the mechanisms of the LINE sequences for their duplication. In fact, they are “parasites” of the LINE sequences and are able to duplicate even without no protein-coding capacity [105, 106].
* LTR (long terminal repeat) Retroelements, which are retrotransposons [107]
LTR Retroelements are more complex than the aforementioned other two categories of transposable elements. Generally, they code for three proteins with enzymatic activity and two proteins with structural functions to form a capsid. The DNA of these LTR Retroelements is transcribed into RNA, which can either be translated into proteins to form a capsid, or remain as RNA which has to be inserted into the capsid. Subsequently, the RNA is reverse transcribed and integrated at a different point in the DNA [108].
A particular type of LTR retrotransposons, present in all types of vertebrates, is constituted by the endogenous retroviruses (ERVs) which are retroviruses integrated into the genome and thus become transposable elements [109]. ERVs have been documented in various vertebrate species, including baboons and mice [110].
Transposon elements can be inserted at points that are indifferent to host cell functions or at points that somehow modify those functions. Similar to mutations, most of the time these changes are harmful, but in some rare cases they can cause improvements or innovations in the functions of the organism. Therefore, their effects are analogous to those of mutations [90].
Furthermore, transposon elements can cause larger modifications of the genetic heritage (e.g., duplications, deletions, rearrangements, inversions, translocations) which are often harmful or lethal but which can be significant elements of non-minimal modifications of the genome “unlikely to arise by any other means (e.g., during DNA replications)” [90], pp. 280-287.
Apart from this, in the long run, as the copies multiply, the DNA of the host cell and the related metabolic demands increase more and more. For plants it has been demonstrated that a greater load of transposable elements is related to a greater risk of extinction [111]. Among vertebrates this has been demonstrated for reptiles and birds, which have the smallest mean genome sizes, but not for fish, amphibians, and mammals, which have higher genome sizes [112]. Therefore, it is important that the rate of copy number expansion of transposon elements is very low. A limiting factor for the expansion of the transposon elements, among other things, is that any mutations of the sequence can invalidate the duplication mechanism and therefore many of the duplicated sequences become inactive sequences, which however remain to unnecessarily burden the length of the DNA [90].
The genetic weight of the transposon elements is not irrelevant, but the cell appears to have mechanisms which greatly slow down the rhythms of their accumulation.
In particular, the estimated numbers of inserts for DNA transposons, LINEs, SINEs, and LTR retroelements are 400,000 (3% of the genome), 1,000,000 (21%), 2,000,000 (14%), and 600,000 (9%), respectively; and the total represents about 46% of human or mouse genome (24% acquired in the last 75 million of years and 22% before) (data for human genome from [113] and for mouse genome from [114]).
(2) Genes increasing their frequencies by drive actions. There are many genes of this category [90]. For the sake of brevity, only two general types will be mentioned:
* HEGs (homing endonuclease genes) (see [90], pp. 198-217). HEG sequence is translated into an endonuclease which, in the heterozygous condition, cleaves DNA at a specific recognition site opposed to HEG sequence. Then, cell repair system, necessary to rejoin the two ends of a broken DNA sequence, uses the template of HEG sequence in a DNA strand to insert it in the middle of the recognition site of the other strand. So, the HEG sequence passes from the heterozygous to the homozygous condition and doubles its frequency [115].
HEGs are transmitted horizontally by bacteriophages [116] and this would explain their greater presence in eukaryotes with simpler organization [90].
* Selfish sex chromosomes (see [90], pp. 60-95). Most species have different sex chromosomes (females XX and males XY, or, in other species, males ZZ and females ZW). This determines different conditions for the transmission of the genes existing in the parts of the chromosomes which are not identical in the two sexes. So, different selection conditions arise which can favor “selfish” genes in these parts of the chromosomes. For example, in some diptera species there are “killer X chromosomes” which reduce male frequency in the offspring [117]. Conversely, in two mosquito species, “killer Y chromosomes” have been described that reduce female frequency in the offspring [118]. As a likely defense against this type of selfish genes, in mammals, transcription from sex chromosomes is blocked early in meiotic prophase and “backup” copies of some X-linked genes are expressed in meiotic and postmeiotic cells [119].
Plasmids. In addition to these “selfish” genes, on the blurred border between transposable elements and viruses, there are the plasmids, genetic elements that are widely diffused among prokaryotes [120], and have a higher level of complexity than the transposable elements: “... our provisional understanding of a plasmid is that it is a genetic element that is extra-chromosomal, yet some plasmids are temporarily integrated into chromosomes; it is a small circular genome, yet some plasmids are linear; it is mobile or self-transmissible, yet about half of the known plasmids are non-mobilizable or not self-transmissible; it is transferred as naked DNA, yet some plasmids have their own packaging; it can have positive or negative effect on the host fitness, yet some plasmids are cryptic and have no clear impact on the host.” [121]
Plasmids have the important effect of allowing the transmission of genes between different individuals, even of different prokaryotic species: “Plasmids are a major driver of horizontal gene transfer in prokaryotes, allowing the sharing of ecologically important accessory traits between distantly related bacterial taxa.” [122]
The effects of plasmids, together with those of bacteriophages, are among the main ways by which antibiotic resistance can spread rapidly among bacteria [123].
Natural selection in an ecosystem. The individuals of a species, or more precisely its holobionts, are not and cannot be organisms living in isolation without contacts and interrelationships with individuals/holobionts of other species. The set of these species, their relationships and the physical environment in which they live constitute an ecosystem. This concept, fundamental for the explanation of innumerable biological phenomena, was for the first time defined as “ecosystem” in a publication in 1935 [124], and was soon refined including in it “not only the organism-complex, but also the whole complex of physical factors forming what we call the environment” [125].
Natural selection is certainly influenced by the innumerable interrelationships between the species of an ecosystem. Numberless cases of natural selection deriving from these interrelationships are fundamental or peculiar characteristics of an ecosystem. For example:
* A basic need for the species of an ecosystem is nitrogen fixation.
Without it, plants cannot survive nor is life possible for species
directly or indirectly depending on plants. Various plants, as many
legume plants, are in symbiosis with nitrogen-fixing bacteria (see [126], Chap. 12). The roots of many plants and trees
are symbiotic with mycorrhizal fungi and particular soil microbes to
which they supply carbohydrates and in return obtain phosphorus and
nitrogen (N), also from dead organic matter [127]: “While plants grown with either soil
microbes or AM [Arbuscular mycorrhizal] fungi acquired twofold and
threefold more N from the organic matter than control plants,
respectively, plants grown with both soil microbes and AM fungi
acquired ten to twelvefold more N from the organic matter than control
plants.” [127];
* “Arbuscular
mycorrhizal (AM) fungi are mutualistic symbionts living in the roots of
80% of land plant species, and developing extensive, below-ground
extraradical hyphae fundamental for the uptake of soil nutrients and
their transfer to host plants. Since AM fungi have a wide host range,
they are able to colonize and interconnect contiguous plants [of the
same species or of different species] by means of hyphae extending from
one root system to another.” [128] This
connection system has also been defined as a “wood-wide
web” [128-130];
* The connection between plants is allowed by symbiotic fungi, through
a link of the type: roots of plant 1 – mycorrhizal network
– roots of plant 2 [131]. In fact, the
connection is multiple and also affects plant microbiomes and dodders
(see below). It takes place through communications both with direct
physical contact (“wired”) and without direct contact
(“wireless”):
“Wired communications involve direct signal transfers between plants mediated by mycorrhizal hyphae and parasitic plant stems. Wireless communications involve plant volatile emissions and root exudates elicited by microbes/insects, which enable inter-plant signaling without physical contact. These producer-plant signals induce microbiome adaptation in receiver plants via facilitative or competitive mechanisms.” [132];
* “Dodders (Cuscuta spp.) are plant holoparasites that acquire
water and nutrients from host plants via the haustorium, which
physically connects the parasite to its host. Dodder species have broad
host range, and can interconnect several plant species or clusters of
the same species ... to generate a common dodder network. The common
dodder network can be considered as an inter-plant highway that
translocates large numbers of proteins, RNA, metabolites, and plant
viruses over a distance of at least 100 cm”. [132-134];
* The removal
of even a single key component of an ecosystem (e.g., an apex predator)
can cause serious alterations of the entire ecosystem. This was
underestimated or not understood until the definition of the concept of
ecosystem and its implications. A well-documented example is that of
the severe ecosystem alterations of the Yellowstone Park, and of their
corrections only since 1995. After a long period of unlimited hunting
for wolves and other predators (e.g., mountain lions, bears and
coyotes), the wolf population was completely wiped out from Yellowstone
Park in 1926. This resulted in an overgrowth of deer population, severe
alterations to vegetation eaten by the elks, severe reductions in bear
and beaver numbers, reductions in numbers of hares, rats, muskrats,
ducks and fish, and also unstable stream banks. These serious
alterations of the ecosystem were only partially reduced by the
continuous elimination of part of the deer population. In 1995, 19
wolves were reintroduced to the Yellowstone Park. In a few years it was
possible to observe the reduction in the number of deer, the
restoration of normal vegetation growth, the return of bears and
beavers, an increase in the population of the species whose reduction
had been observed, and the stabilization of watercourses, with a return
to the previous conditions of the ecosystem [135,
136];
* In ecosystems there are also cases
of cooperation between individuals of the same species, with serious
risk to their lives, to fight against predators. For example, the
predominant males of chacma baboons (Papio ursinus) [137] and of yellow baboons (Papio
cynocephalus) [138] strenuously defend their
herd in case of attack by predators even at the cost of their own
lives, which it is an example of optional phenoptosis [17];
* Another case of optional phenoptosis is
that of bacterial phytoplankton mass suicide to block the spread of a
phage [139];
* A species in general is
unable to directly synthesize or absorb all the substances the organism
needs. E.g., our species needs various vitamins, indispensable amino
acids, and minerals, two essential fatty acids, and choline [140]. Under normal ecological conditions, without
the interaction with many other species, these essential nutrients
would not be available and survival would be impossible.
In short, an ecosystem is not simplistically a set of individuals of different species that continuously struggle to overwhelm each other, but also a very complex intertwining of various types of interrelationships between the holobionts of numberless species and the correlated natural selection.
As with holobiontic selection, it is important to highlight that an ecosystem is not a distinct unit of selection. However, natural selection certainly acts on all the species (or more precisely holobionts) which constitute it, in relation to their characteristics of which the interrelationships with the other species of the ecosystem are certainly an important part. Consequently, natural selection, or more precisely the set of countless selective actions on each component of the ecosystem, indirectly shapes the entire ecosystem. As with other “types” of natural selection, that at the ecosystem level is not a different form of selection but the effects of the same selective mechanisms in a much more complex context, which is difficult to describe in mathematical terms.
DEFINITIONS THAT ARE NECESSARILY ARBITRARY
The previous brief description of the various types of natural selection implicitly shows that some essential biological definitions have uncertain boundaries. Or rather, it is always possible to define limits for them, but these boundaries, however useful and necessary, are often arbitrary and questionable:
About the definition of living being. Entities such as viruses, and other MGEs (Mobile Genetic Elements), including those previously referred to as selfish genes, are generally not considered part of the tree of life [141]. However:
* “MGEs are semiautonomous replicative genomic entities that are
ubiquitous in the natural environment and believed to be an intrinsic
part of cellular evolution [142]. They include
viruses which may encode one or more proteins comprising the viral
particle (virion) encasing the genome of the respective MGE [142]. Categorically, viruses are believed to be the
most abundant biological entities on the planet, shaping ecological and
evolutionary components of the biosphere [143].” [144];
* It
was pointed out in a previous section that in human/mouse genome about
46% of the DNA is made up of mobile elements;
*
“Approximately 8% of the human genome, over four times more than
its protein-coding regions, comprises sequences of viral origin that
are known as human endogenous retroviral elements (HERVs).” [145]
Consequently, by accepting a definition of life excluding viruses, MGEs, and selfish genes, we will also have to accept that over half of our DNA originates from non-living entities and that most of the “biological entities on the planet” [144] are non-living.
The exclusion of viruses from the living world is also undermined by the existence of the so-called giant viruses:
* “Viruses are the most prevalent infectious agents, populating
almost every ecosystem on earth. Most viruses carry only a handful of
genes supporting their replication and the production of capsids. It
came as a great surprise in 2003 when the first giant virus was
discovered and found to have a >1 Mbp genome encoding almost a
thousand proteins. Following this first discovery, dozens of giant
virus strains across several viral families have been reported.”
[146];
* “Different giant viruses
have robust metabolic machinery, especially those in the
Phycodnaviridae and Mimiviridae families.” [147];
* “Can giant viruses get infected?
Yes, they can! Giant viruses with a cytoplasmic replication-cycle, such
as mimivirus, can be targeted by other viruses, called
‘virophages’, which use their host’s viral factory to
replicate. Virophages are small DNA viruses with genomes of ∼20 kb
encoding about 20 proteins. They express their genomes as late genes
using the giant virus transcription machinery.” [148].
About the definition of species. According to classical ideas, the term species defines a precise concept with clear distinctions between the various species. Anyway:
* Each eukaryotic cell is the evolution of a very ancient symbiosis
between a species of the domain archaea and a species of the domain
bacteria, subsequently enriched, in the progenitor of the
photosynthetic species, with a further symbiosis with another species
of the domain bacteria. These various components of the eukaryotic
cell, among other things, have distinct DNA, but cannot live
independently;
* There are also other cases of symbiosis in which
the independent life of the single component species is impossible
(e.g., lichens [149]).
And, as shown before:
* Contact and fusion between genomes of the eukaryotic cell and genomes
of viral origin is frequent;
* The horizontal transfer of genes
between different species is well known;
* There are many cases
in which the definition of species is subordinated to the definition of
life or living beings.
* These facts make the definition of
species difficult to delimit precisely.
About the definition of individual. The definition of individual appears well delineated according to the usual ideas. However:
* As far as multicellular eukaryotic organisms are concerned, each
“individual” is in fact a highly evolved colony of cells
with specialized functions on the basis of different epigenetic
modulations [150], and often capable of
reproducing autonomously. However, only one type of cell has the
function of transmitting the genes that characterize the colony to the
next generation. In the evolutionary past of every multicellular
organism there has been a transition phase from a colony of
undifferentiated individuals to that of a colony with individuals with
differentiated functions;
* In eusocial species, which form
“superorganisms”, we have “individuals” with
differentiated functions. Only two types of individuals (e.g., for
bees, the queen bee, and the drones) have the function of transmitting
the genes of the superorganism;
* In cases of symbiosis, often
the distinction between the individuals that make up the symbiont is
not entirely definable (e.g., see the case of eukaryotic cell);
*
Apart from cases of symbiosis, each individual coexists with a myriad
of other species and constitutes a holobiont.
As with the definition of species, these cases make the definition of individual somewhat imprecise.
About the definition of phenoptosis. The definition of the concept of phenoptosis (see [15-17]) finds a case of uncertain delimitation when considering both eukaryotic multicellular organisms and organizations defined as eusocial [13].
In multicellular eukaryotic organisms, the cells have different functions and, where this is useful for the organism, are subject to forms of programmed cell death (PCD), including apoptosis. These cases of PCD fall outside the definition of phenoptosis, primarily because the individual cell types are not capable of autonomous life or reproduction.
In the case of the eusocial species, the individuals of the single functional types perform different functions, are unable to reproduce (except for the individuals specifically assigned to this function) and in particular cases sacrifice their life for the survival of the superorganism. These cases are regarded as phenoptotic phenomena, even though the single functional types of individuals, as well as the single cells of multicellular organisms, are unable to reproduce themselves or survive long in isolation from the superorganism.
For both multicellular organisms and the organization of eusocial species, one could say that at their origin there is ideally a “phenoptotic pact” in which several individuals form a colony differentiating their functions and with the availability to sacrifice their lives if necessary.
We therefore have two analogous situations described differently. An arbitrary solution is to establish that the definition of phenoptosis does not extend to the case of self-sacrificing cells in a multicellular organism. Here, as in the previous cases, the problem is not so much a lack of definition but that of complicated biological realities not always allowing for perfect delimitations.
CONCLUSION
In the classical conception of Darwinian selection, the individual struggles for its own survival and reproduction and there is little or no room for actions with different outcomes. An exception, highlighted by Darwin himself but not elaborated in its implications, is when tribesmen take mortal risk or sacrifice themselves for the survival of the tribe [9].
In biological reality, as summarily outlined in the previous parts, the cases are more complex and often apparently contradictory to each other.
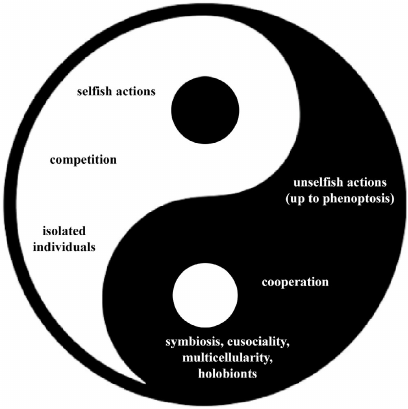
Natural selection favors and models phenomena that represent a complex interweaving between:
– “selfish” and “unselfish” actions (up to the supreme sacrifice of phenoptosis);
– acts of competition and others of cooperation;
– individuals operating as isolated entities and forms of close integration between individuals of various species or of the same species, with phenomena of symbiosis, eusociality, multicellularity, and the condition of holobionts.
This extreme variety of manifestations, summarized in Fig. 2 as a sort of biological yin-yang, does not mean a denial of the original Darwinian vision, but a development of this vision up to paradoxical consequences that perhaps would have astounded Darwin himself.
Fig. 2. Biological phenomena caused by natural selection.
Ethics declarations. The author declares no conflicts of interest in financial or any other area. This article does not contain any studies with human participants or animals performed by the author.
REFERENCES
1.Wilson, D. S., and Wilson, E. O. (2007) Rethinking
the theoretical foundation of sociobiology, Quart. Rev. Biol.,
82, 327-348, doi: 10.1086/522809.
2.Gardner, A. (2015) The genetical theory of
multilevel selection, J. Evol. Biol., 28, 305-319, doi:
10.1111/jeb.12566.
3.Goodnight, C. J. (2016) On the effectiveness of
multilevel selection, Behav. Brain Sci., 39, e99, doi:
10.1017/S0140525X15001053.
4.Szathmáry, E., and Smith, J. M. (1995) The
major evolutionary transitions, Nature, 374, 227-232,
doi: 10.1038/374227a0.
5.West, S. A., Fisher, R. M., Gardner, A., and Kiers,
E. T. (2015) Major evolutionary transitions in individuality, Proc.
Natl. Acad. Sci. USA, 112, 10112-10119, doi:
10.1073/pnas.1421402112.
6.Darwin, C. R. (1859) On the Origin of Species by
Means of Natural Selection, or the Preservation 1 of The Favoured Races
in the Struggle for Life, John Murray, London.
7.Spencer, H. (1864) The Principles of
Biology, Williams and Norgate, London.
8.Darwin, C. R. (1869) Origin of Species, 5th
ed., John Murray, London.
9.Darwin, C. R. (1871) The Descent of Man, and
Selection in Relation to Sex, John Murray, London.
10.Hamilton, W. D. (1964) The genetical evolution of
social behaviour. II, J. Theor. Biol., 7, 1-52, doi:
10.1016/0022-5193(64)90039-6.
11.Hamilton, W. D. (1970) Selfish and Spiteful
Behaviour in an Evolutionary Model, Nature, 228,
1218-1220, doi: 10.1038/2281218a0.
12.Trivers, R. L. (1971) The evolution of reciprocal
altruism, Quart. Rev. Biol., 46, 35-57, doi:
10.1086/406755.
13.Wilson, E. O. (1975). Sociobiology: The New
Synthesis, Belknap Press of Harvard University Press, Harvard.
14.Libertini, G., Corbi, G., Conti, V.,
Shubernetskaya, O., and Ferrara, N. (2021) Evolutionary Gerontology
and Geriatrics – Why and How We Age. Advances in Studies of
Aging and Health, 2, Springer, Switzerland, doi:
10.1007/978-3-030-73774-0.
15.Skulachev, V. P. (1997) Aging is a specific
biological function rather than the result of a disorder in complex
living systems: biochemical evidence in support of Weismann’s
hypothesis, Biochemistry (Moscow), 62, 1191-1195.
16.Skulachev, V. P. (1999) Phenoptosis: programmed
death of an organism, Biochemistry (Moscow), 64,
1418-1426.
17.Libertini, G. (2012) Classification of
phenoptotic phenomena, Biochemistry (Moscow), 77,
707-715, doi: 10.1134/S0006297912070024.
18.Finch, C. E. (1990) Longevity, Senescence, and
the Genome, University of Chicago Press, Chicago.
19.Kirkwood, T. B., and Austad, S. N. (2000) Why do
we age? Nature, 408, 233-238, doi: 10.1038/35041682.
20.Kirkwood, T. B., and Melov, S. (2011) On the
programmed/non-programmed nature of ageing within the life history,
Curr. Biol., 21, R701-R707, doi:
10.1016/j.cub.2011.07.020.
21.Libertini, G. (1988) An adaptive theory of the
increasing mortality with increasing chronological age in populations
in the wild, J. Theor. Biol., 132, 145-162, doi:
10.1016/s0022-5193(88)80153-x.
22.Skulachev, V. P. (2002) Programmed death
phenomena: from organelle to organism, Ann. N. Y. Acad. Sci.,
959, 214-237, doi: 10.1111/j.1749-6632.2002.tb02095.x.
23.LeBlanc, J. G., Milani, C., de Giori, G. S.,
Sesma, F., van Sinderen, D., and Ventura, M. (2013) Bacteria as vitamin
suppliers to their host: a gut microbiota perspective, Curr. Opin.
Biotechnol., 24, 160-168, doi:
10.1016/j.copbio.2012.08.005.
24.Zhan, Q., Wang, R., Thakur, K., Feng, J. Y., Zhu,
Y. Y., Zhang, J. G., and Wei, Z. J. (2022) Unveiling of dietary and
gut-microbiota derived B vitamins: Metabolism patterns and their
synergistic functions in gut-brain homeostasis, Crit. Rev. Food Sci.
Nutr., 22, 1-13, doi: 10.1080/10408398.2022.2138263.
25.Sender, R., Fuchs, S., and Milo, R. (2016)
Revised estimates for the number of human and bacteria cells in the
body, PLoS Biol., 14, e1002533, doi:
10.1371/journal.pbio.1002533.
26.Pride, D. T., Salzman, J., Haynes, M., Rohwer,
F., Davis-Long, C., White, R. A. 3rd, Loomer, P., Armitage, G. C., and
Relman, D. A. (2012) Evidence of a robust resident bacteriophage
population revealed through analysis of the human salivary virome,
ISME J., 6, 915-926, doi: 10.1038/ismej.2011.169.
27.Abeles, S. R., and Pride, D. T. (2014) Molecular
bases and role of viruses in the human microbiome, J. Mol.
Biol., 426, 3892-3906, doi: 10.1016/j.jmb.2014.07.002.
28.Liang, G., and Bushman, F. D. (2021) The human
virome: assembly, composition and host interactions, Nat. Rev.
Microbiol., 19, 514-527, doi:
10.1038/s41579-021-00536-5.
29.Moustafa, A., Xie, C., Kirkness, E., Biggs, W.,
Wong, E., Turpaz, Y., Bloom, K., Delwart, E., Nelson, K. E., Venter, J.
C., and Telenti, A. (2017) The blood DNA virome in 8,000 humans,
PLoS Pathogens, 13, e1006292, doi:
10.1371/journal.ppat.1006292.
30.Ghose, C., Ly, M., Schwanemann, L. K., Shin, J.
H., Atab, K., Barr, J. J., Little, M., Schooley, R. T., Chopyk, J.,
Pride, D. T. (2019) The virome of cerebrospinal fluid: viruses where we
once thought there were none, Front. Microbiol., 10,
2061, doi: 10.3389/fmicb.2019.02061.
31.Pannaraj, P. S., Ly, M., Cerini, C., Saavedra,
M., Aldrovandi, G. M., Saboory, A. A., Johnson, K. M., and Pride, D. T.
(2018) Shared and distinct features of human milk and infant stool
viromes, Front. Microbiol., 9, 1162, doi:
10.3389/fmicb.2018.01162.
32.Deo, P. N., and Deshmukh, R. (2019) Oral
microbiome: unveiling the fundamentals, J. Oral Maxillofac.
Pathol., 23, 122-128, doi: 10.4103/jomfp.JOMFP_304_18.
33.Glendinning, L., and Free, A. (2014)
Supra-organismal interactions in the human intestine, Front. Cell.
Infect. Microbiol., 4, 47, doi:
10.3389/fcimb.2014.00047.
34.Grice, E. A., and Segre, J. A. (2012) The human
microbiome: our second genome, Annu. Rev. Genomics Hum. Genet.,
13, 151-170, doi: 10.1146/annurev-genom-090711-163814.
35.Woese, C. R., and Fox, G. E. (1977) Phylogenetic
structure of the prokaryotic domain: the primary kingdoms, Proc.
Natl. Acad. Sci. USA, 74, 5088-5090, doi:
10.1073/pnas.74.11.5088.
36.Conway de Macario, E., and Macario, A. J. L.
(2009) Methanogenic archaea in health and disease: a novel paradigm of
microbial pathogenesis, Int. J. Med. Microbiol., 299,
99-108, doi: 10.1016/j.ijmm.2008.06.011.
37.Bang, C., and Schmitz, R. A. (2015) Archaea
associated with human surfaces: not to be underestimated, FEMS
Microbiol. Rev., 39, 631-648, doi:
10.1093/femsre/fuv010.
38.Chibani, C. M., Mahnert, A., Borrel, G., Almeida,
A., Werner, A., Brugère, J. F., Gribaldo, S., Finn, R. D.,
Schmitz, R. A., and Moissl-Eichinger, C. (2022) A catalogue of 1,167
genomes from the human gut archaeome, Nat. Microbiol., 7,
48-61, doi: 10.1038/s41564-021-01020-9.
39.Cai, M., and Tang, X. (2022) Human archaea and
associated metabolites in health and disease, Biochemistry,
61, 2835-2840, doi: 10.1021/acs.biochem.2c00232.
40.Mohammadzadeh, R., Mahnert, A., Duller, S., and
Moissl-Eichinger, C. (2022) Archaeal key-residents within the human
microbiome: characteristics, interactions and involvement in health and
disease, Curr. Opin. Microbiol., 67, 102146, doi:
10.1016/j.mib.2022.102146.
41.Litwin, D., Chen, W., Dzika, E., and
Korycińska, J. (2017) Human permanent ectoparasites; recent
advances on biology and clinical significance of demodex mites:
narrative review article, Iran. J. Parasitol., 2,
12-21.
42.Charbonneau, M. R., Blanton, L. V., DiGiulio, D.
B., Relman, D. A., Lebrilla, C. B., Mills, D. A., and Gordon, J. I.
(2016) A microbial perspective of human developmental biology,
Nature, 535, 48-55, doi: 10.1038/nature18845.
43.Baedke, J., Fábregas-Tejeda, A., and
Nieves Delgado, A. (2020) The holobiont concept before Margulis, J.
Exp. Zool. B Mol. Dev. Evol., 334, 149-155, doi:
10.1002/jez.b.22931.
44.Margulis, L., and Fester, R. (1991) Symbiosis
as a Source of Evolutionary Innovation, MIT Press, Cambridge.
45.Urashima, T., Hirabayashi, J., Sato, S., and
Kobata, A. (2018) Human milk oligosaccharides as essential tools for
basic and application studies on galectins, Trends Glycosci.
Glycotechnol., 30, SJ11-SJ24, doi:
10.4052/tigg.1734.1SJ.
46.Salminen, S., Stahl, B., Vinderola, G., and
Szajewska, H. (2020) Infant formula supplemented with biotics: current
knowledge and future perspectives, Nutrients, 12, 1952,
doi: 10.3390/nu12071952.
47.Dunn, J. C., Turner, H. C., Tun, A., and
Anderson, R. M. (2016) Epidemiological surveys of, and research on,
soil-transmitted helminths in Southeast Asia: a systematic review,
Parasit. Vectors, 9, 31, doi:
10.1186/s13071-016-1310-2.
48.Veesenmeyer, A. F. (2022) Important nematodes in
children, Pediatr. Clin. North Am., 69, 129-139, doi:
10.1016/j.pcl.2021.08.005.
49.Mishra, P. K., Palma, M., Bleich, D., Loke, P.,
and Gause, W. C. (2014) Systemic impact of intestinal helminth
infections, Mucosal Immunol., 7, 753-762, doi:
10.1038/mi.2014.23.
50.Gazzinelli-Guimaraes, P. H., and Nutman, T. B.
(2018) Helminth parasites and immune regulation, F1000Research,
7, 1685, doi: 10.12688/f1000research.15596.1.
51.Vieira, S. M., Pagovich, O. E., and Kriegel, M.
A. (2014) Diet, microbiota and autoimmune diseases, Lupus,
23, 518-526, doi: 10.1177/0961203313501401.
52.Weinstock, J. V., and Elliott, D. E. (2014)
Helminth infections decrease host susceptibility to immune-mediated
diseases, J. Immunol., 193, 3239-3247, doi:
10.4049/jimmunol.1400927.
53.Platts-Mills, T. A. E. (2015) The allergy
epidemics: 1870-2010, J. Allergy Clin. Immunol., 136,
3-13, doi: 10.1016/j.jaci.2015.03.048.
54.Martin, W., and Kowallik, K. V. (1999) Annotated
English translation of Mereschkowsky’s 1905 paper
‘Über Natur und Ursprung der Chromatophoren im
Pflanzenreiche’, Eur. J. Phycol., 34, 287-295.
55.Sagan, L. (1967) On the origin of mitosing cells,
J. Theor. Biol., 14, 255-274, doi:
10.1016/0022-5193(67)90079-3.
56.Gray, M. W., and Doolittle, W. F. (1982) Has the
endosymbiont hypothesis been proven? Microbiol. Rev., 46,
1-42, doi: 10.1128/mr.46.1.1-42.1982.
57.Gray, M. W. (2012) Mitochondrial evolution,
Cold Spring Harb. Perspect. Biol., 4, a011403, doi:
10.1101/cshperspect.a011403.
58.Gray, M. W. (2017) Lynn Margulis and the
endosymbiont hypothesis: 50 years later, Mol. Biol. Cell,
28, 1285-1287, doi: 10.1091/mbc.E16-07-0509.
59.Woese, C. R., Kandler, O., and Wheelis, M. L.
(1990) Towards a natural system of organisms: proposal for the domains
Archaea, Bacteria, and Eucarya, Proc. Natl. Acad. Sci. USA,
87, 4576-4579, doi: 10.1073/pnas.87.12.4576.
60.Van Wolferen, M., Pulschen, A. A., Baum, B.,
Gribaldo, S., and Albers, S. V. (2022) The cell biology of archaea,
Nat. Microbiol., 7, 1744-1755, doi:
10.1038/s41564-022-01215-8.
61.Sibbald, S. J., and Archibald, J. M. (2020)
Genomic insights into plastid evolution, Genome Biol. Evol.,
12, 978-990, doi: 10.1093/gbe/evaa096.
62.López-García, P., and Moreira, D.
(2015) Open questions on the origin of eukaryotes, Trends Ecol.
Evol., 30, 697-708, doi: 10.1016/j.tree.2015.09.005.
63.Koonin, E. V. (2015) Origin of eukaryotes from
within archaea, archaeal eukaryome and bursts of gene gain:
eukaryogenesis just made easier? Philos. Trans. R. Soc. Lond. B
Biol. Sci., 370, 20140333, doi: 10.1098/rstb.2014.0333.
64.Lang, B. F., Gray, M. W., and Burger, G. (1999)
Mitochondrial genome evolution and the origin of eukaryotes, Annu.
Rev. Genet., 33, 351-397, doi:
10.1146/annurev.genet.33.1.351.
65.Sutovsky, P., Moreno, R. D., Ramalho-Santos, J.,
Dominko, T., Simerly, C., and Schatten, G. (2000) Ubiquitinated sperm
mitochondria, selective proteolysis, and the regulation of
mitochondrial inheritance in mammalian embryos, Biol. Reprod.,
63, 582-590, doi: 10.1095/biolreprod63.2.582.
66.Niklas, K. J., and Newman, S. A. (2020) The many
roads to and from multicellularity, J. Exp. Bot., 71,
3247-3253, doi: 10.1093/jxb/erz547.
67.Niklas, K. J., and Newman, S. A. (2013) The
origins of multicellular organisms, Evol. Dev., 15,
41-52, doi: 10.1111/ede.12013.
68.Niklas, K. J. (2014) The
evolutionary-developmental origins of multicellularity, Am. J.
Bot., 101, 6-25, doi: 10.3732/ajb.1300314.
69.Bonner, J. T. (2000) First Signals: The
Evolution of Multicellular Development, Princeton University Press,
Princeton, doi: 10.1515/9781400830589.
70.Schirrmeister, B. E., Antonelli, A., and Bagheri,
H. C. (2011) The origin of multicellularity in cyanobacteria, BMC
Evol. Biol., 11, 45, doi: 10.1186/1471-2148-11-45.
71.Brunet, T., and King, N. (2017) The origin of
animal multicellularity and cell differentiation, Dev. Cell,
43, 124-140, doi: 10.1016/j.devcel.2017.09.016.
72.Ros-Rocher, N., Pérez-Posada, A., Leger,
M. M., and Ruiz-Trillo, I. (2021) The origin of animals: an ancestral
reconstruction of the unicellular-to-multicellular transition, Open
Biol., 11, 200359, doi: 10.1098/rsob.200359.
73.Fox, S. I., and Rompolski, K. (2019) Human
Physiology, 15th Edn. McGraw-Hill Education, New York.
74.Kerr, J. F., Wyllie, A. H., and Currie, A. R.
(1972) Apoptosis: a basic biological phenomenon with wide-ranging
implications in tissue kinetics, Br. J. Cancer, 26,
239-257, doi: 10.1038/bjc.1972.33.
75.Jacobson, M. D., Weil, M., and Raff, M. C. (1997)
Programmed cell death in animal development, Cell, 88,
347-354, doi: 10.1016/s0092-8674(00)81873-5.
76.Queller, D. C., and Strassmann, J. E. (2003)
Eusociality, Curr. Biol., 13, R861-R863, doi:
10.1016/j.cub.2003.10.043.
77.Griffin, A. S. (2008) Naked mole-rat, Curr.
Biol., 18, R844-R845, doi: 10.1016/j.cub.2008.07.054.
78.Wong, H. S., Freeman, D. A., and Zhang, Y. (2022)
Not just a cousin of the naked mole-rat: Damaraland mole-rats offer
unique insights into biomedicine, Comp. Biochem. Physiol. B Biochem.
Mol. Biol., 262, 110772, doi:
10.1016/j.cbpb.2022.110772.
79.Nowak, M. A., Tarnita, C. E., and Wilson, E. O.
(2010) The evolution of eusociality, Nature, 466,
1057-1062, doi: 10.1038/nature09205.
80.Hughes, W. O., Oldroyd, B. P., Beekman, M., and
Ratnieks, F. L. (2008) Ancestral monogamy shows kin selection is key to
the evolution of eusociality, Science, 320, 1213-1216,
doi: 10.1126/science.1156108.
81.Hongoh, Y. (2010) Diversity and genomes of
uncultured microbial symbionts in the termite gut, Biosci.
Biotechnol. Biochem., 74, 1145-1151, doi:
10.1271/bbb.100094.
82.Ohkuma, M. (2008) Symbioses of flagellates and
prokaryotes in the gut of lower termites, Trends Microbiol.,
16, 345-352, doi: 10.1016/j.tim.2008.04.004.
83.Ohkuma, M., and Brune, A. (2010) Diversity,
structure, and evolution of the termite gut microbial community, in
Biology of Termites: A Modern Synthesis (Bignell, D. E., Roisin,
Y., and Lo, N., eds) Springer, Dordrecht, pp. 413-438, doi:
10.1007/978-90-481-3977-4_15.
84.Thong-On, A., Suzuki, K., Noda, S., Inoue, J.,
Kajiwara, S., and Ohkuma, M. (2012) Isolation and characterization of
anaerobic bacteria for symbiotic recycling of uric acid nitrogen in the
gut of various termites, Microbes Environ., 27, 186-192,
doi: 10.1264/jsme2.me11325.
85.Ikeda-Ohtsubo, W., and Brune, A. (2009)
Cospeciation of termite gut flagellates and their bacterial
endosymbionts: Trichonympha species and Candidatus Endomicrobium
trichonymphae, Mol. Ecol., 18, 332-342, doi:
10.1111/j.1365-294X.2008.04029.x.
86.Slaytor, M. (1992) Cellulose digestion in
termites and cockroaches: What role do symbionts play? Compar.
Biochem. Physiol. B, 103, 775-784, doi:
10.1016/0305-0491(92)90194-V.
87.Zheng, H., Dietrich, C., Thompson, C. L., Meuser,
K., and Brune, A. (2015) Population structure of Endomicrobia in single
host cells of termite gut flagellates (Trichonympha spp.),
Microbes Environ., 30, 92-98, doi:
10.1264/jsme2.ME14169.
88.Potrikus, C. J., and Breznak, J. A. (1981) Gut
bacteria recycle uric acid nitrogen in termites: a strategy for
nutrient conservation, Proc. Natl. Acad. Sci. USA, 78,
4601-4605, doi: 10.1073/pnas.78.7.4601.
89.Breznak, J. A. (2000) Ecology of prokaryotic
microbes in the guts of wood- and litter-feeding termites, in
Termites: Evolution, Sociality, Symbioses, Ecology (Abe, T.,
Bignell, D. E., and Higashi, M., eds) Kluwer Academic Publishers,
Dordrecht, pp. 209-231, doi: 10.1007/978-94-017-3223-9_10.
90.Burt, A., and Trivers, R. (2006) Genes in
conflict. The biology of selfish genetic elements, The Belknap
Press of Harvard University Press, Cambridge, doi:
10.4159/9780674029118.
91.Green, M. M. (1985) The role of mobile DNA
elements in unequal and intrachromosomal crossing-over in
Drosophila melanogaster, Basic Life Sci.,
36, 353-361, doi: 10.1007/978-1-4613-2127-9_24.
92.McClintock, B. (1951) Chromosome organization and
genic expression, Cold Spring Harb. Symp. Quant. Biol.,
16, 13-47, doi: 10.1101/sqb.1951.016.01.004.
93.McGrayne, S. B. (1998) Nobel Prize Women in
Science: Their Lives, Struggles, and Momentous Discoveries, Carol
Publishing Group, Secaucus (NJ).
94.Craig, N. L., Craigie, R., Gellert, M., and
Lambowitz, A. M. (2002) Mobile DNA II, ASM Press, Washington
(D.C.).
95.Branco, M. R., and de Mendoza Soler, A. (2022)
Transposable Elements. Methods and Protocols, Springer, New
York, doi: 10.1007/978-1-0716-2883-6.
96.Pandita, A., and Pandita D. (2023) Plant
Transposable Elements. Biology and Biotechnology, Apple Academic
Press, Palm Bay (FL), doi: 10.1201/9781003315193.
97.Sun, C., Feschotte, C., Wu, Z., and Mueller, R.
L. (2015) DNA transposons have colonized the genome of the giant virus
Pandoravirus salinus, BMC Biol., 13, 38,
doi: 10.1186/s12915-015-0145-1.
98.Hickman, A. B., and Dyda, F. (2016) DNA
transposition at work, Chem. Rev., 116, 12758-12784, doi:
10.1021/acs.chemrev.6b00003.
99.Tang, Z., Zhang, H. H., Huang, K., Zhang, X. G.,
Han, M. J., and Zhang, Z. (2015) Repeated horizontal transfers of four
DNA transposons in invertebrates and bats, Mob. DNA, 6,
3, doi: 10.1186/s13100-014-0033-1.
100.Paulat, N. S., Storer, J. M.,
Moreno-Santillán, D. D., Osmanski, A. B., Sullivan, K. A. M.,
Grimshaw, J. R., Korstian, J., Halsey, M., Garcia, C. J., Crookshanks,
C., Roberts, J., Smit, A. F. A., Hubley, R., Rosen, J., Teeling, E. C.,
Vernes, S. C., Myers, E., Pippel, M., Brown, T., Hiller, M., Zoonomia
Consortium, Rojas, D., Dávalos, L. M., Lindblad-Toh, K.,
Karlsson, E. K., and Ray, D. A. (2023) Chiropterans are a hotspot for
horizontal transfer of DNA transposons in mammalia, Mol. Biol.
Evol., 40, msad092, doi: 10.1093/molbev/msad092.
101.Ohshima, K., and Okada, N. (2005) SINEs and
LINEs: symbionts of eukaryotic genomes with a common tail,
Cytogenet. Genome Res., 110, 475-490, doi:
10.1159/000084981.
102.Guffanti, G., Bartlett, A., DeCrescenzo, P.,
Macciardi, F., and Hunter, R. (2019) Transposable elements, Curr.
Top. Behav. Neurosci., 42, 221-246, doi:
10.1007/7854_2019_112.
103.Luan, D. D., Korman, M. H., Jakubczak, J. L.,
and Eickbush, T. H. (1993) Reverse transcription of R2Bm RNA is primed
by a nick at the chromosomal target site: a mechanism for non-LTR
retrotransposition, Cell, 72, 595-605, doi:
10.1016/0092-8674(93)90078-5.
104.Boeke, J. D. (1997) LINEs and Alus – the
polyA connection, Nat Genet., 16, 6-7, doi:
10.1038/ng0597-6.
105.Okada, N., Hamada, M., Ogiwara, I., and
Ohshima, K. (1997) SINEs and LINEs share common 3' sequences: a review,
Gene, 205, 229-243, doi:
10.1016/s0378-1119(97)00409-5.
106.Ogiwara, I., Miya, M., Ohshima, K., and Okada,
N. (1999) Retropositional parasitism of SINEs on LINEs: identification
of SINEs and LINEs in elasmobranchs, Mol. Biol. Evol.,
16, 1238-1250, doi: 10.1093/oxfordjournals.molbev.a026214.
107.Cordaux, R., and Batzer, M. A. (2009) The
impact of retrotransposons on human genome evolution, Nat. Rev.
Genet., 10, 691-703, doi: 10.1038/nrg2640.
108.Coffin, J. M., Hughes, S. M., and Varmus, H. E.
(1997) Retroviruses, Cold Spring Harbor Laboratory Press,
NY.
109.Hughes, S. H. (2015) Reverse transcription of
retroviruses and LTR retrotransposons, Microbiol. Spectr.,
3, doi: 10.1128/microbiolspec.MDNA3-0027-2014.
110.Boeke, J. D., and Stoye, J. P. (1997)
Retrotransposons, endogenous retroviruses, and the evolution of
retroelements, in Retroviruses (Coffin, J. M., Hughes, S. M.,
and Varmus, H. E., eds) Cold Spring Harbor Laboratory Press, Cold
Spring Harbor, NY.
111.Vinogradov, A. E. (2003) Selfish DNA is
maladaptive: evidence from the plant Red List, Trends Genet.,
19, 609-614, doi: 10.1016/j.tig.2003.09.010.
112.Vinogradov, A. E. (2004) Genome size and
extinction risk in vertebrates, Proc. Biol. Sci., 271,
1701-1705, doi: 10.1098/rspb.2004.2776.
113.International Human Genome Sequencing
Consortium (2001) Initial sequencing and analysis of the human genome,
Nature, 409, 860-921, doi: 10.1038/35057062.
114.Mouse Genome Sequencing Consortium (2002)
Initial sequencing and comparative analysis of the mouse genome,
Nature, 420, 520-562, doi: 10.1038/nature01262.
115.Colleaux, L., d’Auriol, L., Betermier,
M., Cottarel, G., Jacquier, A., Galibert, F., and Dujon, B. (1986)
Universal code equivalent of a yeast mitochondrial intron reading frame
is expressed into E. coli as a specific double strand
endonuclease, Cell, 44, 521-533, doi:
10.1016/0092-8674(86)90262-x.
116.Barth, Z. K., Dunham, D. T., and Seed, K. D.
(2023) Nuclease genes occupy boundaries of genetic exchange between
bacteriophages, bioRxiv, doi: 10.1101/2023.03.23.533998.
117.Jaenike, J. (2001) Sex chromosome meiotic
drive, Annu. Rev. Ecol. Syst., 32, 25-49, doi:
10.1146/annurev.ecolsys.32.081501.113958.
118.Wood, R. J., and Newton, M. E. (1991) Sex-ratio
distortion caused by meiotic drive in mosquitoes, Am. Nat.,
137, 379-391, doi: 10.1086/285171.
119.Emerson, J. J., Kaessmann, H., Betrán,
E., and Long, M. (2004) Extensive gene traffic on the mammalian X
chromosome, Science, 303, 537-540, doi:
10.1126/science.1090042.
120.Garcillán-Barcia, M. P., Redondo-Salvo,
S., and de la Cruz, F. (2023) Plasmid classifications, Plasmid,
126, 102684, doi: 10.1016/j.plasmid.2023.102684.
121.Wein, T., and Dagan, T. (2022) Plasmid
evolution, Curr. Biol., 30, R1158-R1163, doi:
10.1016/j.cub.2020.07.003.
122.Bottery, M. J. (2022) Ecological dynamics of
plasmid transfer and persistence in microbial communities, Curr.
Opin. Microbiol., 68, 102152, doi:
10.1016/j.mib.2022.102152.
123.Lerminiaux, N. A., and Cameron, A. D. S. (2019)
Horizontal transfer of antibiotic resistance genes in clinical
environments, Can. J. Microbiol., 65, 34-44, doi:
10.1139/cjm-2018-0275.
124.Willis, A. J. (1997) The ecosystem: an evolving
concept viewed historically, Funct. Ecol., 11, 268-271,
doi: 10.1111/j.1365-2435.1997.00081.x.
125.Tansley, A. G. (1935) The use and abuse of
vegetational concepts and terms, Ecology, 16, 284-307,
doi: 10.2307/1930070.
126.Chapin, F. S. 3rd, Matson, P. A., and Vitousek,
P. M. (2011) Principles of Terrestrial Ecosystem Ecology, 2nd
Edn, Springer Nature, Switzerland, doi: 10.1007/978-1-4419-9504-9.
127.Hestrin, R., Hammer, E. C., Mueller, C. W., and
Lehmann, J. (2019) Synergies between mycorrhizal fungi and soil
microbial communities increase plant nitrogen acquisition, Commun.
Biol., 2, 233, doi: 10.1038/s42003-019-0481-8.
128.Giovannetti, M., Avio, L., Fortuna, P.,
Pellegrino, E., Sbrana, C., and Strani, P. (2006) At the root of the
wood wide web: self recognition and non-self incompatibility in
mycorrhizal networks, Plant Signal. Behav., 1, 1-5, doi:
10.4161/psb.1.1.2277.
129.Peter, M. (2006) Ectomycorrhizal fungi –
fairy rings and the wood-wide web, New Phytol., 171,
685-687, doi: 10.1111/j.1469-8137.2006.01856.x.
130.Beiler, K. J., Durall, D. M., Simard, S. W.,
Maxwell, S. A., and Kretzer, A. M. (2010) Architecture of the wood-wide
web: Rhizopogon spp. genets link multiple Douglas-fir cohorts, New
Phytologist, 185, 543-553, doi:
10.1111/j.1469-8137.2009.03069.x.
131.Gorzelak, M. A., Asay, A. K., Pickles, B. J.,
and Simard, S. W. (2015) Inter-plant communication through mycorrhizal
networks mediates complex adaptive behaviour in plant communities,
AoB Plants, 7, plv050, doi: 10.1093/aobpla/plv050.
132.Sharifi, R., and Ryu, C. M. (2021) Social
networking in crop plants: Wired and wireless cross-plant
communications, Plant Cell Environ., 44, 1095-1110, doi:
10.1111/pce.13966.
133.Hettenhausen, C., Li, J., Zhuang, H., Sun, H.,
Xu, Y., Qi, J., Zhang, J., Lei, Y., Qin, Y., Sun, G., Wang, L.,
Baldwin, I. T., and Wu, J. (2017) Stem parasitic plant Cuscuta
australis (dodder) transfers herbivory-induced signals among plants,
Proc. Natl. Acad. Sci. USA, 114, E6703-E6709, doi:
10.1073/pnas.1704536114.
134.Zhuang, H., Li, J., Song, J., Hettenhausen, C.,
Schuman, M. C., Sun, G., Zhang, C., Li, J., Song, D., and Wu, J. (2018)
Aphid (Myzus persicae) feeding on the parasitic plant dodder (Cuscuta
australis) activates defense responses in both the parasite and soybean
host, New Phytol., 218, 1586-1596, doi:
10.1111/nph.15083.
135.Ripple, W. J., and Beschta, R. L. (2004) Wolves
and the ecology of fear: can predation risk structure ecosystems?
BioScience, 54, 755, doi:
10.1641/0006-3568(2004)054[0755:WATEOF]2.0.CO;2.
136.Robbins, J. (2004) Lessons from the wolf,
Sci. Am., 290, 76-81, doi:
10.1038/scientificamerican0604-76.
137.Hall, K. R. L. (1960) Social vigilance
behaviour of the chacma baboon, Papio ursinus, Behaviour,
16, 261-294, doi: 10.1163/156853960X00188.
138.Altmann, S. A., and Altmann, J. (1970)
Baboon Ecology: African Field Research, The University of
Chicago Press, Chicago.
139.Lane, N. (2008) Marine microbiology: origins of
death, Nature, 453, 583-585, doi: 10.1038/453583a.
140.Institute of Medicine (USA) (2006) Dietary
Reference Intakes: The Essential Guide to Nutrient Requirements,
The National Academies Press, Washington (DC), doi: 10.17226/11537.
141.López-García, P. (2012) The place
of viruses in biology in light of the metabolism-
versus-replication-first debate, Hist. Philos. Life Sci.,
34, 391-406.
142.Koonin, E. V., Krupovic, M., and Agol, V. I.
(2021) The Baltimore classification of viruses 50 years later: how does
it stand in the light of virus evolution? Microbiol. Mol. Biol.
Rev., 85, e0005321, doi: 10.1128/MMBR.00053-21.
143.Krupovic, M., Dolja, V. V., and Koonin, E. V.
(2019) Origin of viruses: primordial replicators recruiting capsids
from hosts, Nat. Rev. Microbiol., 17, 449-458, doi:
10.1038/s41579-019-0205-6.
144.Spang, A., Mahendrarajah, T. A., Offre, P., and
Stairs, C. W. (2022) Evolving perspective on the origin and
diversification of cellular life and the Virosphere, Genome Biol.
Evol., 14, evac034, doi: 10.1093/gbe/evac034.
145.Stein, R. A., and DePaola, R. V. (2023) Human
endogenous retroviruses: our genomic fossils and companions,
Physiol. Genomics, 55, 249-258, doi:
10.1152/physiolgenomics.00171.2022.
146.Brandes, N., and Linial, M. (2019) Giant
viruses – big surprises, Viruses, 11, 404, doi:
10.3390/v11050404.
147.De Oliveira, E. G., Carvalho, J. V. R. P.,
Botelho, B. B., da Costa Filho, C. A., Henriques, L. R., de Azevedo, B.
L., and Rodrigues, R. A. L. (2022) Giant Viruses as a Source of Novel
Enzymes for Biotechnological Application, Pathogens, 11,
1453, doi: 10.3390/pathogens11121453.
148.Abergel, C., and Claverie, J. M. (2020) Giant
viruses, Curr. Biol., 30, R1108-R1110, doi:
10.1016/j.cub.2020.08.055.
149.Zonca, V. (2021) Lichens: For Minimal
Resistance [in French], Éditions le Pommier, Paris.
150.Loyfer, N., Magenheim, J., Peretz, A., Cann,
G., Bredno, J., Klochendler, A., Fox-Fisher, I., Shabi-Porat, S.,
Hecht, M., Pelet, T., Moss, J., Drawshy, Z., Amini, H., Moradi, P.,
Nagaraju, S., Bauman, D., Shveiky, D., Porat, S., Dior, U., Rivkin, G.,
Or, O., Hirshoren, N., Carmon, E., Pikarsky, A., Khalaileh, A., Zamir,
G., Grinbaum, R., Gazala, M. A., Mizrahi, I., Shussman, N., Korach, A.,
Wald, O., Izhar, U., Erez, E., Yutkin, V., Samet, Y., et al. (2023) A
DNA methylation atlas of normal human cell types, Nature,
613, 355-364, doi: 10.1038/s41586-022-05580-6.