REVIEW: Recent Developments in Bioprocessing of Recombinant Antibody Fragments
Nevena Zelenovic1, Lidija Filipovic2, and Milica Popovic3,a*
1Center for Chemistry, Institute for Chemistry, Technology, and Metallurgy, National Institute of Republic of Serbia, University of Belgrade, 11000 Belgrade, Serbia2Innovative Centre, Faculty of Chemistry, University of Belgrade, 11000 Belgrade, Serbia
3Department of Biochemistry, Faculty of Chemistry, University of Belgrade, 11000 Belgrade, Serbia
* To whom correspondence should be addressed.
Received May 13, 2023; Revised July 12, 2023; Accepted August 18, 2023
Biotechnological and biomedical applications of antibodies have been on a steady rise since the 1980s. As unique and highly specific bioreagents, monoclonal antibodies (mAbs) have been widely exploited and approved as therapeutic agents. However, the use of mAbs has limitations for therapeutic applications. Antibody fragments (AbFs) with preserved antigen-binding sites have a significant potential to overcome the disadvantages of conventional mAbs, such as heterogeneous tissue distribution after systemic administration, especially in solid tumors, and Fc-mediated bystander activation of the immune system. AbFs possess better biodistribution coefficient due to lower molecular weight. They preserve the functional features of mAbs, such as antigen specificity and binding, while at the same time, ensuring much better tissue penetration. An additional benefit of AbFs is the possibility of their production in bacterial and yeast cells due to the small size, more robust structure, and lack of posttranslational modifications. In this review, we described current approaches to the AbF production with recent examples of AbF synthesis in bacterial and yeast expression systems and methods for the production optimization.
KEY WORDS: antibody fragments, bacterial expression, yeast expression, scFv, VHHDOI: 10.1134/S0006297923090018
Abbreviations: AbF, antibody fragment; mAb, monoclonal antibody; scFv, single-chain variable fragments; VHH, variable domains of heavy-chain antibody.
INTRODUCTION
Antibodies have become a staple in diagnostics, imaging, and therapeutic applications. In particular, they are routinely used in various clinical assays, including immunoblotting, flow cytometry, and immunohistochemistry [1], e.g., for detection of antigens or toxins. Antibodies can be obtained in a monoclonal form, which makes possible their wide application in the treatment of cancer and inflammatory diseases. However, production of monoclonal antibodies (mAbs) using classical hybridoma technology is restricted, as hybridomas are typically low-secreting and genetically unstable [2]. Thus, it takes between 6 to 8 months on average to obtain a reasonable amount of mAbs. Mouse mAbs are immunogenic and have low efficiency due to the xenogeneic Fc structure, so that further humanization is required to utilize them in therapeutic applications, which ramps up the production costs [1].
The development of techniques for recombinant mAb production in the 1980s had enabled manufacturing antibodies on a large scale for utilization in preclinical and clinical trials. Despite obvious benefits of mAbs, there are some shortcomings, such as a high cost and time constraints related to their production. Large molecular mass and heterotetrameric structure of both classical and humanized mAbs render mAbs them inappropriate for some in vivo applications. As an alternative, different types of antibody fragments (AbFs) of different formats have been engineered, with single-chain variable fragments (scFvs), single-domain antibodies (variable domains of heavy-chain antibodies, i.e., VHHs), and variable fragments of immunoglobulin new antigen receptor (VNARs) being the most common of them [3]. Screening of large AbF libraries has been made possible by the phage and yeast display techniques that have significantly accelerated identification of antibodies for specific antigens and enabled optimization of antibody structure to enhance its binding properties. Display techniques, particularly phage display, are a significant driving force in the discovery of antibodies that can be further engineered, optimized, and produced [4].
AbFs can be obtained by proteolysis or genetic engineering (Fig. 1). The most studied AbF types are Fabs (fragment antigen-binding regions), scFvs, and VHHs. Fabs can be obtained by enzymatic digestion of IgGs with papain or pepsin to yield two Fabs or F(ab’)2, respectively. A breakthrough in the production of recombinant Abs came with the development of scFvs by connecting the variable domains of the heavy and light chains (VH and VL, respectively) through a peptide linker to form a single polypeptide chain. AbFs of this type retain the antigen-binding capabilities of mAb, but can be expressed E. coli [5]. VHHs engineered from heavy-chain-only Abs naturally occurring in camelids are stable and can bind an antigen despite the absence of the light chain. VHHs share structure and sequence similarities with the VH domains of conventional Abs except for key mutations in the framework 2 region responsible for the interaction between the variable domains in conventional immunoglobulins. Due to their increased structural stability and reduced propensity for aggregation, VHHs are preferred under conditions that require highly stable Abs (increased detergent concentration, low pH) and can be used for imaging and therapy of oncological, infectious, inflammatory, and neurodegenerative diseases [3]. Despite their shorter half-life in the circulation because of the kidney filtration, AbFs offer a number of unique advantages over classical mAbs, in particular, enhanced tissue penetration, especially in solid tumors [6], higher tumor-to-organ ratio [7], and effective penetration across biological (e.g., epithelial and endothelial) barriers [8].
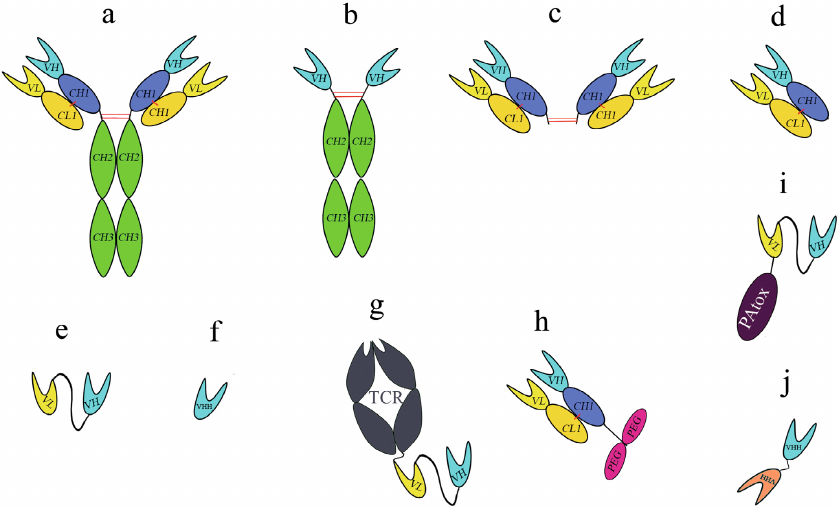
Fig. 1. Structure of different antibody formats: a) conventional mAb; b) heavy-chain only antibody; c) F(ab’)2 fragment; d) Fab fragment; e) scFv; f) VHH; g) bispecific TT cell receptor (TCR)-scFv fusion; h) PEGylated Fab; i) scFv, truncated Pseudomonas exotoxin A fusion; j) tandem VHH. Red lines represent interchain disulfide bridges; intrachain disulfide bridges are not shown.
scFvs and VHHs are emerging as biotherapeutic agents, because their smaller size allows for better tissue penetration [9]. Biodistribution coefficient (BC, a ratio of tissue and plasma mAb concentration) can be used for efficient estimation of tissue concentrations of proteins based on their pharmacokinetics in the plasma. The relation between BC and protein size allowed to derive BC50 values for all tissues. The bigger molecular weight will result in 50% reduction in the tissue uptake of protein. The BC50 value for most tissue was found to be ~35 kDa, indicating that the distribution of AbFs into tissues would be more efficient compare to mAbs [10]. A smaller size of AbFs and better BC50 values can help in AbF dosing and administration, especially in the case of non-systemic application [11]. AbFs have high BC50 values in kidneys, indicating that they will likely be filtered and reabsorbed from urine [10, 12]. In recent years, recombinant AbFs have become one of the most widely used biopharmaceutical products for therapeutic purposes, as well as in immunodetection, purification, and bioseparation [3]. Their biotechnological appeal stems from the possibility of AbF biosynthesis in bacterial and/or yeast cells and, consequently, lower production costs per milligram of protein. In this review, we have focused on recent advances in the AbF production in prokaryotic and eukaryotic expression systems, such as Escherichia coli, Saccharomyces cerevisiae, and Pichia pastoris.
AbF PRODUCTION IN E. coli CELLS
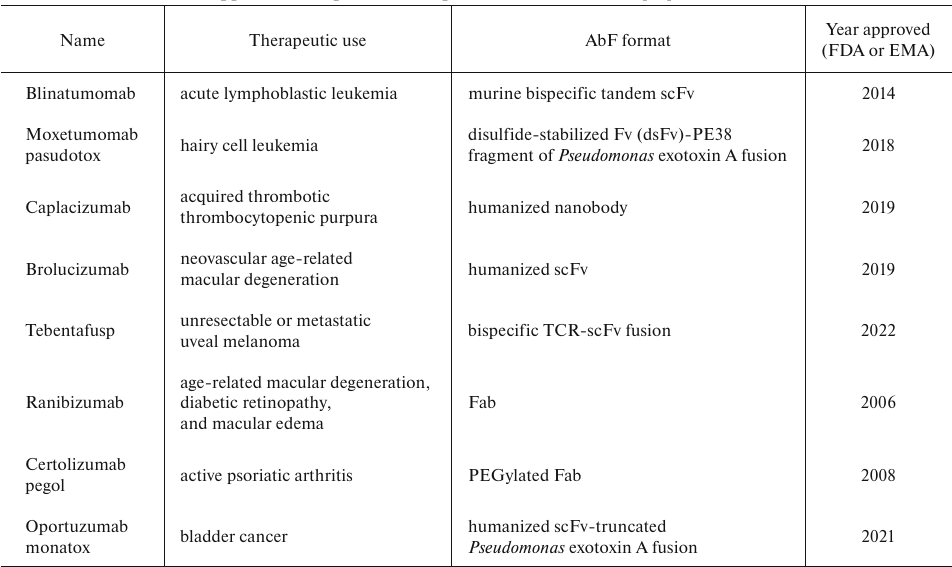
E. coli as an AbF expression host. Prokaryotic expression systems are widely used for biosynthesis of different types of recombinant proteins, including allergens, enzymes, and AbFs [13-15]. The most used bacterial host is E. coli, which has evolved from a model research organism to an industrial producer of heterologous proteins. Around 30% of biopharmaceutical preparations worldwide are produced in E. coli, which is still used as the gold standard in the production of recombinant proteins [9, 14]. This organism is well-studied and thoroughly characterized; it can be easily controlled with various molecular tools. E. coli as a host for the expression of recombinant AbFs has advantages over other expression systems, including rapid growth, minimal medium complexity, low production cost, and easy scaling up. It remains the first choice for a high-amount low-cost production of nonglycosylated recombinant proteins. Since AbFs are not glycosylated, they can be synthesized in E. coli cells without losing their functional properties [16]. Several FDA- and EMA-approved drugs based on AbFs have been successfully produced in E. coli (Table 1). AbFs can be synthesized in different cellular compartments, such as periplasm (Ranibizumab and Certolizumab pegol) and extracellular space (Caplacizumab and Oportuzumab), or produced as inclusion bodies and then refolded (Tebentafusp and Brolucizumab) [9, 17].
Table 1. FDA- and EMA-approved therapeutic
AbFst produced in E. coli cells [18]
Numerous strategies for obtaining high yields of recombinant AbFs have been developed. There are three main aspects of AbF production that should be considered when using E. coli as a producer: strains, vectors, and cultivation conditions.
Conventional approaches for AbF production in E. coli cells. AbFs can be successfully produced in the cytoplasm and periplasm, although production in each of these compartments has its limitations [19]. For example, it is possible to achieve high levels of AbFs synthesis in the cytoplasm the proteins can misfold and form insoluble inclusion bodies. Secretion to the periplasm allows formation of natural disulfide bonds, but the periplasmic space is limited, which results in lower yields or formation of aggregates. The periplasm offers a more straight-forward approach for protein purification, as it can be lysed under very mild conditions. The outer membrane can be disrupted by osmotic shock, mechanical stress, or mild heat treatment. Also, unlike the cytoplasm, the periplasmic space contains only 4-8% of native E. coli proteins, which simplifies the purification process [20].
The most common strategy of AbFs production is protein expression in a soluble form in the cytoplasm, which is simple, typically provides a high protein yield, and requires no introduction of the signal sequence into the vector [21]. However, protein expression in the cytoplasm of bacterial cells can lead to inadequate formation of disulfide bonds resulting in the accumulation of recombinant antibodies mainly in a form of inclusion bodies. Inclusion bodies can be subjected to in vitro refolding, disulfide bond formation, and purification to restore their functional activity [22]. Recombinant AbFs can also be produced in the cytosol using mutant E. coli strains overexpressing disulfide isomerase for the enhanced formation of disulfide bonds [23, 24].
To ensure the synthesis of recombinant proteins in a soluble form, it is better to aim for the optimal rather than maximal protein production. Some research groups have shown that the activity of disulfide isomerase DsbC can increase the yield of functional AbFs. For example, combination of this approach with co-expression of oxidoreductases was shown to be the best strategy for the high-yield production in the cytoplasm (up to 72 mg/liter) [25, 26]. Fusion with other proteins (e.g., GFP) can increase the solubility of the target protein. Others fusion tags, like SNAP and SORT, might provide better expression results but can complicate the purification process [27].
Although AbFs are not glycosylated, other post-translational modifications are important for their functional activity. Thus, scFv contains two disulfide bonds, while Fab contains five or six disulfide bonds. Producing AbFs with natural disulfide bonds is challenging, especially on a large scale. An example of successful biosynthesis of AbFs with correctly formed disulfide bonds in the cytoplasm of E. coli is the CyDisCo system based on the co-expression of the disulfide-bond-forming enzyme Erv1p and protein disulfide isomerase (PDI). This system was tested for expression and purification of eleven scFv and eleven Fabs expressed from identical vectors under identical conditions. The use of CyDisCo yielded large amounts of naturally folded and biologically active AbFs, as was shown by the circular dichroism spectra. The yield of the purified scFvs and Fab fragments was up to 240 and42 mg/liter, respectively [28].
Compared to the cytoplasm (reducing environment), the periplasm is a better choice for the disulfide bond formation in AbFs because it provides a more oxidizing environment. Furthermore, the presence of many chaperons and isomerases, such as the Dsb system can facilitate correct formation of disulfide bond. Most proteins are first produced in the cytoplasm and then secreted to the periplasm through different pathways (Sec, SRP, and Tat). For trafficking into the periplasm, proteins need the presence of the signal peptide (SpA, PhoA, PelB, OmpA, OmpT, DsbA, TorT, and TolT) at the N-terminus that along with molecular chaperons and disulfide isomerases (Dsb system) help proteins to achieve proper folding and disulfide bond formation. Around 20 signal peptides can be used for direct protein secretion to the periplasm [29]. Periplasmic targeting has been used in many cases, for example, for expression of scFv PGT135 in the amounts up to ~1.2 g/liter [8] or several humanized Fab` with the yield up to 2.4 g/liter [30].
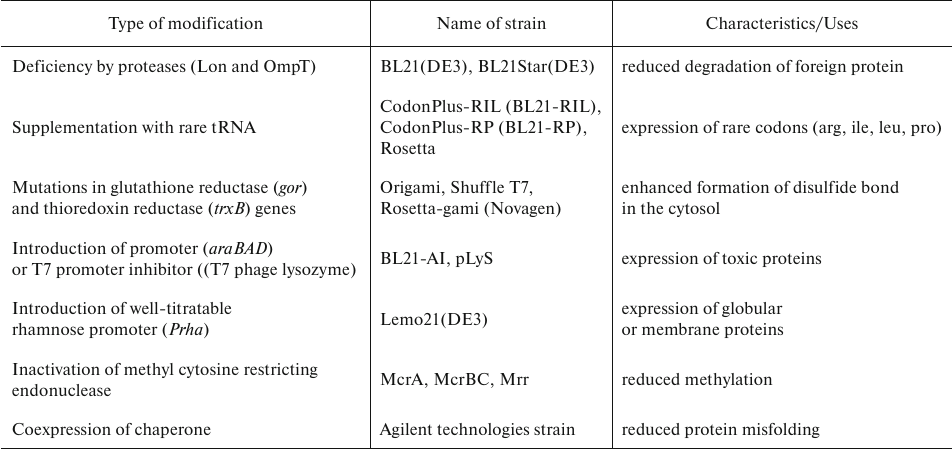
E. coli strains used for AbF production. There are several strains commonly used in biotechnological application (Table 2), in particular, E. coli K-12 (MG1655, JM109, W3110, BW25113, DH5α, DH1, WK6) and B [BL21, BL21(DE3), BL21(DE3) pLysS, BL21(DE3) Rosetta] [31]. K strains are used more often, although they produce high amounts of acetate which can negatively affect both protein production and cell growth [32]. On the opposite, B strains, which have been derived from K strains, produce less acetate allowing to grow the cells at higher glucose concentrations [33]. Therefore, K strains are a better choice for screening, while B strains are better for protein production in biopharmaceutical applications [34].
VHHs are usually produced in E. coli WK6 expression system [35] that has been developed from the K12 strain and used for inducible expression of VHH genes. These cells have a high replication rate and turned out to be the best choice providing correct disulfide bond formation. VHHs expressed in the periplasm space are typically modified with various tags, which significantly reduces the possibility inclusion body formation [35, 36]. E. coli WK6 cells have been successfully used for aa high-yield production of VHH against SARS-CoV-2 spike protein and food allergens [37, 38].
Table 2. Modifications of E. coli
strains [39]
VECTORS FOR AbF PRODUCTION
The choice of promoter is important for the regulation of protein expression and depends on the nature of the target protein [40-43]. Beside the most commonly used T7 promoter, there are several other promoters that have certain advantages. For example, araBAD is a strong, tightly regulated, and well-titratable promoter mostly used for the expression of toxic proteins. Also, the cspA promoter is a cold-shock promoter, meaning that protein expression can take place at low temperatures (with the optimum at 10-25°C) [44].
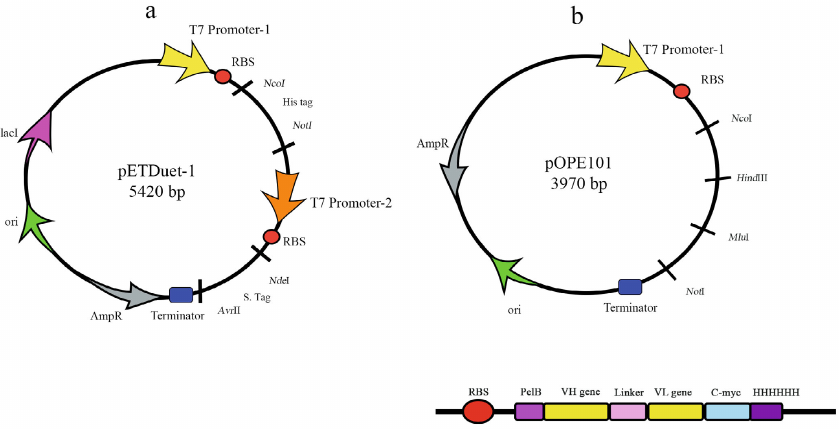
There are several vectors typically used for the AbF production. Progen has developed a new cloning vector, pOPE101, that ensures biosynthesis of scFv in a soluble form (Fig. 2) in E. coli cells. It contains strong IPTG-inducible T7 promoter, ampicillin resistance gene, leader sequence that provides protein expression in the periplasm, and C-myc/His6 tag for easier purification. In the case of scFv, the heavy chain was cloned by the NcoI and HindIII sites and the light chain was cloned by the MluI and NotI sites. The two chains were connected with 18-residue linker that enabled co-expression of the two target proteins. New pETDUET-1 vector (Fig. 2) designed by Novagen, has two multiple cloning sites (MCSs), each with the T7 promoter. The MSC2 ends with the T7 terminator. The vector also contains the His6 tag in MSC1 and S tag in MSC2 and can be used with any compatible E. coli strain [40]. For example, pETDUET-1 was used for the production of rHu (Ranibizumab biosimilar) with the yields up to 2.8 mg/liter (in this case, the light and heavy chains were cloned into 2 MCS) [41].
Fig. 2. pETDUET-1 (a) and pOPE101 (b) plasmids used for bacterial expression of AbFs.
CULTIVATION CONDITONS FOR HIGH-YIELD PRODUCTION OF AbFs
Attaining high AbF yield in E. coli cells requires optimization of numerous production parameters, such as temperature, agitation, cultivation time, aeration, carbon sources, medium, and type of induction [42, 43].
The effect of temperature on the AbF yield has been well studied. Although the maximum growth rate of E. coli is observed at 37°C, expression of recombinant antibodies at lower temperatures (25°C) proved to be a better solution, as it reduces, as its protein degradation and formation of inclusion bodies [45].
Several studies have reported that complex media, such as terrific broth (TB) or EnPresso, promote cell growth leading to the product overexpression [46]. However, the richness of growth medium has no significant effect on the protein yield that is affected by the protein sequence rather that the medium composition [47].
Optimization of the carbon source concentration (glucose, glycerol, lactose, glycine) can have a beneficial effect on the AbF expression. Generally, lowering the concentration of glucose and increasing the content of glycerol in the medium can increase the amount of extracellular VHHs [48]. Glycine affects the peptidoglycan layer of bacterial cell wall, thus increasing cell permeability, so that proteins can easily leak into the culture medium. Since the discovery that thirteen percent of VHH sequence is represented by glycine residues, glycine added to the cultivation medium reduces the metabolic burden and acts as a precursor for the production of AbFs [49].
Reducing the concentrations of IPTG can sometimes be beneficial for the solubility and activity of produced proteins by decreasing the level of protein expression. The optimal concentration of IPTG depends on a protein and should be optimized for each AbF. Abroad range of IPTG concentrations (from 0.005 to 5 mM) have been reported, although the most typical IPTG concentration for the T7 lac promoter is 1 mM. In some cases, there is no need for using the maximum IPTG concentration as this can burden the cell and induce the formation of inclusion bodies. On the opposite, low IPTG amounts often reduce inclusion body formation and improve protein folding and stability [46].
EXPRESSION OF AbFs IN YEAST
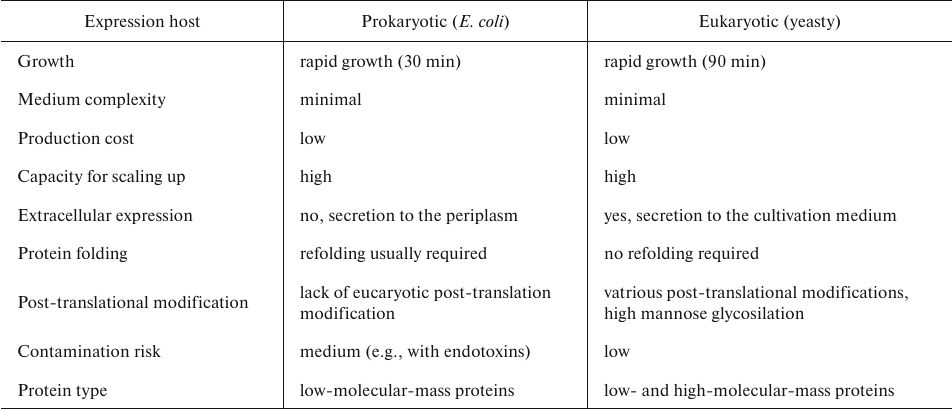
Although production of AbF in E. coli has its benefits (rapid cell growth, easy manipulation, and minimal medium complexity), there are also disadvantages, including formation of inclusion bodies due to the intracellular oxidative environment. Inclusion bodies require additional refolding to obtain soluble and functional antibodies [50]. This has led to the development of eukaryotic expression systems. The most accessible eukaryotic organism for the production of recombinant proteins is yeast. The advantages of this single-cell eukaryotic organism are numerous. Yeast easily grows on simple media and reach high cell density upon small- and large-scale cultivation. Biochemistry, genetics, and cell biology of yeast are well-studied, allowing for easy genetic manipulation. As a recombinant protein expression system, yeast support production of small and large (over 50 kDa) proteins with high yields and low levels of contamination with endogenous proteins [51]. Yeast secretes only a few of their own proteins, which facilitates purification of extracellularly secreted recombinant protein the medium. Furthermore, yeasty ensure glycosylation, methylation, acylation, disulfide bridge formation, and other post-translational modification in heterologous expressed proteins of eukaryotic origin [52, 53]. Yeast provides low production costs and easy scaling up of the production process in fermenters. Also, yeast is much more tolerable to the fermentation conditions, such as pH range, presence of fermentation inhibitors, high sugar and ethanol concentrations, etc., compared to E. coli cells (Table 3) [54].
Table 3. Characteristics of prokaryotic and
eukaryotic expression systems [55, 56]
Common yeast species (Table 4) used as hosts for the expression of recombinant proteins can be non-methylotrophic Saccharomyces cerevisiae, Yarrowia lipolytica, Kluyveromyces lactis) and methylotrophic (Pichia pastoris, Hansenula polymorpha, Candida boidinii). S. cerevisiae and P. pastoris are representatives of the two groups, respectively, that are most commonly used for the production of therapeutic proteins [57].
Table 4. Most commonly used strains of S.
cerevisiae and P. pastoris
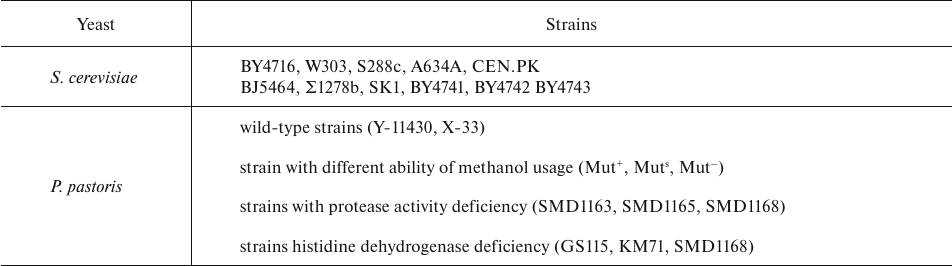
S. cerevisiae is a generally recognized as safe (GRAS) non-pathogenic organism, which has promoted its use in food industry. The genome of S. cerevisiae strain S288C was the first fully sequenced eukaryotic genome [58] that provided a wealth of information on the genetics and cell biology of yeast and enabled easy genetic manipulation of this species. Advances in genetic engineering, stability of the expression system, and easy cultivation have made S. cerevisiae an attractive host for the biosynthesis of mAbs and AbFs. Production of llama VHHs in S. cerevisiae is a well-established industrial process with a yield up to hundreds mg/liter [59]. Since then, a wide range of yeast strains have been employed for industrial and research applications, with BJ5464 as the strain used most often for recombinant protein expression.
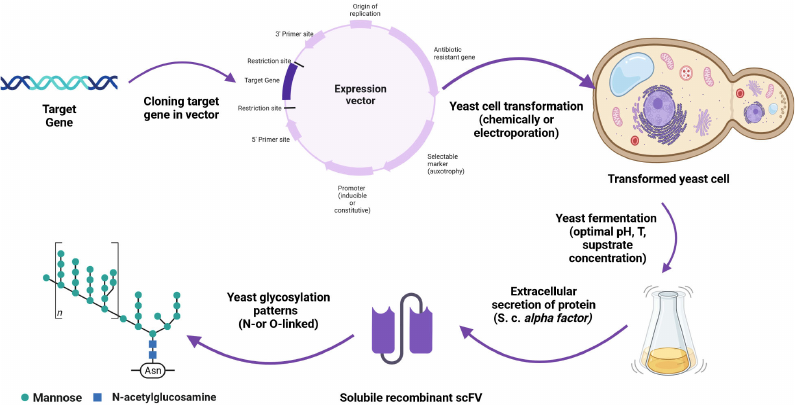
Expression of recombinant proteins requires yeast cells to be transformed chemically or by electroporation (Fig. 3) to introduce a vector carrying a gene of interest into the cells. Due to rapid proliferation of yeast cells, the vector is multiplied along with the target gene. Many eukaryotic vectors are the so-called shuttle vectors that can exist in both bacterial and eukaryotic cells. They have two replication origins and two sets of marker gene, one of which functions in E. coli and the other functions in a eukaryotic expression host, most commonly, yeast. There are three types of S. cerevisiae expression vectors: episomal plasmids (YEps), integrating plasmids (YIps), and centromeric plasmids (YCps).
Fig. 3. AbF expression in yeast: a gene of interest is cloned in the expression vector; yeast cells are transformed with the resulting construct and fermentation process is optimized; AbFs are produced at a large scale and their glycosylation profiles are analyzed. Created with BioRender.com with permission.
YEps are the most commonly used vectors for both intracellular and extracellular production of heterologous proteins. These vectors are derived from a small (2-µm) circular plasmid multiple copies of which are present in many naturally occurring strains of S. cerevisiae. The plasmid replicates independently of the yeast chromosome because it contains its own origin of replication. Selection of S. cerevisiae clones relies on mutant (auxotrophic) strains that require specific amino acid (histidine, tryptophan, leucine) or nucleotide (uracil) for growth and can grow on the minimal medium only if the medium is supplemented with this specific nutrient. The vector contains a functional wild-type version of the gene complementing the mutant gene in the host.
YIps lack the origin of replication and integrate into the host genome, while YACs can autonomously replicate in a small number of copies in the host cell.
Heterologous genes are placed under control of special promoters to increase their expression levels [50, 60]. These promoters can be constitutive, i.e., permanently active, or inducible, i.e., activated by specific stimuli. Promoters stimuli derived from S. cerevisiae genes enable efficient transcription of heterologous genes inserted into episomal vectors. Easily regulated inducible promoters are essential for producing several heterologous proteins simultaneously. For example, galactose-induced promoters respond rapidly to the addition of galactose and increase transcription up to 1000 times [61]. In S. cerevisiae, galactose is metabolized in a series of reactions to glucose 6-phosphate, which enters glycolysis or pentose phosphate pathway. Such transformation is necessary because glycolytic enzymes do not recognize galactose. This principle is used to express heterologous proteins in yeast using the GAL1 and GAL10 promoters that strictly regulate expression of galactose metabolism genes. Galactose is an unconventional nutrient for yeast that can be used as the only carbon source in the absence of glucose in the medium [62].
Production of recombinant llama VHHs in sufficient quantities is a well-established industrial process that is possible because VHH molecule is stable and soluble. The solubility of VHH comes from its hydrophilicity, which enables VHH secretion from the endoplasmic reticulum (ER). Unlike VHHs, scFv derivatives are more hydrophobic and, therefore, less soluble and might accumulate in the ER [63]. This problem can be solved by the development of yeast strains overexpressing chaperones and foldases, i.e., proteins ensuring correct protein folding with subsequent scFv secretion [64]. Also, new strains are being developed to increase the overall secretory capacity of the yeast, which can be achieved by engineering intracellular protein traffic, reducing proteolytic degradation, and engineering heat shock response enabling higher protein production levels [50].
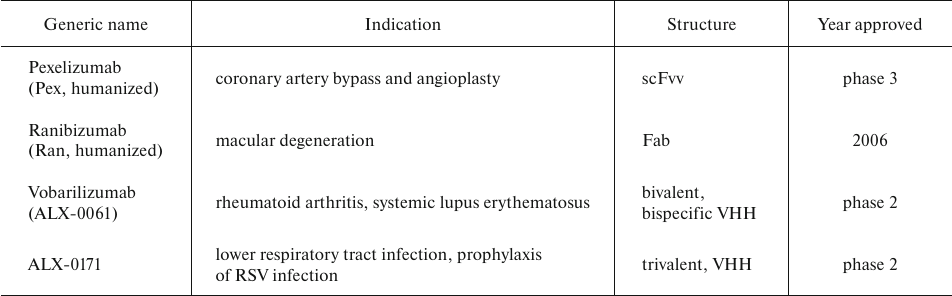
AbFs are not glycosylated and, hence, can be expressed in yeast. Thus, S. cerevisiae was used to and, hence, produce AbF-based drugs Ranibizumab (Ran; Fab fragment), Pexelizumab (Pex; scFv peptide), and others (Table 5) using the B184 strain with significantly elevated α-amylase production that was derived from the AAC strain through random mutagenesis and microfluidic screening. Elimination of the amylase plasmid from the B184 strain yielded the HA strain that was transformed with the plasmids constructed by inserting Nan (nanobody consisting of a single V-type domain), Pex, and Ran genes in the CPOTud vector, respectively. Although the three antibody fragments had different molecular masses and post-translational modifications, they were successfully expressed in the S. cerevisiae HA strain [65].
Table 5. Therapeutic AbFs produced in S.
cerevisiae and approved by FDA or tested in clinical trial
Hypermannosylation can result in a lower activity, higher immunogenicity, decreased serum half-life, and reduced therapeutic efficacy of glycoproteins (Fig. 3). However, in the case of AbFs, hypermannosylation can be avoided by inactivation of Mnn2p and Mnn11p genes for mannosyltransferases responsible for this type of glycosylation. The knockdown of the Och1p gene coding for α-1,6-mannosyltransferase also increased production of recombinant proteins, including AbFs. Hence, modification of N-glycosylation can lead to the increased secretion of proteins [66].
Many heterologous genes are cloned with a signal sequence coding for a peptide facilitating the transport of a recombinant protein through the cell membrane and its release to the external medium. Addition of the signaling sequence might significantly simplify the purification process. The signal sequence most commonly used in yeast was derived from the mating factor α1 (MFα1) gene (Fig. 3). Typically, it is inserted immediately upstream of the gene of interest [67]. In addition to the signal sequence, expressed gene may contain a tag (mostly at the C-terminus) that enables easy purification and quantification of the recombinant protein. The most commonly used tags are GFP are polyhistidine tag [62]. Thus, a scFv fragment with the C-terminally fused GFP was produced in S. cerevisiae BJ5464 strain, with GFP fluorescence used to monitor its intracellular processing [68].
However, S. cerevisiae yeast have several limitations, including plasmid instability, hyperglycosylation, and low protein yield, which have led to the development of alternative expression systems, including methylotrophic yeast P. pastoris.
Like S. cerevisiae, P. pastoris is a single-cell GRAS eukaryotic organism [50] that can be easily genetically manipulated and cultivated. P. pastoris cells provide many post-translational modifications, such as proper protein folding, glycosylation, and disulfide bond formation. Compared to insect or mammalian cells, P. pastoris offers faster, easier, and more cost-efficient production of recombinant proteins. The main advantage of P. pastoris compared to S. cerevisiae is a lower degree of heterologous protein glycosylation [52]. The use of P. pastoris for controlled expression of heterologous proteins is possible due to the existence of well-regulated and efficient promoters, ability to grow to a high density in bioreactors on simple media, and secretion of a small number of endogenous proteins, which significantly simplifies purification of the protein product [67].
The vector for P. pastoris transformation should be linearized for efficient integration into the yeast genome by homologous recombination with the formation of stable transformants. Inducible promoter of the AOX1 (alcohol oxidase) gene is frequently employed for the expression of heterologous proteins using methanol as an inducer. This promoter is inhibited by glucose, glycerol, and most other carbon sources [62, 69]. Alternatively, constitutive promoters can be used, as they ensure similar expression levels as the inducible ones. The most commonly used constitutive promoter is pGAP normally regulating expression of the glyceraldehyde-3-phosphate dehydrogenase gene [62].
Some P. pastoris expression vectors contain an additional sequence (e.g., the pre-pro sequence of S. cerevisiae MFα1) immediately after the promoter that serves as a signal for secretion of the expressed protein. [69]. Purification of secreted proteins is mainly achieved by separation of the fermentation medium from the yeast cell biomass by centrifugation. Next, recombinant proteins can concentrate and purified by various methods, such as ultrafiltration, precipitation, or chromatography, depending on the properties of the target protein. As an example, we can mention production of the anti-CEACAM5 nanobody in P. pastoris GS115 under control of the pGAP promoter with a yield of ~50 mg/liter. Purified nanobodies were used in the early detection of carcinoembryonic antigen (CEA) in the serum of patients with gastrointestinal tumors [70].
Selection of the transformed host clones is provided by introduction of antibiotic resistance genes. There are two groups of vectors that confer host resistance to kanamycin (pPIC3K and pPIC9K) and zeocin (pPICZ), respectively [57]. These vectors also contain unique restriction endonucleases sites used for the target gene cloning, as well as tag sequences (6×His or GFP) that allow more straightforward purification of heterologous proteins.
A new P. pastoris protein expression system named PichiaPink that can produce large amounts of protein (up to 12 g/liter) has been introduced by Thermo Fischer Scientific, USA. The difference between the strain used in this system and conventional strains is the selection method based on complementation of adenine auxotrophs (and not on antibiotic resistance). At present, there are four strains available for the transformation and extracellular protein expression. Proteins are transported to the extracellular medium with the help of eight secretion sequences. The advantage of this expression system is that it can be easily scaled-up, from small to large fermentation volumes [52].
For the first time, P. pastoris was used as an expression system for production of LR and 10FG2 human AbFs that were cloned into the pPICZαA vector and expressed in P. pastoris strain X33. The vector contained the -factor secretion signal sequence enabling extracellular expression of the recombinant proteins. Both expressed AbFs were able to bind scorpion toxins, indicating that their biochemical and functional properties were not modified. Also, the yield of recombinant proteins in P. pastoris was higher than in E. coli cells [71].
STRATEGIES FOR IMPROVING AbF EXPRESSION IN YEAST
The strategies for improving heterologous protein production are based on varying conditions (temperature, incubation time, substrate concentration) of S. cerevisiae and P. pastoris cultivation. As a carbon source, S. cerevisiae yeast use 2% glucose or 2% raffinose; protein expression is induced by addition of 2% galactose to the medium. For expression of recombinant AbFs, S. cerevisiae cells are typically grown at 28-30°C [72]. Methanol that serves as a carbon source, is another component essential for cell growth. It also induces gene expression thus increasing protein production. However, excessive concentrations of methanol (over 5%) can be harmful because of the formation of toxic products, while low methanol concentrations (below 1%) cause proteolytic degradation of heterologous proteins. The optimal concentration of methanol is 2-2.5% [55]. Protein expression might be improved by introducing 0.5-1.0% sorbitol in in addition to methanol. Sorbitol is a carbon source that does not induce or suppress the AOX promoter, hence, growing yeast on a methanol/sorbitol mixture reduces the toxic effects of methanol and increases cell density and process productivity [55, 73].
Temperature is another significant factor in the cultivation of P. pastoris. The optimal temperature for these yeasts is 30°C. Increasing temperature to 32°C results in cell death and decreased protein expression, while decreasing growth temperature below 28°C ensures cell viability, reduces proteolytic degradation of expressed protein, and increases protein yield [55, 74]. Another critical factor affecting protein production is expression time. In order to obtain sufficient amount of cell biomass, P. pastoris cells are grown for 96 h. However, it was reported that growing cells for long periods of time affects proteolytic degradation of recombinant proteins. Therefore, successful protein production requires optimization of the incubation time. It was found that protein expression occurs mostly between 72 and 96 h, although it can start as early as 48 h after induction [55, 75].
P. pastoris proved to be an excellent expression host for AbF production. Two recombinant nanobodies, ALX061 and ALX00171, obtained in P. pastoris have found application in the treatment of rheumatoid arthritis and respiratory syncytial virus (RSV) infection, respectively [50]. Some AbFs have been used in molecular imaging, e.g., an scFv targeting human ether-a-go-go-related gene 1 (hERG1) potassium channel, a hallmark of emerging tumors. Native anti-hERG1 scFv and its mutant anti-hERG1 scFv-Cys variant were produced in P. pastoris GS115 cells. The highest protein expression level was reached 72 h after induction. The mutant variant in which cysteine residue replaced phenylalanine in the framework 3 of the VH domain was more stable in the serum and had a higher affinity to the cell-surface hERG1 antigen, which makes scFv-hERG1-Cis a convenient tool for molecular imaging in the in vivo diagnostics of cancer [76].
CONCLUSION REMARKS AND FUTURE PROSPECTS
Production of AbFs has been tested in various eukaryotic and prokaryotic expression systems (e.g., mammalian cells, transgenic animals, transgenic plants, insect cells, filamentous fungi, algae, protozoa, yeast, and bacteria). However, E. coli has several benefits, especially in terms of large-scale production, that have not been matched by any other system. Without a doubt, E. coli cells offer ample advantages, including rapid growth, high expression levels, possibilities of possibilities of genetic engineering due to vast knowledge on the genetics and molecular biology of this organism, large number of expression vectors and mutant host strains, and cost-efficient cultivation media/conditions.
During the last two decades, yeast-based expression systems have become common in the production of recombinant AbFs. Yeast expression systems have several advantages over bacteria because they are non-pathogenic, easy to grow, and ensure high expression levels of recombinant proteins. Unlike prokaryotic expression systems, yeasty can produce antibodies with human-like glycosylation patterns. Expression in bacterial cells often results in the formation of inclusion bodies, while yeast ensure proper protein folding and generation of functional proteins. Signal sequences enable direct protein secretion into the medium, which facilitates further purification process. The yield of recombinant products in yeast is high and can reach milligrams to grams per liter. Therefore, yeast ensure fast and cost-efficient production of recombinants proteins.
Other systems that have been developed for expression of therapeutic AbFs are mammalian cells, plants, and baculoviruses. Mammalian cells are the only expression system capable of producing native human proteins with correct post-translational modification patterns. Expression in mammalian cells is costly, difficult, and time-consuming, while the yields are often low. Purification of AbFs produced in mammalian systems requires removal of viruses, which adds another level of complexity to the procedure [9].
Plant expression systems offer a number of advantages, including low production costs and high protein yields. Despite the fact that proteins can be correctly folded in plant systems, post-translational modifications in these cells, especially glycosylation, differ from modifications in human cells and might lead to a potential immunogenicity of such AbFs, which limits the use of transgenic plants. Depending on the used promotors, proteins produced in plants can be stored in leaves, seeds, or both. While protein expression levels can be high, protein preparations might be contaminated with other substances, e.g., polyphenols and proteases, that can hinder purification [9].
Baculoviral expression systems are an excellent platform for the production and post-translational modification of human proteins. Compared to other systems, the flexibility and capacity for the “plug-and-play” production in baculovirus/insect cell systems are truly outstanding. While the development of AbF expression in insect cells and plants is still at the early stage in both research and clinical settings, these systems have a good potential to revolutionize AbF production and engineering, as they can provide fast- and cost-effective expression of AbFs.
Contributions. M.P. supervised the study; L.F., N.Z., and M.P. wrote and edited the manuscript.
Funding. This work was funded by the Ministry of Education, Science, and Technological Development of Republic of Serbia (contracts nos. 451-03-47/2023-01/200168, 451-03-47/2023-01/200288, and 451-03-47/2023-01/200026).
Ethics declarations. The authors declare no conflict of interest. This article does not contain description of studies with the involvement of humans or animal subjects performed by any of the authors.
REFERENCES
1.Mitra, S., and Tomar, P. C. (2021) Hybridoma
technology; advancements, clinical significance, and future aspects,
J. Genet. Eng. Biotechnol., 19, 159, doi:
10.1186/s43141-021-00264-6.
2.Liu, J. K. (2014) The history of monoclonal
antibody development – progress, remaining challenges and future
innovations, Ann. Med. Surg. (Lond), 3, 113-116, doi:
10.1016/j.amsu.2014.09.001.
3.De Marco, A. (2011) Biotechnological applications
of recombinant single-domain antibody fragments, Microb. Cell
Fact., 10, 44, doi: 10.1186/1475-2859-10-44.
4.Geyer, C. R., McCafferty, J., Dubel, S., Bradbury,
A. R., and Sidhu, S. S. (2012) Recombinant antibodies and in vitro
selection technologies, Methods Mol. Biol., 901, 11-32,
doi: 10.1007/978-1-61779-931-0_2.
5.Ahmad, Z. A., Yeap, S. K., Ali, A. M., Ho, W. Y.,
Alitheen, N. B., and Hamid, M. (2012) scFv antibody: principles and
clinical application, Clin. Dev. Immunol., 2012, 980250,
doi: 10.1155/2012/980250.
6.Pluen, A., Boucher, Y., Ramanujan, S., McKee, T.
D., Gohongi, T., di Tomaso, E., Brown, E. B., Izumi, Y., Campbell, R.
B., Berk, D. A., and Jain, R. K. (2001) Role of tumor-host interactions
in interstitial diffusion of macromolecules: cranial vs. subcutaneous
tumors, Proc. Natl. Acad. Sci. USA, 98, 4628-4633, doi:
10.1073/pnas.081626898.
7.Khawli, L. A., Biela, B., Hu, P., and Epstein, A.
L. (2003) Comparison of recombinant derivatives of chimeric TNT-3
antibody for the radioimaging of solid tumors, Hybrid
Hybridomics, 22, 1-9, doi: 10.1089/153685903321538026.
8.Thurber, G. M., and Wittrup, K. D. (2008)
Quantitative spatiotemporal analysis of antibody fragment diffusion and
endocytic consumption in tumor spheroids, Cancer Res.,
68, 3334-3341, doi: 10.1158/0008-5472.Can-07-3018.
9.Pirkalkhoran, S., Grabowska, W. R., Kashkoli, H.
H., Mirhassani, R., Guiliano, D., Dolphin, C., and Khalili, H. (2023)
Bioengineering of antibody fragments: challenges and opportunities,
Bioengineering (Basel), 10, 122, doi:
10.3390/bioengineering10020122.
10.Li, Z., Krippendorff, B. F., Sharma, S., Walz, A.
C., Lavé, T., and Shah, D. K. (2016) Influence of molecular size
on tissue distribution of antibody fragments, MAbs, 8,
113-119, doi: 10.1080/19420862.2015.1111497.
11.Balthasar, J., and Fung, H. L. (1994) Utilization
of antidrug antibody fragments for the optimization of intraperitoneal
drug therapy: studies using digoxin as a model drug, J. Pharmacol.
Exp. Ther., 268, 734-739.
12.Dugel, P. U., Koh, A., Ogura, Y., Jaffe, G. J.,
Schmidt-Erfurth, U., Brown, D. M., Gomes, A. V., Warburton, J.,
Weichselberger, A., Holz, F. G., and HAWK and HARRIER Study
Investigators (2020) HAWK and HARRIER: phase 3, multicenter,
randomized, double-masked trials of brolucizumab for neovascular
age-related macular degeneration, Ophthalmology, 127,
72-84, doi: 10.1016/j.ophtha.2019.04.017.
13.Popovic, M., Andjelkovic, U., Burazer, L.,
Lindner, B., Petersen, A., and Gavrovic-Jankulovic, M. (2013)
Biochemical and immunological characterization of a
recombinantlyproduced antifungal cysteine proteinase inhibitor from
green kiwifruit (Actinidia deliciosa),
Phytochemistry, 94, 53-59, doi:
10.1016/j.phytochem.2013.06.006.
14.Abughren, M., Popović, M.,
Dimitrijević, R., Burazer, L., Grozdanović, M.,
Atanasković-Marković, M., and
Gavrović-Jankulović, M. (2012) Optimization of the
heterologous expression of banana glucanase in Escherichia
coli, J. Serb. Chem. Soc., 77, 43-52, doi:
10.2298/JSC110309158A.
15.Arbabi-Ghahroudi, M., Tanha, J., and MacKenzie,
R. (2005) Prokaryotic expression of antibodies, Cancer Metastasis
Rev., 24, 501-519, doi: 10.1007/s10555-005-6193-1.
16.Charlton, K. A. (2004) Expression and
Isolation of Recombinant Antibody Fragments in E. coli. in
Antibody Engineering: Methods and Protocols (Lo, B. K. C., ed)
Humana Press, Totowa, NJ, pp. 245-254, doi:
10.1385/1-59259-666-5:245.
17.Walsh, G. (2010) Biopharmaceutical benchmarks
2010, Nat. Biotechnol., 28, 917-924, doi:
10.1038/nbt0910-917.
18.Kang, T. H., and Seong, B. L. (2020) Solubility,
stability, and avidity of recombinant antibody fragments expressed in
microorganisms, Front. Microbiol., 11, 1927, doi:
10.3389/fmicb.2020.01927.
19.Choi, J., and Lee, S. (2004) Secretory and
extracellular production of recombinant proteins using
Escherichia coli, Appl. Microbiol. Biotechnol.,
64, 625-635, doi: 10.1007/s00253-004-1559-9.
20.Sandomenico, A., Sivaccumar, J. P., and Ruvo, M.
(2020) Evolution of Escherichia coli expression system in
producing antibody recombinant fragments, Int. J. Mol. Sci.,
21, 6324, doi: 10.3390/ijms21176324.
21.Challener, C. A. (2015) Fermentation for the
future, BioPharm Int., 28, 30-31.
22.Basu, A., Li, X., and Leong, S. S. J. (2011)
Refolding of proteins from inclusion bodies: rational design and
recipes, Appl. Microbiol. Biotechnol., 92, 241-251, doi:
10.1007/s00253-011-3513-y.
23.Popovic, M., Mazzega, E., Toffoletto, B., and de
Marco, A. (2018) Isolation of anti-extra-cellular vesicle single-domain
antibodies by direct panning on vesicle-enriched fractions, Microb.
Cell Factor., 17, 6, doi: 10.1186/s12934-017-0856-9.
24.De Marco, A. (2022) Cytoplasmic production of
nanobodies and nanobody-based reagents by co-expression of sulfhydryl
oxidase and DsbC isomerase, Methods Mol. Biol., 2446,
145-157, doi: 10.1007/978-1-0716-2075-5_7.
25.Veggiani, G., and de Marco, A. (2011) Improved
quantitative and qualitative production of single-domain intrabodies
mediated by the co-expression of Erv1p sulfhydryl oxidase, Protein
Express. Purif., 79, 111-114, doi:
10.1016/j.pep.2011.03.005.
26.Rahbarizadeh, F., Rasaee, M. J.,
Forouzandeh-Moghadam, M., and Allameh, A.-A. (2005) High expression and
purification of the recombinant camelid anti-MUC1 single domain
antibodies in Escherichia coli, Protein Express.
Purif., 44, 32-38, doi: 10.1016/j.pep.2005.04.008.
27.Djender, S., Schneider, A., Beugnet, A., Crepin,
R., Desrumeaux, K. E., Romani, C., Moutel, S., Perez, F., and de Marco,
A. (2014) Bacterial cytoplasm as an effective cell compartment for
producing functional VHH-based affinity reagents and Camelidae IgG-like
recombinant antibodies, Microb. Cell Factor., 13, 140,
doi: 10.1186/s12934-014-0140-1.
28.Gaciarz, A., Veijola, J., Uchida, Y., Saaranen,
M. J., Wang, C., Hörkkö, S., and Ruddock, L. W. (2016)
Systematic screening of soluble expression of antibody fragments in the
cytoplasm of E. coli, Microb. Cell Fact.,
15, 22, doi: 10.1186/s12934-016-0419-5.
29.Humphreys, D. P., Sehdev, M., Chapman, A. P.,
Ganesh, R., Smith, B. J., King, L. M., Glover, D. J., Reeks, D. G., and
Stephens, P. E. (2000) High-level periplasmic expression in Escherichia
coli using a eukaryotic signal peptide: importance of codon usage at
the 5′ end of the coding sequence, Protein Express.
Purif., 20, 252-264, doi: 10.1006/prep.2000.1286.
30.Ellis, M., Patel, P., Edon, M., Ramage, W.,
Dickinson, R., and Humphreys, D. P. (2017) Development of a high
yielding E. coli periplasmic expression system for the production of
humanized Fab' fragments, Biotechnol. Prog., 33, 212-220,
doi: 10.1002/btpr.2393.
31.Liu, M., Feng, X., Ding, Y., Zhao, G., Liu, H.,
and Xian, M. (2015) Metabolic engineering of Escherichia
coli to improve recombinant protein production, Appl.
Microbiol. Biotechnol., 99, 10367-10377, doi:
10.1007/s00253-015-6955-9.
32.Selas Castiñeiras, T., Williams, S. G.,
Hitchcock, A. G., and Smith, D. C. (2018) E. coli strain
engineering for the production of advanced biopharmaceutical products,
FEMS Microbiol. Lett., 365, fny162, doi:
10.1093/femsle/fny162.
33.Sørensen, H. P., and Mortensen, K. K.
(2005) Advanced genetic strategies for recombinant protein expression
in Escherichia coli, J. Biotechnol., 115,
113-128, doi: 10.1016/j.jbiotec.2004.08.004.
34.Hayat, S. M., Farahani, N., Golichenari, B., and
Sahebkar, A. (2018) Recombinant protein expression in
Escherichia coli (E. coli): what we need to know,
Curr. Pharmaceut. Design, 24, 718-725, doi:
10.2174/1381612824666180131121940.
35.Pardon, E., Laeremans, T., Triest, S., Rasmussen,
S. G., Wohlkönig, A., Ruf, A., Muyldermans, S., Hol, W. G.,
Kobilka, B. K., and Steyaert, J. (2014) A general protocol for the
generation of Nanobodies for structural biology, Nat. Protocols,
9, 674-693, doi: 10.1038/nprot.2014.039, doi:
10.1038/nprot.2014.039.
36.Jember, T. F. (2021) Molecular cloning,
expression and purification of recombinant VHH proteins expressed in
E. coli, Am. J. Mol. Biol., 11, 129-141, doi:
10.4236/ajmb.2021.114011.
37.Su, Q., Shi, W., Huang, X., Wan, Y., Li, G.,
Xing, B., Xu, Z. P., Liu, H., Hammock, B. D., Yang, X., Yin, S., and
Lu, X. (2022) Screening, expression, and identification of nanobody
against SARS-CoV-2 spike protein, Cells, 11, 3355, doi:
10.3390/cells11213355.
38.Hu, Y., Wang, Y., Lin, J., Wu, S., Muyldermans,
S., and Wang, S. (2022) Versatile application of nanobodies for food
allergen detection and allergy immunotherapy, J. Agricult. Food
Chem., 70, 8901-8912, doi: 10.1021/acs.jafc.2c03324.
39.Hemamalini, N., Ezhilmathi, S., and Mercy, A. A.
(2020) Recombinant protein expression optimization in
Escherichia coli: a review, Ind. J. Animal Res.,
54, 653-660.
40.Gupta, S. K., and Shukla, P. (2017) Microbial
platform technology for recombinant antibody fragment production: a
review, Crit. Rev. Microbiol., 43, 31-42, doi:
10.3109/1040841X.2016.1150959.
41.Mehta, D., Chirmade, T., Tungekar, A. A., Gani,
K., and Bhambure, R. (2021) Cloning and expression of antibody fragment
(Fab) I: Effect of expression construct and induction strategies on
light and heavy chain gene expression, Biochem. Engin. J.,
176, 108189, doi: 10.1016/j.bej.2021.108189.
42.Henry, K. A., Sulea, T., van Faassen, H.,
Hussack, G., Purisima, E. O., MacKenzie, C. R., and Arbabi-Ghahroudi,
M. (2016) A rational engineering strategy for designing protein
A-binding camelid single-domain antibodies, PLoS One, 11,
e0163113, doi: 10.1371/journal.pone.0163113.
43.Bossi, S., Ferranti, B., Martinelli, C., Capasso,
P., and de Marco, A. (2010) Antibody-mediated purification of
co-expressed antigen–antibody complexes, Protein Express.
Purif., 72, 55-58, doi: 10.1016/j.pep.2010.01.003.
44.Jia, B., and Jeon, C. O. (2016) High-throughput
recombinant protein expression in Escherichia coli:
current status and future perspectives, Open Biol., 6,
160196, doi: 10.1098/rsob.160196.
45.Farshdari, F., Ahmadzadeh, M., Nematollahi, L.,
and Mohit, E. (2020) The improvement of anti-HER2 scFv soluble
expression in Escherichia coli, Braz. J. Pharm.
Sci., 56, e17861, doi: 10.1590/s2175-97902019000317861.
46.Zarschler, K., Witecy, S., Kapplusch, F.,
Foerster, C., and Stephan, H. (2013) High-yield production of
functional soluble single-domain antibodies in the cytoplasm of
Escherichia coli, Microb. Cell Factor., 12,
97, doi: 10.1186/1475-2859-12-97.
47.Farasat, A., Rahbarizadeh, F., Ahmadvand, D., and
Yazdian, F. (2017) Optimization of an anti-HER2 nanobody expression
using the Taguchi method, Prepar. Biochem. Biotechnol.,
47, 795-803, doi: 10.1080/10826068.2017.1342259.
48.Studier, F. W. (2005) Protein production by
auto-induction in high-density shaking cultures, Protein Express.
Purif., 41, 207-234, doi: 10.1016/j.pep.2005.01.016.
49.Zou, C., Duan, X., and Wu, J. (2014) Enhanced
extracellular production of recombinant Bacillus
deramificans pullulanase in Escherichia coli
through induction mode optimization and a glycine feeding strategy,
Biores. Technol., 172, 174-179, doi:
10.1016/j.biortech.2014.09.035.
50.Spadiut, O., Capone, S., Krainer, F., Glieder,
A., and Herwig, C. (2014) Microbials for the production of monoclonal
antibodies and antibody fragments, Trends Biotechnol.,
32, 54-60, doi: 10.1016/j.tibtech.2013.10.002.
51.Demain, A. L., and Vaishnav, P. (2009) Production
of recombinant proteins by microbes and higher organisms,
Biotechnol. Adv., 27, 297-306, doi:
10.1016/j.biotechadv.2009.01.008.
52.Gupta, S. K., and Shukla, P. (2017) Sophisticated
cloning, fermentation, and purification technologies for an enhanced
therapeutic protein production: a review, Front. Pharmacol.,
8, 419, doi: 10.3389/fphar.2017.00419.
53.Vieira Gomes, A. M., Souza Carmo, T., Silva
Carvalho, L., Mendonça Bahia, F., and Parachin, N. S. (2018)
Comparison of yeasts as hosts for recombinant protein production,
Microorganisms, 6, 38, doi:
10.3390/microorganisms6020038.
54.Hong, M. S., Velez-Suberbie, M. L., Maloney, A.
J., Biedermann, A., Love, K. R., Love, J. C., Mukhopadhyay, T. K., and
Braatz, R. D. (2021) Macroscopic modeling of bioreactors for
recombinant protein producing Pichia pastoris in defined medium,
Biotechnol. Bioeng., 118, 1199-1212, doi:
10.1002/bit.27643.
55.Karbalaei, M., Rezaee, S. A., and Farsiani, H.
(2020) Pichia pastoris: A highly successful expression
system for optimal synthesis of heterologous proteins, J. Cell.
Physiol., 235, 5867-5881, doi: 10.1002/jcp.29583.
56.De Sá Magalhães, S., and
Keshavarz-Moore, E. (2021) Pichia pastoris
(Komagataella phaffii) as a cost-effective tool for
vaccine production for low- and middle-income countries (LMICs),
Bioengineering, 8, 119, doi: 10.3390/bioengineering8090119.
57.Baghban, R., Farajnia, S., Rajabibazl, M.,
Ghasemi, Y., Mafi, A., Hoseinpoor, R., Rahbarnia, L., and Aria, M.
(2019) Yeast expression systems: overview and recent advances, Mol.
Biotechnol., 61, 365-384, doi:
10.1007/s12033-019-00164-8.
58.Goffeau, A., Barrell, B. G., Bussey, H., Davis,
R. W., Dujon, B., Feldmann, H., Galibert, F., Hoheisel, J. D., Jacq,
C., Johnston, M., Louis, E. J., Mewes, H. W., Murakami, Y., Philippsen,
P., Tettelin, H., and Oliver, S. G. (1996) Life with 6000 genes,
Science, 274, 546-567, doi:
10.1126/science.274.5287.546.
59.Gorlani, A., de Haard, H., and Verrips, T. (2012)
Expression of VHHs in Saccharomyces cerevisiae,
Methods Mol. Biol., 911, 277-286, doi:
10.1007/978-1-61779-968-6_17.
60.Chee, M. K., and Haase, S. B. (2012) New and
redesigned pRS plasmid shuttle vectors for genetic manipulation of
Saccharomyces cerevisiae, G3 (Bethesda, Md.),
2, 515-526, doi: 10.1534/g3.111.001917.
61.Berlec, A., and Strukelj, B. (2013) Current state
and recent advances in biopharmaceutical production in Escherichia
coli, yeasts and mammalian cells, J. Indust. Microbiol.
Biotechnol., 40, 257-274, doi:
10.1007/s10295-013-1235-0.
62.Carlesso, A., Delgado, R., Ruiz Isant, O.,
Uwangue, O., Valli, D., Bill, R. M., and Hedfalk, K. (2022) Yeast as a
tool for membrane protein production and structure determination,
FEMS Yeast Res., 22, foac047, doi:
10.1093/femsyr/foac047.
63.Joosten, V., Lokman, C., van den Hondel, C. A.,
and Punt, P. J. (2003) The production of antibody fragments and
antibody fusion proteins by yeasts and filamentous fungi, Microb.
Cell Factor., 2, 1, doi: 10.1186/1475-2859-2-1.
64.Xu, P., Raden, D., Doyle, F. J., 3rd, and
Robinson, A. S. (2005) Analysis of unfolded protein response during
single-chain antibody expression in Saccaromyces cerevisiae reveals
different roles for BiP and PDI in folding, Metab. Engin.,
7, 269-279, doi: 10.1016/j.ymben.2005.04.002.
65.Wang, Y., Li, X., Chen, X., Nielsen, J.,
Petranovic, D., and Siewers, V. (2021) Expression of antibody fragments
in Saccharomyces cerevisiae strains evolved for enhanced
protein secretion, Microb. Cell Fact., 20, 134, doi:
10.1186/s12934-021-01624-0.
66.Tang, H., Wang, S., Wang, J., Song, M., Xu, M.,
Zhang, M., Shen, Y., Hou, J., and Bao, X. (2016) N-hypermannose
glycosylation disruption enhances recombinant protein production by
regulating secretory pathway and cell wall integrity in
Saccharomyces cerevisiae, Scientific reports,
6, 25654, doi: 10.1038/srep25654.
67.Ahmad, M., Hirz, M., Pichler, H., and Schwab, H.
(2014) Protein expression in Pichia pastoris: recent
achievements and perspectives for heterologous protein production,
Appl. Microbiol. Biotechnol., 98, 5301-5317, doi:
10.1007/s00253-014-5732-5.
68.Huang, D., and Shusta, E. V. (2006) A yeast
platform for the production of single-chain antibody-green fluorescent
protein fusions, Appl. Environ. Microbiol., 72,
7748-7759, doi: 10.1128/aem.01403-06.
69.Frenzel, A., Hust, M., and Schirrmann, T. (2013)
Expression of recombinant antibodies, Front. Immunol., 4,
217, doi: 10.3389/fimmu.2013.00217.
70.Chen, Q., Zhou, Y., Yu, J., Liu, W., Li, F.,
Xian, M., Nian, R., Song, H., and Feng, D. (2019) An efficient
constitutive expression system for Anti-CEACAM5 nanobody production in
the yeast Pichia pastoris, Protein Expr. Purif.,
155, 43-47, doi: 10.1016/j.pep.2018.11.001.
71.Gómez-Ramírez, I. V.,
Corrales-García, L. L., Possani, L. D., Riaño-Umbarila,
L., and Becerril, B. (2023) Expression in Pichia pastoris
of human antibody fragments that neutralize venoms of Mexican
scorpions, Toxicon, 223, 107012, doi:
10.1016/j.toxicon.2022.107012.
72.Guthrie, C., and Fink, G. R. (2002) Guide to
Yeast Genetics and Molecular and Cell Biology, Part C, Gulf
Professional Publishing.
73.Gao, M.-J., Li, Z., Yu, R.-S., Wu, J.-R., Zheng,
Z.-Y., Shi, Z.-P., Zhan, X.-B., and Lin, C.-C. (2012) Methanol/sorbitol
co-feeding induction enhanced porcine interferon-α production by
P. pastoris associated with energy metabolism shift,
Bioprocess Biosystems Engin., 35, 1125-1136, doi:
10.1007/s00449-012-0697-1.
74.Gao, M. J., Zhan, X. B., Gao, P., Zhang, X.,
Dong, S. J., Li, Z., Shi, Z. P., and Lin, C. C. (2015) Improving
performance and operational stability of porcine interferon-α
production by Pichia pastoris with combinational
induction strategy of low temperature and methanol/sorbitol co-feeding,
Appl. Biochem. Biotechnol., 176, 493-504, doi:
10.1007/s12010-015-1590-6.
75.Farsiani, H., Mosavat, A., Soleimanpour, S.,
Sadeghian, H., Akbari Eydgahi, M. R., Ghazvini, K., Sankian, M., Aryan,
E., Jamehdar, S. A., and Rezaee, S. A. (2016) Fc-based delivery system
enhances immunogenicity of a tuberculosis subunit vaccine candidate
consisting of the ESAT-6:CFP-10 complex, Mol. bioSystems,
12, 2189-2201, doi: 10.1039/c6mb00174b.
76.Duranti, C., Carraresi, L., Sette, A., Stefanini,
M., Lottini, T., Crescioli, S., Crociani, O., Iamele, L., De Jonge, H.,
Gherardi, E., and Arcangeli, A. (2018) Generation and characterization
of novel recombinant anti-hERG1 scFv antibodies for cancer molecular
imaging, Oncotarget, 9, 34972-34989, doi:
10.18632/oncotarget.26200.