REVIEW: Discovery and Study of Transmembrane Rotary Ion-Translocating Nano-Motors: F-ATPase/Synthase of Mitochondria/Bacteria and V-ATPase of Eukaryotic Cells
Vladimir Marshanskya
Neuro-Horizon Pharma, Sharon, MA, 02067, USA
Received April 19, 2022; Revised July 8, 2022; Accepted July 8, 2022
This review discusses the history of discovery and study of the operation of the two rotary ion-translocating ATPase nano-motors: (i) F-ATPase/synthase (holocomplex F1FO) of mitochondria/bacteria and (ii) eukaryotic V-ATPase (holocomplex V1VO). Vacuolar adenosine triphosphatase (V-ATPase) is a transmembrane multisubunit complex found in all eukaryotes from yeast to humans. It is structurally and functionally similar to the F-ATPase/synthase of mitochondria/bacteria and the A-ATPase/synthase of archaebacteria, which indicates a common evolutionary origin of the rotary ion-translocating nano-motors built into cell membranes and invented by Nature billions of years ago. Previously we have published several reviews on this topic with appropriate citations of our original research. This review is focused on the historical analysis of the discovery and study of transmembrane rotary ion-translocating ATPase nano-motors functioning in bacteria, eukaryotic cells and mitochondria of animals.
KEY WORDS: F-ATPase/synthase, V-ATPase, transmembrane ATPase, rotary ion-translocating nano-motorDOI: 10.1134/S000629792208003X
Abbreviations: ΔµH+, proton-motive force of transmembrane electrochemical transmembrane proton gradient; EP, elementary particles; factor F1, cytoplasmic part of holocomplex F1FO; factor FO, transmembrane part of holocomplex F1FO; factor V1, cytoplasmic part of holocomplex V1VO; factor VO, transmembrane part of holocomplex V1VO; holocomplex F1FO, F-ATPase/synthase of mitochondria/bacteria; holocomplex V1VO, eukaryotic vacuolar adenosine triphosphatase (V-ATPase); SMP, submitochondrial particles.
INTRODUCTION
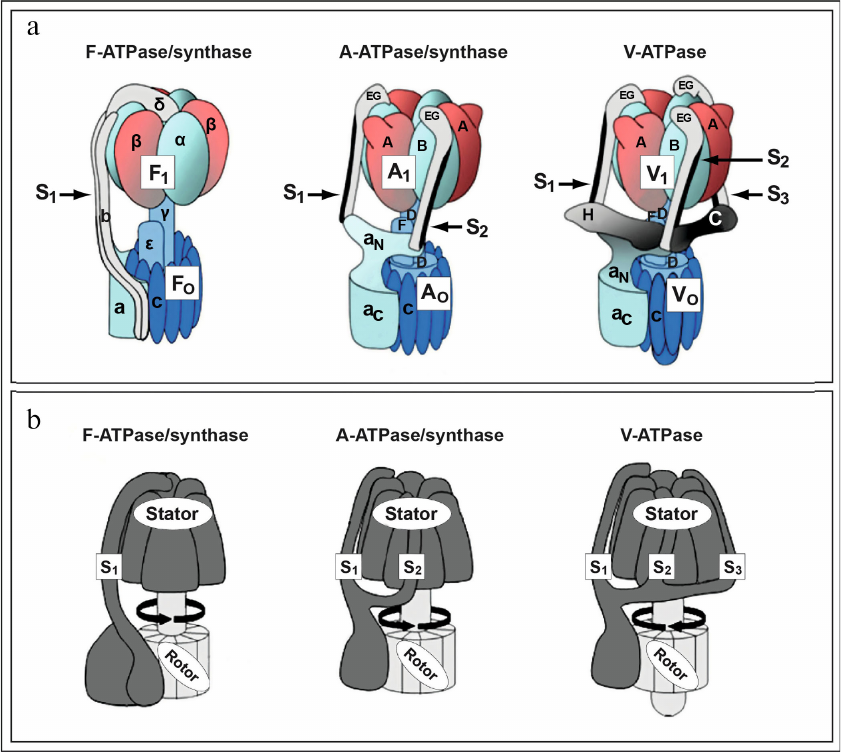
Enzymatic complexes of F-ATPase/synthase of mitochondria/bacteria, A-ATPase/synthase of archaebacteria, and eukaryotic V-ATPase (Fig. 1, a and b) have common principle of operation, which indicates a common evolutionary origin as rotary ion-translocating nano-motors built into cell membranes and invented by Nature billions of years ago (Fig. 1) [1-5].
Fig. 1. Rotary ion-translocating ATPase nano-motors invented by Nature billions of years ago. a) Structure and composition of subunits of the rotary ion-translocating F-ATPase of mitochondria/bacteria (left), A-ATPase/synthase of archaebacteria (center), and V-ATPase of eukaryotes (right). Peripheral stalk structures, stalks S1, S2, and S3 linking: 1) factors F1 and FO F-ATPase of mitochondria/bacteria (left, S1,); 2) factors A1 and AO A-ATPase/synthase of archaebacteria (center, S1 and S2); 3) factors V1 and VO of V-ATPase of eukaryotes (right, S1, S2, and S3) are indicated with arrows. b) Rotary ion-translocating ATPase nano-motors consist of immobile part (stator) and rotating part (rotor). Figure is adapted from [5] with permission from the Publisher.
They represent rotary nano-motors performing translocation of ions (protons or sodium) across membranes of cells and intracellular organelles, which is coupled with large change in free energy and with synthesis/hydrolysis of ATP. In this review focus is on the comparative analysis of F-ATPase/synthase of mitochondria/bacteria (Fig. 1, a and b, left; Fig. 2a) and V-ATPase of eukaryotes (Fig. 1, a and b, right; Fig. 2b) [2, 5] without discussing A-ATPase/synthase of archaebacteria.
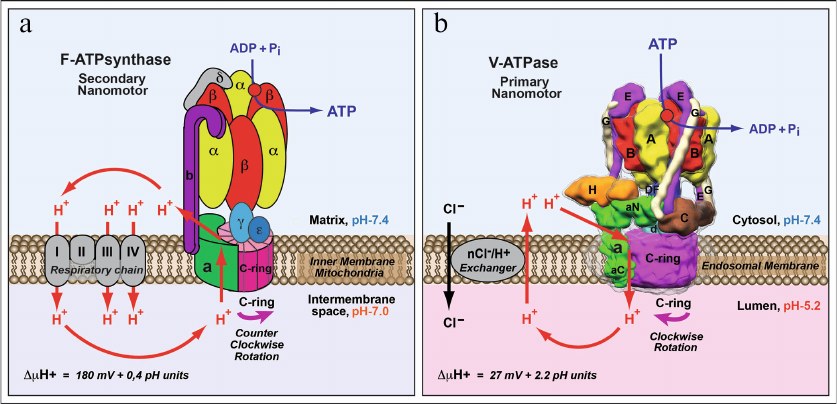
Fig. 2. Comparison of structure and functions of F-ATPase/synthase of mitochondria/bacteria and V-ATPase of eukaryotes. a) F-ATPase/synthase of mitochondria/bacteria is a secondary rotary nano-motor synthesizing ATP by using energy of the proton gradient generated by the respiratory chain enzymes. b) V-ATPase of eukaryotes is a primary ion-translocating nano-motor with hydrolysis of ATP molecules causing rotation and proton transport across the membrane. Proton translocation by V-ATPase is facilitated by generation of a neutralizing current mediated by electrogenic nCl–/H+-ion exchanger. This process results in acidification of intracellular organelles and extracellular medium. Figure is adapted from [2] with permission from the Publisher.
Discoveries and further investigation of the structures and functions of transmembrane ATPases (F-ATPase/synthase of mitochondria/bacteria and V-ATPase of eukaryotes) as rotary ion-translocating nano-motors are closely related complementing each other. Owing to the fact that the principles of structural organization and functioning of the eukaryotic V-ATPase are very similar to those of F-ATPase/synthase of mitochondria/bacteria, historically they were described mainly according to the well-known paths of investigation of the F-ATPase/synthase of mitochondria of animals/bacteria.
DISCOVERY AND INVESTIGATION OF FUNCTIONING OF F-ATPase/SYNTHASE
OF MITOCHONDRIA OF ANIMALS/BACTERIA
The fundamental process of oxidative phosphorylation was discovered in 1930 by the outstanding Soviet biochemist, academician, Professor Vladimir Aleksandrovich Engelhardt [6-8]. According to the memoirs of Professor Engelhardt (“Life and Science”) published in 1982 in the journal Annual Review of Biochemistry [9], this discovery was made possible due to the use of a unique and appropriate object in these studies – avian erythrocytes. Unlike the mammalian erythrocytes, these eukaryotic cells contain nuclei and mitochondria, and owing to this they exhibit high respiratory activity and contain high levels of ATP. As mentioned by V. A. Engelhardt in his autobiographic publication, the choice of this research object was made in accordance with principle proposed by outstanding Danish biologist Professor August Krogh, a Nobel Prize Laureate in Physiology and Medicine 1920 “for his discovery of the capillary motor regulating mechanism” [10]. This is citation from the V. A. Engelhardt’s manuscript: “Experimenters neglected the valuable advice given by the remarkable biologist, Dr. Krogh, in his address to one of the Physiological Congresses. He said that Nature has been generous toward naturalists, by creating some special object particularly suited for the study of the more important problems. The condition of success for a scientist attacking a new problem is to find and use the appropriate object” [9]. Thus, by using a research object corresponding to the goal of the study V. A. Engelhardt discovered a phenomenon of aerobic esterification of inorganic phosphate with formation of ATP, which he termed “respiratory resynthesis of ATP” currently known as the process of oxidative phosphorylation [6-8]. This fundamental biochemical process was further investigated and quantitatively characterized in 1939 by V. A. Engelhardt in his following studies [11, 12] and in the studies by academician, Professor Vladimir Aleksandrovich Belitser [13].
Later in the second half of the last century the main focus of research in bioenergetics was elucidation of molecular mechanisms of coupling of oxidation processes and phosphorylation in mitochondria and bacteria. History of the discoveries and research in this area have been presented and analyzed in detail in the publications of the outstanding Russian biochemist, academician, Professor Vladimir Petrovich Skulachev [14-23]. A cornerstone step in this regard was the publication by Professor Peter Mitchell in 1961 in the journal Nature of the article entitled: “Coupling of phosphorylation to electron and hydrogen transfer by a chemi-osmotic type of mechanism” [24-29]. In 1978, P. Mitchell was awarded the Nobel Prize in Chemistry “for his contribution to the understanding of biological energy transfer through the formulation of the chemiosmotic theory”.
It is important to emphasize that experimental confirmation of this theory and its general acceptance as a chemiosmotic mechanism of coupling of oxidation with phosphorylation was to a great extent provided by V. P. Skulachev. The pivotal studies conducted in his laboratory used the mitochondria-targeted penetrating cations [14-18, 30-35]. Based on the discoveries made by the V. P. Skulachev’s research group an antioxidant molecule was developed later that was named “Skulachev ions” (SkQ). It was demonstrated in a series of important studies that this unique molecule could switch off the programmed death pathway in eukaryotic cells, slow down phenoptosis and aging in mammals [36-39]. These innovative studies and development of Skulachev ions resulted in its application as therapeutic agent, which is used successfully nowadays in medical practice [40-43].
Another important task of bioenergetics was elucidation of the mechanism of ATP synthesis/hydrolysis, which was observed in animal mitochondria [14-20, 44, 45]. The history of discovery and investigation of F-ATPase/synthase starts with its discovery in 1960 by Professor Efraim Racker [46, 47]. It was shown in his studies that the inner mitochondrial membrane contains a protein responsible for coupling of electron transport with ATP synthesis and required for oxidative phosphorylation. This protein was named by E. Racker “coupling factor F1” (factor F1). It was shown in the later studies that the mitochondrial F-ATPase/synthase consists of: (i) hydrophilic factor F1 capable of catalyzing ATP hydrolysis, and (ii) hydrophobic/membrane factor FO capable of restoring ATP synthesis together with the factor F1, which is sensitive to the specific inhibitor of oxidative phosphorylation oligomycin (hence, subscript “o” in the name of the factor) [46-51].
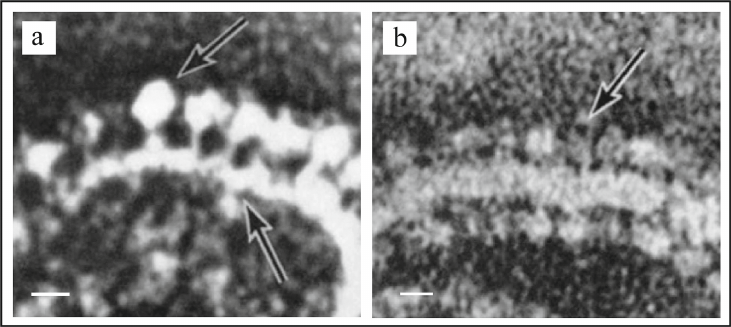
In 1963 using electron microscopy (EM), Professor Humberto Fernandez-Moran had discovered repeating spherical structures with 80-100 Å diameter on the inner mitochondrial membrane, which he named elementary particles (EP) [52]. In the subsequent collaboration with the Professor David Green, it was shown that the EP particles consisted of three components: (i) headpiece or knob; diameter of 80-100 Å; (ii) stalk; size of 30 × 50 Å; and (iii) basepiece incorporated into the membrane; size of 110 × 40 Å (Fig. 3, a and b) [53, 54].
Fig. 3. Electron microscopy images of intact mitochondria and submitochondrial particles (SMP) isolated from the bovine heart muscle. a) Fragment of mitochondrial inner membrane with elementary particles on it, scale bar – 100 Å. b) Fragment of SMP with elementary particles on it, scale – 100 Å. Figure is adapted from [53] with permission from the Publisher.
Based on these studies a hypothesis was proposed by D. Green suggesting that elementary particles and, in particular, their spherical headpiece comprised of the components (cytochromes) of the mitochondrial electron transport chain. However, this hypothesis was refuted in the further studies. On the one hand, it was shown in the independent investigations by Professors B. Chance and D. F. Parsons [55] and Professors J. T. Stasny and F. L. Crane [56, 57] that the headpieces separated from the submitochondrial particles (SMP) with ultrasound treatment did not contain cytochromes. On the other hand, it was shown in the Racker’s laboratory that the headpieces could be separated from the membrane by urea treatment. The membranes with removed headpieces did not exhibit ATPase activity, and the soluble fraction in this case contained the protein identical to the soluble part of the coupling factor F1 [49].
Final validation of the Racker’s hypothesis that the elementary particles are comprised of F-ATPase/synthase was obtained in the experiments on reconstruction of oxidative phosphorylation. Addition of the purified factor F1 to the SMP membranes treated with urea resulted in re-assembly of the elementary particles on the SMP membrane, and, what is more important, resulted in restoration of the oligomycin-sensitive ATPase activity [49-51]. These data signify experimental evidence of the fact that EPs represent a mitochondrial F1FO F-ATPase/synthase holocomplex. Hence, the spherical headpiece in an elementary particle comprises a soluble catalytic factor F1, while the basepiece contains factor FO that imparts oligomycin sensitivity to ATP hydrolysis and is responsible for proton transfer across the mitochondrial membrane.
Relatively slow time-dependent changes of the ATPase activity properties of the purified factor F1 were demonstrated in the P. Mitchell’s laboratory [27]. This phenomenon was found to be universal as it was also observed for the mitochondrial F1-ATPase in yeast [58, 59]. The phenomenon of slow kinetic properties of F1-ATPase was investigated in detail, and its mechanism was elucidated in the laboratory of the outstanding Russian biochemist, Professor Andrei Dmitrievich Vinogradov [60-63]. In particular, it was shown that ATP hydrolysis by the isolated factor F1 is suppressed in an unusual manner by ADP, which is a product of this enzymatic activity and, hence, explains a number of its properties [64-70]. The isolated factor F1 is also sensitive to azides and sulfites [71]. It has been also established that the removal of ADP from the active center of factor F1 and inhibition of hydrolysis by the reaction product in the intact F-ATP synthase is controlled by ΔµH+ [72]. Furthermore, the mechanism of regulation of the F-ATPase enzyme by the specific protein inhibitor was investigated in detail [73-75]. Hence, the results of these studies showed for the first time that the product of ATP hydrolysis by the intact F-ATP synthase as well as protein inhibitor ensure suppression of ATPase activity and preservation of ATP with decreasing of the proton gradient in mitochondria [64-75]. This phenomenon was named ADP-Mg2+-dependent inhibition of factor F1 and holocomplex of F1FO F-ATPase/synthase of mitochondria [60-63]. The similar mechanism of slow ADP-Mg+2-dependent activation/inactivation has been also described for regulation of V-ATPase from Thermus thermophilus [76].
Cornerstone studies carried out in 1960-1990 in the laboratories of Professor Paul D. Boyer and Professor John E. Walker resulted in the proposal of the mechanism of the F-ATPase/synthase functioning as a rotary ion-translocating nano-motor driven by ATP hydrolysis/synthesis. Both scientists were awarded the Nobel Prize in Chemistry in 1997 “for their elucidation of the enzymatic mechanism underlying the synthesis of adenosine triphosphate (ATP)”.
In particular, the reaction of O18 and P32 isotope exchange in the course of ATP synthesis was investigated in the P. D. Boyer laboratory [77]. These studies demonstrated that contrary to other well-known enzymes, mitochondrial F-ATP synthase uses energy of ΔµH+ not for the synthesis of ATP from ADP and Pi, but for binding of ADP, Pi and release of the synthesized ATP from its active center. It was shown in the further studies that ATP hydrolysis occurs as a result of cooperative interaction of nucleotide-binding centers located on three β-subunits of factor F1, each of which sequentially changes affinity to the substrate (ATP) and products (ADP and Pi) of the reaction. This mechanism of functioning of ATP synthase was first suggested by the Professors K. Repke and R. Schön. [78]. This mechanism was investigated in detail in the later studies by P. D. Boyer with the trimer of the pairs of α,β-subunits (3α,3β-hexamer) of the factor F1 and was termed “rotational alternative binding site mechanism” [79-82]. Considering symmetric structure of the pairs of α,β-subunits, P. D. Boyer suggested a hypothesis that alternation of the affinity is mediated through rotation of the γ-subunit located inside the cavity formed by these pairs [44, 45, 83-85]. In the following studies conducted in the J. E. Walker laboratory atomic structure of the 3α,3β-hexamer of factor F1 of mitochondrial F-ATP synthase was resolved with resolution 2.4-2.8 Å [86, 87]. This atomic structure was in perfect agreement with the model of rotational alternative binding site mechanism suggested earlier by P. D. Boyer. At present several dozens of atomic structures of the key components of F-ATPase/synthase have been determined using X-ray crystallography [44, 45, 88-90], as well as the structures of intact holocomplex F1FO of F-ATPases/synthases isolated from bacteria, archaea, and eukaryotes resolved with the help of cryo-electron microscopy (cryo-EM) [91-94]. It is important to mention that the structures of all investigated enzymes were also in agreement with the “rotational alternative binding site mechanism” proposed by P. D. Boyer, which allowed to finally recognize the mechanism of functioning of F-ATPase/synthase of mitochondria/bacteria as a transmembrane rotary ion-translocating nano-motor.
DISCOVERY AND INVESTIGATION OF FUNCTIONING OF EUKARYOTIC
V-ATPase
Discovery of vacuolar adenosine triphosphatase or vacuolar H+-ATPase (vacuolar-ATPase or V-ATPase) have not been accomplished in a single experiment, as was the case of discovery of mitochondrial F-ATPase/synthase from bovine heart by the researchers from the E. Racker laboratory in 1960 [46, 47]. Discovery and investigation of V-ATPase in eukaryotic cells was made as a result of numerous efforts in many laboratories working in different areas of cell biology in 1960-1980.
In retrospect it can be stated that the gradual discovery of V-ATPase was warranted due to both multiple locations of V-ATPase in different organelles and cell membrane and also due to diverse functions of V-ATPase in eukaryotic cells [1-4, 95-98]. These properties and functions of V-ATPase are fundamentally different from the properties and function of F-ATPase/synthase, which is located solely on the inner membrane of mitochondria and performs very important but only one function – synthesis of ATP in the cells (Fig. 2a) [14-18, 44, 45, 49-51, 60-63].
The history of discovery and investigation of V-ATPase starts with publication in 1962 of the results of experiments by Professor Norman Kirshner, which demonstrated ATP-dependent accumulation of catecholamines in chromaffin granules isolated from the endocrine cells of adrenal gland. This was the first indication of the existence of a novel H+-ATPase in these vesicles [99, 100]. However, only thirteen years later in 1975 important experiments were carried out that used uncouplers, which showed that a novel H+-ATPase participates in accumulation of catecholamines in chromaffin granules functioning as a proton pump [101, 102]. Later, in the beginning of 1980s, owing to the efforts of many research groups, it was experimentally proven that acidification of lumen of different intracellular organelles such as (i) vacuoles of eukaryotic Saccharomyces cerevisiae [103] and Neurospora crassa [104] yeast cells, (ii) clathrin-coated vesicles isolated from bovine brain [105], and (iii) lysosomes isolated from rat kidney [106] occurs with participation of a novel H+-ATPase different from the mitochondrial F-ATPase/synthase. Therefore, based on localization of the novel H+-ATPase different from the F-ATPase/synthase of mitochondria in various organelles (vacuoles, granules, vesicles, and lysosomes), this new H+-ATPase was named vacuolar H+-ATPase (vacuolar-ATPase) or V-ATPase [95, 107].
Studies employing EM conducted in 1960-1980 provided significant contribution to discovery of V-ATPase. Images visualizing location of V-ATPase on the membranes of cells from different organisms and tissues under physiological conditions were published as a result of these experiments. In particular, in 1966 Professors E. Anderson and W. Harvey published the first EM-image of V-ATPase of insects. The spike-like units or projections on the surface of cytoplasmic membrane of epithelial goblet cells from the midgut of Hyalophora cecropia [108] insect were for the first time described in this work. Similar morphological structures were also described in 1968 in the epithelial cells from blow flies Calliphora erythrocephala [109]. Functions of these structures remained unknown, but it was suggested for the first time in this study that they are associated with an ion-translocating ATPase, which is not sensitive to ouabain inhibitor and, therefore, is different from the Na+/K+-ATPase.
Twelve years later, in 1980-1981, EM examinations of V-ATPase were continued and were successfully completed in the detailed studies performed by Professors D. Brown and R. Montesano. In particular, existence of the so-called “road-shaped particles”, transmembrane structures located both at the apical plasma membrane and on the membrane of vesicles and vacuoles in the cytoplasm of the mitochondria-rich clear cells of the male rat epididymis was shown using freeze-fracture EM technique [110, 111].
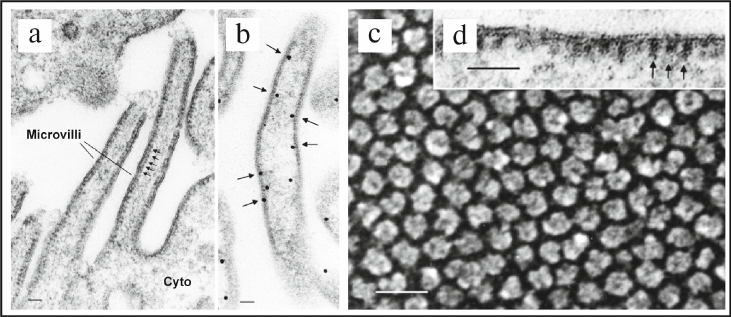
Later, in 1986-1987 in the Professor Dennis Brown’s laboratory presence of the road-shaped particles or intramembranous particles (IPMs) was observed on the membrane of vesicles located in the cytoplasm of the rat kidney collecting duct intercalated cells, and it was shown that these vesicles did not have clathrin coating [112, 113]. Further studies revealed the presence of IPMs in the kidney epithelial cells of the loop of Henle, which corresponded to the transmembrane factor VO of V-ATPase [95]. Furthermore, presence of the stud-like structures on the cytoplasmic membrane of intercalated cells of the rat kidney collecting duct (Fig. 4, a, b, and d) and on the mitochondria-rich cells of the toad bladder (Fig. 4 c) was shown in 1987 using EM [114]. It was clearly demonstrated in the same study by reconstruction of the purified stud-like structures into phospholipid liposomes that these particles actually represent enzymatic complex of V-ATPase. The following works of D. Brown demonstrated critical role of V-ATPase in the kidney physiology [115, 116].
Fig. 4. Discovery and identification of V-ATPase in epithelial cells from different tissues of eukaryotes using standard and freeze-fracture EM techniques. a) EM image of the plasma membrane of the rat kidney collecting duct intercalated cells. Arrows point to the repeating stem-like structures (studs) located on the cytoplasmic side of the membrane. These stud-like structures correspond to the factor V1 of the V1VO V-ATPase holocomplex; scale bar – 60 nm. b) Identification of location of V-ATPase using immunogold EM technique. Arrows point to the locations of gold particles after immunocytochemical identification of V-ATPase on the cytoplasmic side of the membrane of the brush border of intercalated cells; scale bar – 40 nm. c) EM image produced using freeze-fracture technique showing location of the stud-like structures corresponding to the factor V1 of V-ATPase on the cytoplasmic membrane of mitochondria-rich cells of the toad bladder; scale bar – 30 nm. d) High-resolution EM image showing location of the stud-like structures on the cytoplasmic side of the membrane; scale bar – 60 nm. Figure is adapted from [95] with permission from the Publisher.
It is important to emphasize that location of V-ATPase on the cytoplasmic membrane is essential for functioning of the specialized proton-secreting epithelial cells of eukaryotic tissues in general, and kidneys, in particular [95, 96]. Fundamental contribution to investigation of this physiological process was made in the further investigations conducted in the laboratories of Professor Dennis Brown and Professor Sylvie Breton [117-123]. In particular, in 1996 in the S. Breton laboratory final confirmation of localization of V-ATPase on the cytoplasmic membrane of the male rat sperm duct and its importance was obtained [117, 118]. These key studies presented a road-map of further investigation of the role of V-ATPase in functioning of the specialized proton-secreting epithelial cells in organs of animals and humans under normal and pathological conditions [95, 96, 119-123].
In 1981-1983 EM examination of the epithelial cells of midgut of Manduca sexta insects have been conducted in the Professor William Harvey laboratory [124-126]. In these studies, also, the repeated elongated morphological structures were observed on the cytoplasmic membrane of the midgut epithelial cells, which were named “K+-portasomes”. It was suggested that they function as K+-transporting ATPases. However, biochemical studies conducted later in 1989 in the laboratory of Professor Helmut Wieczorek revealed that the portasomes of M. sexta insects were not K+-transporting ATPases, but represent enzymatic complexes of V-ATPase [97, 127, 128].
One of the important contributions in investigation of V-ATPase at that time was provided by the author of this review [129-131]. In particular, the studies conducted in our laboratory in 1994-1996 were devoted to investigation of the role of V-ATPase in physiological function of kidney proximal tubule cells. A new approach was developed in these studies for isolation and purification of early endosomes from the epithelial cells of kidney proximal tubules of animals and humans [129, 131]. Later we demonstrated the presence of transmembrane V-ATPase in these organelles, and the mechanism of ATPase-dependent lumen acidification in the early endosomes was investigated in detail [95, 96, 130-133].
COMMON EVOLUTIONARY ORIGIN OF F-ATPase/SYNTHASE OF
MITOCHONDRIA/BACTERIA AND V-ATPase OF EUKARYOTES INVENTED BY NATURE
BILLION YEARS AGO
Hence, as a result of intensive studies conducted in 1960-1990 it was shown that two types of transmembrane rotary ion-translocating enzyme complexes are expressed and operate in eukaryotes: (i) F-ATPase/synthase located on the inner mitochondrial membrane [14-20, 44, 45, 48-50, 60-63] (Fig. 1, a and b, left; Fig. 2a), and (ii) V-ATPase, located both on cytoplasmic membrane and on the membranes of intracellular organelles of eukaryotic cells including vacuoles, secretory vesicles, endosomes, lysosomes, and complex Golgi (Fig. 1, a and b, right; Fig. 2b) [1-4, 95-98]. The transmembrane enzymatic complexes of F-ATPase/synthase of mitochondria/bacteria (Fig. 1, a and b, left), A-ATPase/synthase of archaebacteria (Fig. 1, a and b, center), and V-ATPase of eukaryotes (Fig. 1, a and b, right) are structurally very similar and function according to the same mechanism, which indicates their common evolutionary origin. In particular, they consist of two structurally and functionally different parts: (i) hydrophilic, cytoplasmic factor (F1/A1/V1) catalyzing hydrolysis/synthesis of ATP, and (ii) hydrophobic, transmembrane factor (FO/AO/VO) mediating translocation of ions across phospholipid membranes (Fig. 1a).
“SODIUM WORLD” HYPOTHESIS AND PHYLOGENOMIC
INVESTIGATION OF EVOLUTIONARY ORIGIN AND FUNCTIONING OF ROTARY
ION-TRANSLOCATING ATPase NANO-MOTORS
The hypothesis of “Sodium World” was first suggested and published by academician and Professor V. P. Skulachev in 1984 [134]. The new concept of “Sodium bioenergetics” was presented by Skulachev in 1985 in his plenary lecture during the FEBS Special Meeting [135]. In this lecture he stated: “In this paper, I would like to summarize recent observations at my own and other laboratories indicating that non-protonic coupling really exists and it is Na+ that can replace H+. A concept will be developed assuming that sodium bioenergetics is a phenomenon of major significance for living organisms and not a curiosity specific for rare ecological niches only” (cited from [135]).
In 1977-1979 Professor Tsutomu Unemoto and colleagues demonstrated activating effect of Na+ ions on functioning of NADH oxidase in Vibrio alginolyticus bacteria [136, 137]. Later, in 1982-1984, it was also shown in this laboratory that the electron transfer from NADH to quinone could generate ΔµNa+ in these bacteria [138] and that reconstruction of NADH oxidase was sufficient for ΔµNa+ generation in proteoliposomes in vitro [139]. It is important to note that the studies conducted in 1985-1986 in the Skulachev’s laboratory showed that Na+-potential (ΔµNa+) generated in the V. alginolyticus bacterium could be used as a driving force for rotation of its flagella [140-142].
Simultaneously, in 1980-1984, it was shown in the laboratory of Professor Peter Dimroth that decarboxylation of oxaloacetate to pyruvate with the help of oxaloacetate decarboxylase, which is a primary Na+-pump located in the membranes of inverted vesicle prepared from the Krebsiella aerogenes bacteria, resulted in generation of ΔµNa+ in these vesicles in vitro [143]. Similar experiments were conducted with the inverted vesicles prepared from Veillonella alcalescens bacteria. It was shown that in these bacteria methylmalonyl-CoA decarboxylase also served as a primary Na+-pump capable of generating ΔµNa+ [144]. Presence of Na+-dependent methylmalonyl-CoA decarboxylase was also demonstrated in the Propionigenum modestum bacteria in which the generated ΔµNa+ is used for ATP synthesis with the help of Na+-translocating F-ATPase/synthase of these bacteria [145].
Based on the results of research in his laboratory investigating the role of sodium cycle [134, 140-142], as well as his studies of evolutionary tree of eubacteria using 5S rRNA nucleotide sequences [135, 142] V. P. Skulachev introduced a hypothesis of “Sodium World” [134, 135] and suggested a new concept of “Sodium Cycle” and “Sodium Bioenergetics” as a cornerstone phenomenon in Nature. Fundamental character of this hypothesis must be emphasized as it changed our views on membrane bioenergetics and presenting it as not exclusively “proton-based” but also “sodium-based” bioenergetics [146-148].
These directions of research were further developed in detail in the laboratory of Professor Armen Y. Mulkidjanian [149-154]. Using bioinformatics approaches A. Y. Mulkidjanian with his co-workers investigated expansion of the Sodium World through the evolutionary time and taxonomic space. It must be mentioned that the results obtained in these studies indicate primacy of the Na+-dependent over H+-dependent bioenergetics in the course of evolution of life on Earth [150, 154].
The hypothesis of “Sodium World” also pointed to new research directions in the area of origin and evolution of the membrane rotary ion-translocating ATPase nano-motors [149, 151, 152]. In particular, phylogenomic studies of the rotary ATPases of bacteria and archaea showed their origin from the Last Universal Cellular Ancestor (LUCA) of the rotary ATPases [154-158]. According to these studies the ancestral rotary ATPase LUCA was invented by Nature four billion years ago as an exclusively Na+-translocating ATPase. In the course of further evolution, F-ATPase/synthase of mitochondria/bacteria (Fig. 1, a and b, left), A-ATPase/synthase of archaebacteria (Fig. 1, a and b, center), and V-ATPase of eukaryotes (Fig. 1, a and b, right) emerged, which are rotary ion-translocating ATPase/synthases that switched to transporting protons across the impermeable phospholipid membranes and adapted to be able of generation/utilization of the ΔµH+ electrochemical gradient. It is currently known that the majority of investigated rotary ATPase/synthase nano-motors transport protons, however, there are F-ATPases, A-ATPases, and V-ATPases transporting sodium ions [150, 151]. It is important to underline in conclusion that V. P. Skulachev also was the first to suggest the key role of formation of proton-impermeable phospholipid membranes in the process of evolutionary transition from the “sodium-based” to “proton-based” bioenergetics in the cells [146-148].
MOLECULAR MECHANISM OF PROTON TRANSPORT BY THE ROTARY
ION-TRANSLOCATING ATPase NANO-MOTORS ACROSS PHOSPHOLIPID
MEMBRANES
In 1971-1974 first in vitro experiments were conducted in the laboratory of E. Racker on reconstruction of factors F1 and FO of F-ATPase/synthase (isolated from bovine heart) in proteoliposomes (prepared from the plant-derived phospholipids, soya bean azolectin [159]), as well as on combined reconstruction with the light-dependent ΔµH+ generator, rhodopsin (isolated from bacteria) [160]. These key experiments demonstrated essential role of the intact, reconstructed from factors F1 and FO of F-ATP synthase of mitochondria in the ΔµH+-driven ATP synthesis, as well as key role of the transmembrane factor FO in the proton transfer across the phospholipid membranes of proteoliposomes. Elucidation of the molecular mechanism of proton transport by factor FO and its coupling with ATP synthesis by factor F1 was an important task of bioenergetics.
Initially two potential mechanisms of coupling of proton transport with ATP synthesis were considered: (i) direct coupling mechanism and (ii) indirect coupling mechanism [83, 90, 161]. In his publications, P. Mitchell proposed the direct mechanism according to which protons are translocated by the factor FO across the phospholipid membrane and are directed to the catalytic center of the factor F1, where they participate directly in ATP synthesis [161]. However, in the present time the indirect mechanism of coupling is generally accepted, which was suggested and validated through the cornerstone studies conducted in 1970-1990 in the laboratories of P. D. Boyer [44, 45, 79-85] and J. E. Walker [86, 87, 90]. As has been described above, according to this mechanism F-ATPase/synthase of mitochondria/bacteria functions as a rotary ion-translocating nano-motor in which proton translocation is coupled with the “rotor” rotation resulting in the hydrolysis/synthesis of ATP in the nano-motor “stator” (Fig. 1, a and b, left; Fig. 2a).
It is important to underline that in 1978 Professors A. N. Glagolev and V. P. Skulachev proposed the mechanism of “indirect coupling” for the operation of the rotary flagellar motor and rotation of flagella in Rhodospirilium rubrum [162]. The proposed model was described in detail according to which: (i) the transmembrane translocation of protons is realized with the help of two non-coaxial semichannels; (ii) entrance of protons through the access semichannel is a driving force of rotation of the M-ring of the basal body, which is followed by the exit of protons from the release semichannel; and (iii) translocation of protons via two separate semichannels is coupled with the stepwise rotation of the basal body of the flagellar motor in the R. rubrum bacteria flagella. Later, in 1988, V. P. Skulachev also suggested the similar mechanism of coupling of protons translocation with ATP hydrolysis/synthesis in F-ATPase/synthase of mitochondria/bacteria [21].
Despite the significant progress achieved in 1970-1990, the detailed mechanism of “indirect coupling” and, in particular, of translocation of protons by the rotary ATPase/synthase nano-motors across phospholipid membranes remained poorly understood [90]. Nevertheless, owing to the further structural studies conducted in 1999-2013, the main principles of operation of the rotary ion-translocating ATPases/synthases were determined [163-167]. At present the following model of translocation of protons in the course of indirect coupling mediated by F-ATPase/synthase of mitochondria/bacteria has been generally recognized [90]: (i) protons cross phospholipid membrane with the help of two non-coaxial semichannels formed by α-helices of the transmembrane a-subunit; (ii) protons entering the access semichannel interact and neutralize conserved glutamate (or aspartate in E. coli) located on each c-subunit forming c-ring (consisting in different organisms from 8-15 c-subunits); (iii) the protonated in such a manner glutamate on each c-subunit is incorporated into the hydrophobic lipid bilayer and causes stepwise rotation of the c-ring; (iv) when the release semichannel that is formed by other α-helices of the transmembrane a-subunit is reached due to c-ring rotation, the protons are released through this semichannel to the opposite side of the phospholipid membrane; (v) the number of protons required for complete (360°) rotation of the c-ring corresponds to the number of c-subunits forming this ring; (vi) furthermore, c-ring is anchored on the γ-subunit (forming rotor), which is located inside the cavity of the hexamer of 3α,3β-subunits (forming stator) (Fig. 1b, left); (vii) complete rotation (360°) of the c-ring ensures rotation of the γ-subunit which results in hydrolysis/synthesis of ATP by the factor F1.
Specific molecular details of this mechanism were elucidated in 2014-2021 due to the so-called cryo-EM resolution revolution [168, 169]. Significant progress has been made in high-resolution cryo-electron microscopy at that time, which allowed resolving atomic protein structures with resolution up to 2.8-3.5 Å, which was comparable with the resolution of structures determined with the help X-ray crystallography. The results of these breakthrough studies have been described in detail in the original works and reviews published by Professor John L. Rubinstein [91, 92, 170-172], Professor Werner Kühlbrandt [93, 94], and Professor Stephan Wilkens [173-175]. Important matter in the progress in this area was the fact that the method of cryo-EM allowed determination of the structures of intact transmembrane protein complexes including holocomplex F1FO (F-ATPase/synthase of mitochondria/bacteria) and holocomplex V1VO (V-ATPase of eukaryotes) isolated from bacteria and eukaryotic cells with this high resolution. It is worth mentioning that owing to these important studies the structures of V-ATPase isolated from the mammalian brain [171] and human renal cells [175] were determined for the first time in 2020-2021.
EXPERIMENTAL PROOF OF THE ROTATIONAL MECHANISM OF ACTION OF THE
ION-TRANSLOCATING F-ATPase/SYNTHASE NANO-MOTORS OF
MITOCHONDRIA/BACTERIA AND OF V-ATPase OF EUKARYOTES
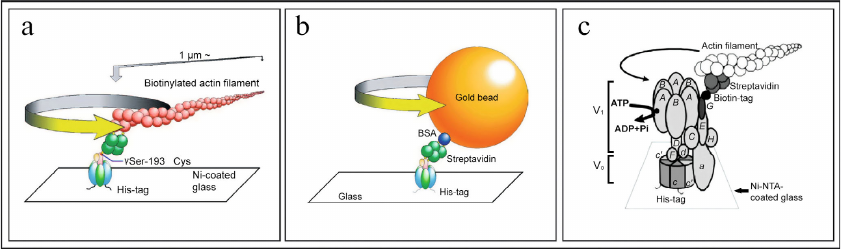
Experimental proof of the rotational mechanism of operation of F-ATPase/synthase was obtained in 1997-1999 owing to the elegant experiments conducted in the laboratories of Professor Masasuke Yoshida [176-178] and Professor Masamitsu Futai [179-183]. In particular, in 1997 M. Yoshida for the first time demonstrated ATP-dependent rotation of the γ-subunit of F1-ATPase from Bacillus PS3 bacteria to which actin filament was attached [176, 177]. In 1999, rotation of the γ-subunit of F1-ATPase from Escherichia coli bacteria with attached actin filament was also shown simultaneously in two laboratories of M. Yoshida [178] and M. Futai (Fig. 5a) [179]. It is important to note that M. Futai was also the first to demonstrate rotation of the intact holocomplex F1FO of F-ATPase of E. coli [180]. These results proved unambiguously physiological significance of the rotational mechanism of the F-ATPase nano-motor of bacteria. Further studies focused on investigation of the stepwise mechanism of rotation of the F-ATPase nano-motor using experimental models with both attached actin filament (Fig. 5a) [181-183], and with attached gold bead (Fig. 5b) [184-187].
Fig. 5. Experiments demonstrating functioning of F-ATPase/synthase of bacteria and V-ATPase of yeast as rotary nano-motors. a) ATP-dependent rotation of the γ-subunit (with attached actin filament) of factor F1 of F-ATPase/synthase isolated from bacteria E. coli. b) ATP-dependent rotation of the γ-subunit (with attached gold bead) of factor F1 of F-ATPase/synthase isolated from bacteria E. coli. c) ATP-dependent rotation of the G-subunit (with attached actin filament) of the intact holocomplex V1VO of V-ATPase isolated from yeast S. cerevisiae. Figure is adapted from [185, 188] with permission from the Publisher.
In 2003 experimental proof of the intact holocomplex V1VO of V-ATPase of yeast functioning as a rotary nano-motor was also obtained in the research conducted by the M. Futai laboratory (Fig. 5c) [188-192]. These results provided clear proof of physiological significance of the rotation mechanism of action of the V-ATPase nano-motor in eukaryotic cells. Hence, these studies confirmed experimentally that the transmembrane F-ATPase/synthase of mitochondria/bacteria and V-ATPases of eukaryotic cells are indeed rotary ion-translocating nano-motors invented by Nature billions of years ago [185-195].
STRUCTURE AND UNIQUE NOVEL FUNCTIONS OF V-ATPase OF EUKARYOTIC
CELLS ACQUIRED IN THE COURSE OF EVOLUTION
Eukaryotic V-ATPase has molecular weight around 900 kDa and consists of 14 subunits (A, B, C, D, E, F, G, H, a, c, c′, c″, d, and e) and two auxiliary subunits: Ac45 (or ATP6AP1) and M8-9 (or ATP6AP2/PRR – (pro)renin receptor) [1-4]. V-ATPase exhibits two-component structure including a cytoplasmic factor V1 consisting of subunits A3, B3, C, D, E3, F, G3, and H, as well as a transmembrane factor VO consisting of subunits a, cx, c′, c′′, d, and e (Fig. 1a, right; Fig. 2b). Factors V1 and VO are linked with each other by three peripheral connecting stalk structures, S1, S2, and S3 (Fig. 1a, right). These stalks are essential for coupling the processes of ATP hydrolysis, occurring in factor V1, and proton translocation, occurring in factor VO (Fig. 2b). It is important to note that structurally the eukaryotic V-ATPase is significantly more complex than F-ATPase/synthase of mitochondria/bacteria and A-ATPase/synthase of archaebacteria (Fig. 1). In particular, the number of connecting stalk structures in these nano-motors represents the most striking difference: one (S1) – in the F-ATPase/synthase of mitochondria/bacteria (Fig. 1, a and b, left); two (S1 and S2) – in A-ATPase/synthase of archaebacteria (Fig. 1, a and b, center), and three (S1, S2, and S3) – in V-ATPase of eukaryotes (Fig. 1, a and b, right) [1-5].
Below, the main functional differences between the F-ATPase/synthase of mitochondria/bacteria and V-ATPase of eukaryotes emerged as a result of divergent evolution will be briefly analyzed. An important difference in functioning of F-ATPase/synthase of mitochondria/bacteria and eukaryotic V-ATPase is due to their fundamentally different roles in cell biology. The main purpose of mitochondrial F-ATP synthase is synthesis of ATP. To perform this function F-ATP synthase uses the energy of transmembrane electrochemical proton gradient (ΔµH+) formed by the electron transport chain (Fig. 2a). The F-ATP synthase nano-motor rotates in counter-clockwise direction (if viewed from the side of factor F1 at factor FO) (Fig. 2a). However, this process is reversible, and under certain conditions F-ATPase/synthase, rotating in the opposite direction, functions as ATPase and hydrolyses ATP. At the same time, the main, generally recognized role of V-ATPase is a reverse function – generation of transmembrane electrochemical gradient (ΔµH+) using the energy of ATP hydrolysis (Fig. 2b). Under physiological conditions its function is irreversible, and nano-motor of V-ATPase always rotates clockwise (if viewed from the side of factor V1 at factor VO) (Fig. 2b) and creates a proton gradient: (i) across the cytoplasmic membrane with acidification of extracellular medium or (ii) across the membranes of intracellular organelles of endocytic (vacuoles, endosomes, and lysosomes) and exocytic (Golgi complex and secretory vesicles) pathways of vesicular transport with acidification of their inner space. Transformation of the energy of ΔµH+ into ΔpH is due to operation of electrogenic nCl–/H+-ion exchangers, hence, hydrolysis of ATP by V-ATPase is accompanied by acidification and accumulation of HCl in the extracellular space or inside the organelles of eukaryotic cells (Fig. 2b).
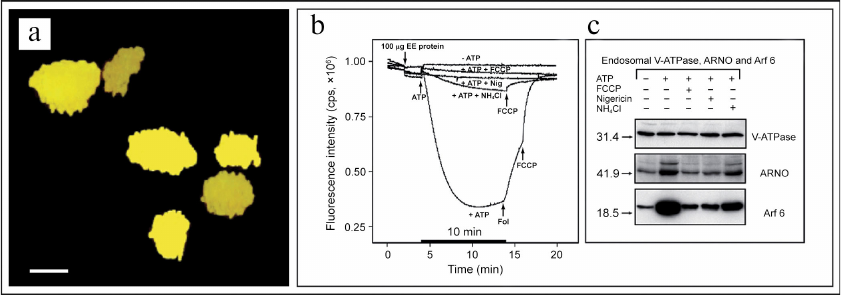
It must be noted in conclusion that up to the recent times the alternative role of V-ATPase in regulation of the signaling function of cytoplasmic small GTPases remained unknown. This important issue was addressed in the studies conducted in our laboratory in 2001-2006 [1-4, 132, 196-202]. In particular, it was shown that the V-ATPase-dependent acidification of the lumen of early endosomes (Fig. 6, a and b) resulted in recruiting of the Arf6 small GTPase (Arf6, ADP-ribosylation factor 6) and its activator cytohesin-2 (CTH2) (or ARNO, ADP-ribosylation nucleotide-side opener) (CTH2/ARNO) from cytosol to the outer side of the endosomal membrane (Fig. 6c) [132, 133].
Fig. 6. V-ATPase nano-motor is a novel pH-sensor and signaling bioenergetic receptor regulating function of small GTPases. a) Confocal microscopy image showing early endosomes with co-localization of V-ATPase and EEA1 marker; scale bar – 100 nm. b) V-ATPase-dependent acidification of the endosomal lumen is prevented by uncouplers FCCP, nigericin, and NH4Cl. c) Recruiting of Arf6 small GTPase and its activator CTH2/ARNO from cytosol to the outer side of endosomal membrane depends on acidification of endosomal lumen and is also prevented by uncouplers FCCP, nigericin, and NH4Cl. Figure is adapted from [132] with permission from the Publisher.
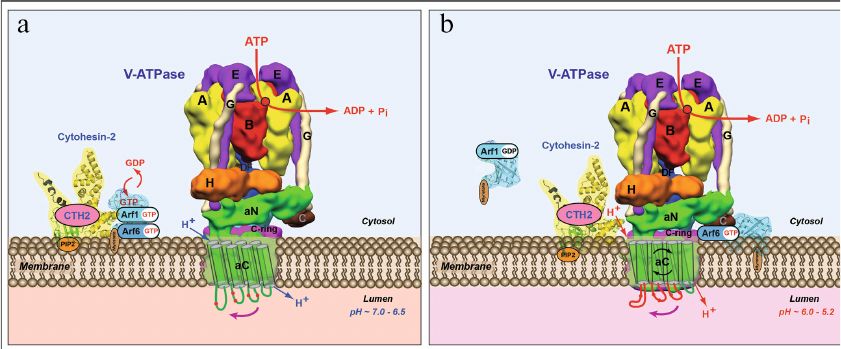
Further investigation of the mechanism of this process led to discovery of a new role of V-ATPase in 2003 (Fig. 7) [196-200]. In these studies, we have shown that in addition to its main role as nano-motor, V-ATPase also functions as a pH-sensor on the membrane of these organelles (Fig. 7, a and b) [198-200]. Moreover, it was demonstrated that V-ATPase directly interacts with Arf6 and CTH2/ARNO and regulates their enzymatic activity as well as modulates their signaling function in the cells [198-202]. In its turn, this biochemical signaling process between the endosomal V-ATPase and cytosolic small GTPases controls vesicular transport of the endosomal-lysosomal protein degradation pathway [198-200].
Fig. 7. Model of operation of V-ATPase as a novel pH-sensor and signaling bioenergetic receptor regulating function of CTH2/ARNO and Arf6 small GTPase. a) Recruiting of Arf6 small GTPase and its activator CTH2/ARNO from cytosol to the outer side of endosomal membrane depends on V-ATPase-mediated acidification of endosomal lumen. b) Transmembrane V-ATPase directly interacts with CTH2/ARNO and Arf6 small GTPase, regulates their function, and controls the process of vesicular transport in eukaryotic cells. Figure is adapted from [201] with permission from the Publisher.
Hence, it was demonstrated for the first time that in addition to its main function of a rotary ion-translocating nanomotor, V-ATPase performs two additional functions: (i) pH-sensing – measuring the level of acidification of extracellular medium and measuring pH of intracellular organelles; and (ii) signaling – transducing this information across membranes and modulation of activity of Arf family small GTPases [198-202]. These cornerstone studies resulted in the discovery and investigation of a novel role of V-ATPase nanomotor as a novel pH-sensor and signaling bioenergetic receptor. In this role the transmembrane V-ATPase regulates function of cytoplasmic small GTPases and controls an important physiological process of vesicular transport in eukaryotic cells (Fig. 7) [1-4, 132, 196-202]. In our experiments, it was also shown that the holocomplex V1VO of yeast V-ATPase is capable of interacting with the activator of human small GTPases CTH2/ARNO [3, 4, 201, 202]. These results indicate that the role of V-ATPase as a pH-sensor and signaling bioenergetic receptor could represent a fundamental feature of eukaryotic cells biology [3, 201, 202].
Ethics declarations. The author declares no conflicts of interest in financial or any other sphere. This article does not contain any studies with human participants or animals performed by the author.
REFERENCES
1.Marshansky, V. (2007) The V-ATPase a2-subunit as a
putative endosomal pH-sensor, Biochem. Soc. Trans., 35,
1092-1099, doi: 10.1042/BST0351092.
2.Marshansky, V., and Futai, M. (2008) The V-type
H(+)-ATPase in vesicular trafficking: targeting, regulation
and function, Curr. Opin. Cell Biol., 20, 415-426, doi:
10.1016/j.ceb.2008.03.015.
3.Marshansky, V., Rubinstein, J. L., and Grüber,
G. (2014) Eukaryotic V-ATPase: Novel structural findings and functional
insights, Biochim. Biophys. Acta Bioenerg., 1837,
857-879, doi: 10.1016/j.bbabio.2014.01.018.
4.Marshansky, V., Futai, M., and Grüber, G.
(2016) Eukaryotic V-ATPase and its super-complexes: from structure and
function to disease and drug targeting, in Advances in Biochemistry
in Health and Disease: Regulation of Ca2+-ATPases, V-ATPases
and F-ATPases (Chakraborti, S., and Dhalla, N. S., eds) Springer
International Publishing, Switzerland, pp. 301-335.
5.Muench, S. P., Trinick, J., and Harrison, M. A.
(2011) Structural divergence of the rotary ATPases, Q. Rev.
Biophys., 44, 311-356, doi: 10.1017/S0033583510000338.
6.Engelhardt, W. A (1930) Pyrophosphate metabolism in
avian erythorocytes, Klein. Wochenschr., 9,
1550-1554.
7.Engelhardt, W. A. (1930) Ortho- and pyrophosphate
in aerobic and anaerobic metabolism of erythrocytes, Biochem.
Zeitschrift, 227, 16-38.
8.Engelhardt, W. A. (1932) Correlation between
respiration and conversion of pyrophosphate in avian erythrocytes,
Biochem. Zeitschrift, 251, 343-368.
9.Engelhardt, W. A. (1982) Life and science, Annu.
Rev. Biochem., 51, 1-19, doi:
10.1146/annurev.bi.51.070182.000245.
10.Krogh, A. (1929) The progress of physiology,
Am. J. Physiol., 90, 243-251.
11.Engelhardt, W. A., and Lyubimova, M. N. (1939)
Myosin and adenosine triphosphatase, Nature, 144,
668-669.
12.Engelgardt, V. A. (1984) Knowledge of the
Phenomenon of Life [in Russian] (Baev, A. A., ed.) Akademia Nauk
SSSR, Moscow.
13.Belitser, V. A., and Tsybakova, E. T. (1939) On
mechanism of phosphorylation couples with respiration,
Biokhimiya, 4, 516-535.
14.Skulachev, V. P. (1972) Solution of the problem
of energy coupling in terms of chemiosmotic theory, J.
Bioenerg., 3, 25-38, doi: 10.1007/BF01515994.
15.Skulachev, V. P. (1974) Enzymic generators of
membrane potential in mitochondria, Ann. N. Y. Acad. Sci.,
227, 188-202, doi: 10.1111/j.1749-6632.1974.tb14384.x.
16.Skulachev, V. P. (1974) Mechanism of oxidative
phosphorylation and general principles of bioenergetics, Usp.
Sovrem. Biol., 77, 125-154.
17.Skulachev, V. P. (1977) Adenosine triphosphate
and transmembrane potential of hydrogen ions: Converting forms of
energy of live cells, Usp. Sovrem. Biol., 84,
165-175.
18.Skulachev, V. P. (1979) Membrane potential and
reconstitution, Methods Enzymol., 55, 586-603, doi:
10.1016/0076-6879(79)55068-x.
19.Skulachev, V. P. (1982) Rasskazy o
bioenergetike. Uchebnoye posobiye (Stories about bioenergetics.
Tutorial book), Moscow: Molodaya Gvardiya.
20.Skulachev, V. P. (1989) Bioenergetics.
Membrane Transformers of Energy [in Russian], Vysshaya Shkola,
Moscow.
21.Skulachev, V. P. (1988) Membrane
Bioenergetics, Springer Verlag, Berlin, New York, doi:
10.1007/978-3-642-72978-2.
22.Skulachev, V. P. (1994) Chemiosmotic concept of
the membrane bioenergetics: what is already clear and what is still
waiting for elucidation? J. Bioenerg. Biomembr., 26,
589-598, doi: 10.1007/BF00831533.
23.Skulachev, V. P., Bogachev, A. V., and
Kasparinskii, F. O. (2010) Membrane Bioenergetics.
Tutorial Book [in Russian], Izdatelstvo Moskovskogo
Universiteta, Moscow.
24.Mitchell, P. (1961) Coupling of phosphorylation
to electron and hydrogen transfer by a chemi-osmotic type of mechanism,
Nature, 91, 144-148, doi: 10.1038/191144a0.
25.Mitchell, P. (1966) Chemiosmotic coupling in
oxidative and photosynthetic phosphorylation, Biol. Rev. Camb.
Philos. Soc., 41, 445-502, doi:
10.1111/j.1469-185x.1966.tb01501.x.
26.Mitchell, P., and Moyle, J. (1967) Chemiosmotic
hypothesis of oxidative phosphorylation, Nature, 213, 137-139,
doi: 10.1038/213137a0.
27.Mitchell, P., and Moyle, J. (1971) Activation and
inhibition of mitochondrial adenosine triphosphatase by various anions
and other agents, J. Bioenerg., 2, 1-11, doi:
10.1007/BF01521319.
28.Mitchell, P. (1972) Chemiosmotic coupling in
energy transduction: a logical development of biochemical knowledge,
J. Bioenerg., 3, 5-24, doi: 10.1007/BF01515993.
29.Mitchell, P. (1976) Vectorial chemistry and the
molecular mechanics of chemiosmotic coupling: power transmission by
proticity, Biochem. Soc. Trans., 4, 399-430, doi:
10.1042/bst0040399.
30.Skulachev, V. P., Sharaf, A. A., and Liberman, E.
A. (1967) Proton conductors in the respiratory chain and artificial
membranes, Nature, 216, 718-719, doi:
10.1038/216718a0.
31.Skulachev, V. P., Sharaf, A. A., Yagujzinsky, L.
S., Jasaitis, A. A., Liberman, E. A., et al. (1968) The effect of
uncouplers on mitochondria, respiratory enzyme complexes and artificial
phospholipid membranes, Curr. Mod. Biol., 2, 98-105, doi:
10.1016/0303-2647(68)90014-2.
32.Liberman, E. A., Topaly, V. P., Tsofina, L. M.,
Yasaitis, A. A., and Skulachev, V. P. (1969) Ion transport and
electrical potential of mitochondrial membranes, Biokhimiya,
34, 1083-1087.
33.Liberman, E. A., Topaly, V. P., Tsofina, L. M.,
Jasaitis, A. A., and Skulachev, V. P. (1969) Mechanism of coupling of
oxidative phosphorylation and the membrane potential of mitochondria,
Nature, 222, 1076-1078, doi: 10.1038/2221076a0.
34.Drachev, L. A., Jasaitis, A. A., Kaulen, A. D.,
Kondrashin, A. A., Liberman, E. A., et al. (1974) Direct measurement of
electric current generation by cytochrome oxidase, H+-ATPase
and bacteriorhodopsin, Nature, 249, 321-324, doi:
10.1038/249321a0.
35.Drachev, L. A., Jasaitis, A. A., Mikelsaar, H,
Nemecek, I. B., Semenov, A. Y., et al. (1976) Reconstitution of
biological molecular generators of electric current
H+-ATPase, J. Biol. Chem., 251, 7077-7082,
doi: 10.1016/S0021-9258(17)32943-5.
36.Skulachev, V. P. (1999) Phenoptosis: programmed
death of an organism, Biochemistry (Moscow), 64,
1418-1426.
37.Skulachev, M. V., and Skulachev, V. P. (2015)
Phenoptosis-programmed death of an organism, in Apoptosis and
Beyong (Radosevich, J., ed.) Springer-Verlag, Berlin,
Heidelberg.
38.Skulachev, M. V., and Skulachev, V. P. (2017)
Programmed aging of mammals: proof of concept and prospects of
biochemical approaches for anti-aging therapy, Biochemistry
(Moscow), 82, 1403-1422, doi: 10.1134/S000629791712001X.
39.Vyssokikh, M. Y., Holtze, S., Averina, O. A.,
Lyamzaev, K. G., Panteleeva, A. A., et al. (2020) Mild depolarization
of the inner mitochondrial membrane is a crusial component of an
anti-aging program, Proc. Natl. Acad. Sci. USA, 117,
6491-6501, doi: 10.1073/pnas.1916414117.
40.Skulachev, M., Antonenko, Y. N., Anisimov, V. N.,
Chernyak, B. V., Cherepanov, D. A., et al. (2011)
Mitochondrial-targeted plastoquinone derivatives. Effect on senescence
and acute age-related pathologies, Curr. Drug Targets,
12, 800-826, doi: 10.2174/138945011795528859.
41.Skulachev, V. P. (2012) Mitochondria-targeted
antioxidants as promising drugs for treatment of age-related brain
diseases, J. Alzheimer’s Dis., 28, 283-289, doi:
10.3233/JAD-2011-111391.
42.Plotnikov, E. Y., Morosanova, M. A., Pevzner, I.
B., Zorove, L. D., Manskikh, V. N., et al. (2013) Protective effect of
mitochondria-targeted antioxidants in an acute bacterial infection,
Proc. Natl. Acad. Sci. USA, 110, E3100-E3108, doi:
10.1073/pnas.1307096110.
43.Skulachev, V. P., Skulachev, M, V., and Fenyuk,
B. A. (2016) Life without Aging: Popular Illustrated Publication
[in Russian], Eksmo, Moscow.
44.Boyer, P. D. (1997) The ATP synthase – a
splendid molecular machine, Annu. Rev. Biochem., 66,
717-749, doi: 10.1146/annurev.biochem.66.1.717.
45.Boyer, P. D. (1998) ATP synthase-past and future,
Biochim. Biophys. Acta, 1365, 3-9, doi:
10.1016/s0005-2728(98)00066-8.
46.Pullman, M. E., Penefsky, H. S., Datta, A., and
Racker, E. (1960) Partial resolution of the enzymes catalyzing
oxidative phosphorylation. I. Purification and properties of soluble
dinitrophenol-stimulated adenosine triphosphatase, J. Biol.
Chem., 235, 3322-3329.
47.Penefsky, H. S., Pullman, M. E., Datta, A., and
Racker, E. (1960) Partial resolution of the enzymes catalyzing
oxidative phosphorylation. II. Participation of a soluble adenosine
triphosphatase in oxidative phosphorylation, J. Biol. Chem.,
235, 3330-3336.
48.Racker, E. (1962) Studies of factors involved in
oxidative phosphorylation, Proc. Natl. Acad. Sci. USA,
48, 1659-1663, doi: 10.1073/pnas.48.9.1659.
49.Racker, E. (1964) A reconstituted system of
oxidative phosphorylation, Biochem. Biophys. Res. Commun.,
14, 75-78, doi: 10.1016/0006-291x(63)90214-6.
50.Racker, E. (1977) Mechanisms of energy
transformations, Annu. Rev. Biochem., 46, 1006-1014, doi:
10.1146/annurev.bi.46.070177.005042.
51.Racker, E., Violand, B., O’Neal, S.,
Alfonzo, M., and Telford, J. (1979) Reconstitution, a way of
biochemical research; some new approaches to membrane-bound enzymes,
Arch. Biochem. Biophys., 198, 470-477, doi:
10.1016/0003-9861(79)90521-6.
52.Fernandez-Moran, H. (1963) Subunit organization
of mitochondrial membranes, Science, 140, 381, doi:
10.1126/science.140.3565.381.
53.Fernandez-Moran, H., Oda, T., Blair, P. V., and
Green, D. E. (1964) A macromolecular repeating unit of mitochondrial
structure and function. Correlated electron microscopic and biochemical
studies of isolated mitochondria and submitochondrial particles of beef
heart muscle, J. Cell Biol., 22, 63-100, doi:
10.1083/jcb.22.1.63.
54.Webster, G., and Green, D. E. (1964) The enzymes
and the enzyme-catalyzed reactions of mitochondrial oxidative
phosphorylation, Proc. Natl. Acad. Sci. USA, 52,
1170-1176, doi: 10.1073/pnas.52.5.1170.
55.Chance, B., and Parsons, D. F. (1963) Cytochrome
function in relation to inner membrane structure of mitochondria,
Science, 142, 1176-1180, doi: 10.1126/science.142.3596.1176.
56.Stasny, J. T., and Crane, F. L. (1964) Separation
of elementary particles from mitochondrial cristae, Exp. Cell
Res., 34, 423-426, doi: 10.1016/0014-4827(64)90382-9.
57.Stasny, J. T., and Crane, F. L. (1964) The effect
of sonic oscillation on the structure and function of beef heart
mitochondria, J. Cell. Biol., 22, 49-62, doi:
10.1083/jcb.22.1.49.
58.Moyle, J., and Mitchell, P. (1975)
Active/inactive state transitions of mitochondrial ATPase molecules
influenced by Mg+, anions and aurovertin, FEBS Lett.,
56, 55-61, doi: 10.1016/0014-5793(75)80110-4.
59.Recktenwald, D, and Hess, B. (1977) A slow
conformation change in the transient state kinetics of soluble ATPase
of yeast mitochondria, FEBS Lett., 80, 187-189, doi:
10.1016/0014-5793(77)80436-5.
60.Vinogradov, A. D. (1984) Catalytic properties of
mitochondrial ATP-synthase, Biokhimiya, 49,
1220-1238.
61.Vinogradov, A. D. (1999) Mitochondrial
ATP-synthase: fifteen years later, Biochemistry (Moscow),
64, 1219-1229.
62.Vinogradov, A. D. (2000) Steady-state and
pre-steady-state kinetics of the mitochondrial FoF1 ATPase: is
ATP synthase a reversible molecular machine? J. Exp. Biol.,
203, 41-49, doi: 10.1242/jeb.203.1.41.
63.Vinogradov A.D. (2019) New Perspective on the
reversibility of ATP synthesis and hydrolysis by
FOF1-ATP synthase (hydrolase), Biochemistry
(Moscow), 84, 1247-1255, doi: 10.1134/s0006297919110038.
64.Akimenko, V. K., Minkov, I. B., and Vinogradov,
A. D. (1971) On effects of uncouplers on the soluble ATPase of
mitochondria, Biokhimiya, 36, 655-658.
65.Akimenko, V. K., Minkov, I. B., Bakeeva, L. E.,
and Vinogradov, A. D. (1972) Isolation and properties of soluble ATPase
from bovine heart mitochondria, Biokhimiya, 37,
348-359.
66.Fitin, A. F., Vasilyeva, E. A., and Vinogradov,
A. D. (1979) An inhibitory high affinity binding site for ADP in the
oligomycin-sensitive ATPase of beef heart submitohondrial particles,
Biochem. Biophys. Res. Commun., 86, 434-439, doi:
10.1016/0006-291x(79)90884-2.
67.Minkov, I. B., Fitin, A. F., Vasilyeva, E. A.,
and Vinogradov, A. D. (1979) Mg2+-induced ADP-dependent
inhibition of the ATPase activity of beef heart mitochondrial coupling
factor F1, Biochem. Biophys. Res. Commun., 89, 1300-1306,
doi: 10.1016/0006-291x(79)92150-8.
68.Vasilyeva, E. A., Fitin, A. F., Minkov, I. B.,
and Vinogradov, A. D. (1980) Kinetics of interaction of adenosine
diphosphate and adenosine triphosphate with adenosine triphosphatase of
bovine heart submitochondrial particles, Biochem. J.,
188, 807-815, doi: 10.1042/bj1880807.
69.Vasilyeva, E. A., Minkov, I. B., Fitin, A. F.,
and Vinogradov, A. D. (1982) Kinetic mechanism of mitochondrial
adenosine triphosphatase. ADP-specific inhibition as revealed by the
steady-state kinetics, Biochem. J., 202, 9-14, doi:
10.1042/bj2020009.
70.Bulygin, V. V., Syroeshkin, A. V., and
Vinogradov, A. D. (1993) Nucleotide/H(+)-dependent change in
Mg2+ affinity at the inhibitory site of the mitochondrial
F1-Fo ATP synthase, FEBS Lett.,
328, 193-196, doi: 10.1016/0014-5793(93)80991-3.
71.Vasilyeva, E. A., Minkov, I. B., Fitin, A. F.,
and Vinogradov, A. D. (1982) Kinetic mechanism of mitochondrial
adenosine triphosphatase. Inhibition by azide and activation by
sulphite, Biochem. J., 202, 15-23, doi:
10.1042/bj2020015.
72.Galkin, M. A., and Vinogradov, A. D. (1999)
Energy-dependent transformation of the catalytic activities of the
mitochondrial FoF1-ATP synthase, FEBS
Lett., 448, 123-126, doi: 10.1016/s0014-5793(99)00347-6.
73.Minkov, I. B., and Vinogradov, A. D. (1973)
Isolation and properties of a mitochondrial protein inhibiting
adenosine triphosphatase, Biokhimiya, 38, 542-547.
74.Panchenko, M. V., and Vinogradov, A. D. (1989)
Kinetics of interaction of submitochondrial fragments of ATPase with
protein inhibitor, Biokhimiya, 54, 569-579.
75.Panchenko, M. V., and Vinogradov, A. D. (1985)
Interaction between the mitochondrial ATP synthetase and ATPase
inhibitor protein. Active/inactive slow pH-dependent transitions of the
inhibitor protein, FEBS Lett., 184, 226-230, doi:
10.1016/0014-5793(85)80611-6.
76.Yokoyama, K., Muneyuki, E., Amano, T., Mizutani,
S., Yoshida, M., et al. (1998) V-ATPase of Thermus thermophilus
is inactivated during ATP hydrolysis but can synthesize ATP, J.
Biol. Chem., 273, 20504-20510, doi:
10.1074/jbc.273.32.20504.
77.Dempsey, M. E., and Boyer, P. D. (1961) Catalysis
of an inorganic phosphate-H2O18 exchange by
actomysin and myosin, J. Biol. Chem., 236, PC6-PC7.
78.Repke, K. R., and Schön, R. (1974) Flip-flop
model of energy interconversion by ATP synthase, Acta Biol. Med.
Ger., 33, K27-K38.
79.Boyer, P. D. (1975) Energy transduction and
proton translocation by adenosine triphosphatases, FEBS Lett.,
5, 91-94, doi: 10.1016/0014-5793(75)80464-9.
80.Smith, D. J., and Boyer, P. D. (1976)
Demonstration of a transitory tight binding of ATP and of committed
P(i) and ADP during ATP synthesis by chloroplasts, Proc. Natl. Acad.
Sci. USA, 73, 4314-4318, doi: 10.1073/pnas.73.12.4314.
81.Rosing, J., Kayalar, C., and Boyer, P. D. (1977)
Evidence for energy-dependent change in phosphate binding for
mitochondrial oxidative phosphorylation based on measurements of medium
and intermediate phosphate-water exchanges, J. Biol. Chem.,
252, 2478-2485.
82.Kayalar, C., Rosing, J., and Boyer, P. D. (1977)
An alternating site sequence for oxidative phosphorylation suggested by
measurement of substrate binding patterns and exchange reaction
inhibitions, J. Biol. Chem., 252, 2486-2491, doi:
10.1016/S0021-9258(17)40484-4.
83.Boyer, P.D. (1993) The binding change mechanism
for ATP synthase-some probabilities and possibilities, Biochim.
Biophys. Acta, 1140, 215-250, doi:
10.1016/0005-2728(93)90063-l.
84.Boyer, P. D. (2000) Catalytic site forms and
controls in ATP synthase catalysis, Biochim. Biophys. Acta,
1458, 252-262, doi: 10.1016/s0005-2728(00)00077-3.
85.Boyer, P. D. (2002) Catalytic site occupancy
during ATP synthase catalysis, FEBS Lett., 512, 29-32,
doi: 10.1016/s0014-5793(02)02293-7.
86.Lutter, R., Abrahams, J. P., van Raaij, M. J.,
Todd, R. J., Lundqvist, T., et al. (1993) Crystallization of
F1-ATPase from bovine heart mitochondria, J. Mol.
Biol., 229, 787-790, doi: 10.1006/jmbi.1993.1081.
87.Abrahams, J. P., Leslie, A. G., Lutter, R., and
Walker, J. E. (1994) Structure at 2.8 Å resolution of
F1-ATPase from bovine heart mitochondria, Nature,
370, 621-628, doi: 10.1038/370621a0.
88.Boyer, P. D., Chance, B., Ernster, L., Mitchell,
P., Racker, E., et al. (1977) Oxidative phosphorylation and
photophosphorylation, Annu. Rev. Biochem., 46, 955-1026,
doi: 10.1146/annurev.bi.46.070177.004515.
89.Stock, D., Leslie, A. G., and Walker, J. E.
(1999) Molecular architecture of the rotary motor in ATP synthase,
Science, 286, 1700-1705, doi:
10.1126/science.286.5445.1700.
90.Walker, J. E. (2013) The ATP synthase: the
understood, the uncertain and the unknown, Biochem. Soc. Trans.,
41, 1-16, doi: 10.1042/BST20110773.
91.Guo, H., Bueler, S. A., and Rubinstein, J. L.
(2017) Atomic model for the dimeric FO region of
mitochondrial ATP synthase, Science, 358, 936-940, doi:
10.1126/science.aao4815.
92.Guo, H., Suzuki, T., and Rubinstein, J. L. (2019)
Structure of a bacterial ATP synthase, eLife, 8, e43128,
doi: 10.7554/eLife.43128.
93.Murphy, B. J., Klusch, N., Langer, J., Mills, D.
J., Yildiz, Ö., et al. (2019) Rotary substrates of mitochondrial
ATP synthase reveal the basis of flexible F1-FO
coupling, Science, 364, eaaw9128, doi:
10.1126/science.aaw9128.
94.Kühlbrandt, W. (2019) Structure and
mechanisms of F-type ATP synthases, Annu. Rev. Biochem.,
88, 515-549, doi:
doi.org/10.1146/annurev-biochem-013118-110903.
95.Wagner, C. A., Finberg, K. E., Breton, S.,
Marshansky, V., Brown, D., et al. (2004) Renal vacuolar
H+-ATPase, Physiol. Rev., 84, 1263-1314, doi:
10.1152/physrev.00045.2003.
96.Brown, D., and Marshansky, V. (2004) The renal
V-ATPase: physiology and pathophysiology, in Handbook of ATPases:
Biochemistry, Cell Biology, Pathophysiology (Futai, M., Wada, Y.,
and Kaplan, J., eds) Wiley-VCH, pp. 413-442.
97.Beyenbach, K. W., and Wieczorek, H. (2006) The
V-type H+-ATPase: molecular structure and function,
physiological roles and regulation, J. Exp. Biol., 209,
577-589, doi: 10.1242/jeb.02014.
98.Forgac, M. (2007) Vacuolar ATPases: rotary proton
pumps in physiology and pathophysiology, Nat. Rev. Mol. Cell
Biol., 8, 917-929, doi: 10.1038/nrm2272.
99.Kirshner, N. (1962) Uptake of catecholamines by a
particular fraction of the adrenal medulla, J. Biol. Chem.,
237, 2311-2317.
100.Kirshner, N., Kirshner, A. G., and Kamin, D. L.
(1966) Adenosine triphosphotase activity of adrenal medulla
catecholamine granules, Biochim. Biophys. Acta, 113,
332-335, doi: 10.1016/s0926-6593(66)80072-3.
101.Bashford, C. L., Casey, R. P., Radda, G. K.,
and Ritchie, G. A. (1975) The effect of uncouplers on catecholamine
incorporation by vesicles of chromaffin granules, Biochem. J.,
148, 153-155, doi: 10.1042/bj1480153.
102.Kanner, B. I., and Schuldner, S. (1987)
Mechanism of transport and storage of neurotransmitters, CRC Crit.
Rev. Biochem., 22, 1-38, doi: 10.3109/10409238709082546.
103.Kakinuma, Y., Ohsumi, Y., and Anraku, Y. (1981)
Properties of H+-translocating adenosine triphosphatase in
vacuolar membranes of Saccharomyces cerevisiae, J. Biol.
Chem., 256, 10859-10863.
104.Bowman, E. J., and Bowman, B. J. (1982)
Identification and properties of an ATPase in vacuolar membranes of
Neurospora crassa, J. Bacteriol., 151,
1326–1337, doi: 10.1128/jb.151.3.1326-1337.1982.
105.Forgac, M., Cantley, L., Wiedenmann, B.,
Altstiel, L., and Branton, D. (1983) Clathrin-coated vesicles contain
an ATP-dependent proton pump, Proc. Natl. Acad. Sci. USA,
80, 1300-1303, doi: 10.1073/pnas.80.5.1300.
106.Harikumar, P., and Reeves, J. P. (1983) The
lysosomal proton pump is electrogenic, J. Biol. Chem.,
258, 10403-10410.
107.Nelson, N., and Harvey, W. R. (1999) Vacuolar
and plasma membrane proton adenosinetriphosphatases, Physiol.
Rev., 79, 361-385, doi: 10.1152/physrev.1999.79.2.361.
108.Anderson, E., and Harvey, W. R. (1966) Active
transport by the cecropia midgut. II. Fine structure of the midgut
epithelium, J. Cell Biol., 31, 107-134, doi:
10.1083/jcb.31.1.107.
109.Berridge, M. J., and Gupta, B. L. (1968)
Fine-structural localization of adenosine triphosphatase in the rectum
of Caliphora, J. Cell Sci., 3, 17-31, doi:
10.1242/jcs.3.1.17.
110.Brown, D., and Montesano, R. (1980) Membrane
specialization in the rat epididymis. I. Rod-shaped intramembrane
particles in the apical (mitochondria-rich) cell, J. Cell. Sci.,
45, 187-198, doi: 10.1242/jcs.45.1.187.
111.Brown, D., and Montesano, R. (1981) Membrane
specialization in the rat epididymis. II. The clear cell, Anat.
Rec., 201, 477-483, doi: 10.1002/ar.1092010305.
112.Brown, D., and Orci, L. (1986) The
“coat” of kidney intercalated cell tubulovesicles does not
contain clathrin, Am. J. Physiol. Cell. Physiol., 250,
C605-C608, doi: 10.1152/ajpcell.1986.250.4.C605.
113.Brown, D., Weyer, P., and Orci, L. (1987)
Nonclathrin-coated vesicles are involved in endocytosis in kidney
collecting dusct intercalated cells, Anat. Rec., 218,
237-242, doi: 10.1002/ar.1092180303.
114.Brown, D., Gluck, S., and Hartwig, J. (1987)
Structure of the novel membrane-coating material in proton-secreting
epithelial cells and identification as an H+ ATPase, J.
Cell Biol., 105, 1637-1648, doi: 10.1083/jcb.105.4.1637.
115.Brown, D., Hirsch, S., and Gluck, S. (1988) An
H+-ATPase in opposite plasma membrane domains in kidney
epithelial cell subpopulations, Nature, 331, 622-624,
doi: 10.1038/331622a0.
116.Brown, D., Hirsch, S., and Gluck, S. (1988)
Localization of a proton-pumping ATPase in rat kidney, J. Clin.
Invest., 82, 2114-2126, doi: 10.1172/JCI113833.
117.Breton, S., Smith, P., Lui, B., and Brown, D.
(1996) Acidification of male reproductive tract by a proton pumping
H+ ATPase, Nat. Med., 2, 470-472, doi:
10.1038/nm0496-470.
118.Brown, D., and Breton, S. (1996)
Mitochondria-rich, proton-secreting epithelial cells, J. Exp.
Biol., 199, 2345-2358, doi: 10.1242/jeb.199.11.2345.
119.Brown, D., and Breton, S. (2000) H+
V-ATPase-dependent luminal acidification in the kidney collecting duct
and the epididymis/vas deferens: vesicle recycling and transcytotic
pathways, J. Exp. Biol., 203, 137-145, doi:
10.1242/jeb.203.1.137.
120.Brown, D., and Breton, S. (2007) New insights
into the regulation of V-ATPase-dependent proton secretion, Am. J.
Physiol. Renal Physiol., 292, F1-F10, doi:
10.1152/ajprenal.00340.2006.
121.Brown, D., Paunescu, T. G., Breton, S., and
Marshansky, V. (2009) Regulation of the V-ATPase in kidney epithelial
cells: dual role in acid-base homeostasis and vesicle trafficking,
J. Exp. Biol., 212, 1762-1772, doi:
10.1242/jeb.028803.
122.Brown, D., Breton, S., Ausiello, D. A., and
Marshansky, V. (2009) Sensing, signaling and sorting events in kidney
epithelial cell physiology, Traffic, 10, 275-284, doi:
10.1111/j.1600-0854.2008.00867.x.
123.Eaton, A.F., Merkulova, M., and Brown, D.
(2021) The H+-ATPase (V-ATPase): from proton pump to
signaling complex in health and disease, Am. J. Physiol. Cell
Physiol., 320, C392-C414.
doi:10.1152/ajpcell.00442.2020.
124.Harvey, W. R., Cioffi, M., and Wolfersberger,
M. G. (1981) Portasomes as coupling factors in active transport and
oxidative phosphorylation, Am. Zool., 21, 775-791.
125.Harvey, W. R., Cioffi, M., and Wolfersberger,
M. G. (1983) Chemiosmotic potassium ion pump of insect epithelia,
Am. J. Physiol., 244, R163-R175, doi:
10.1152/ajpregu.1983.244.2.R163.
126.Harvey, W. R., Cioffi, M., Dow, J. A. T., and
Wolfersberger, M. G. (1983) Potassium ion transport ATPase in insect
epithelia, J. Exp. Biol., 106, 91-117, doi:
10.1242/jeb.106.1.91.
127.Schweikl, H., Klein, U., Schindlbeck, M., and
Wieczorek, H. (1989) A vacuolar-type ATPase, partially purified from
potassium transporting plasma membranes of tobacco hornworm midgut,
J. Biol. Chem., 264, 11136-11142.
128.Harvey W. R., and Wieczorek, H. (1997) Animal
plasma membrane energization by chemiosmotic H+ V-ATPases,
J. Exp. Biol., 200, 203-216, doi:
10.1242/jeb.200.2.203.
129.Marshansky, V., Fleser, A., Noël, J.,
Bourgoin, S., and Vinay, P. (1996) Isolation of heavy endosomes from
proximal dog proximal tubules in suspension, J. Membr. Biol.,
153, 59-73, doi: 10.1007/s002329900110.
130.Marshansky, V., and Vinay, P. (1996) Proton
gradient formation in early endosomes from proximal tubules,
Biochim. Biophys. Acta Biomembr., 1284, 171-180, doi:
10.1016/s0005-2736(96)00123-x.
131.Marshansky, V., Bourgoin, S., Londoño,
I., Bendayan, M., Maranda, B., et al. (1997) Receptor-mediated
endocytosis in kidney proximal tubules: recent advances and hypothesis,
Electrophoresis, 18, 2661-2676, doi:
10.1002/elps.1150181423.
132.Maranda, B., Brown, D., Bourgoin, S., Casanova,
J. E., Vinay, P., et al. (2001) Intra-endosomal pH-sensitive
recruitment of the Arf-nucleotide exchange factor ARNO and Arf6 from
cytoplasm to proximal tubule endosomes, J. Biol. Chem.,
276, 18540-18550, doi: 10.1074/jbc.M011577200.
133.Marshansky, V., Ausiello, D. A., and Brown, D.
(2002) Physiological importance endosomal acidification: potential role
in proximal tubulopathies, Curr. Opin. Nephrol. Hypertens.,
11, 527-537, doi: 10.1097/00041552-200209000-00009.
134.Skulachev, V. P. (1984) Sodium bioenergetics,
Trends Biochem. Sci., 9, 483-485, doi:
10.1016/0968-0004(84)90317-7.
135.Skulachev, V. P. (1985) Membrane linked
energy transductions. Bioenergetic functions of sodium: H+
is not unique as a coupling ion, Eur. J. Biochem., 151,
199-208, doi: 10.1111/j.1432-1033.1985.tb09088.x.
136.Unemoto, T., Hayashi, M., and Hayashi, M.
(1977) Na+-dependent activation of NADH oxidase in membrane
fractions from halophilic Vibrio alginolyticus and V.
costicolus, J. Biochem., 82, 1389-1395, doi:
10.1093/oxfordjournals.jbchem.a131826.
137.Unemoto, T., and Hayashi, M. (1979) NADH:
quinone oxidoreductase as a site of Na+-dependent activation
in the respiratory chain of marine Vibrio alginolyticus, J.
Biochem., 85, 1461-1467, doi:
10.1093/oxfordjournals.jbchem.a132474.
138.Tokuda, H., and Unemoto, T. (1982)
Characterization of the respiration-dependent Na+ pump in
the marine bacterium Vibrio alginolyticus, J. Biol.
Chem., 257, 10007-10014.
139.Tokuda, H. (1984) Solubilization and
reconstitution of the Na+-motive NADH oxidase activity from
the marine bacterium Vibrio alginolyticus, FEBS Lett.,
176, 125-128, doi: 10.1016/0014-5793(84)80925-4.
140.Dibrov, P. A., Kostryko, V. A., Lazarova, R.
L., Skulachev, V. P., and Smirnova, I. A. (1986) The sodium cycle. I.
Na+-dependent motility and modes of membrane energization in
the marine alkalotolerant Vibrio alginolyticus, Biochim.
Biophys. Acta, 850, 449-457, doi:
10.1016/0005-2728(86)90113-1.
141.Dibrov, P. A., Lazarova, R. L., Skulachev, V.
P., and Verkhovskaya, M. L. (1986) The sodium cycle. II. Na+
coupled oxidative phosphorylation in Vibrio alginolyticus cells,
Biochim. Biophys. Acta, 850, 458-465, doi:
10.1016/0005-2728(86)90114-3.
142.Bakeeva, L. E., Chumakov, K. M., Drachev, A.
L., Meltina, A. L., and Skulachev, V. P. (1986) The sodium cycle. III.
Vibrio alginolyticus resembles Vibrio cholerae and some
other vibriones by flagellar motor and ribosomal 5S-RNA structures,
Biochim. Biophys. Acta, 850, 466-472, doi:
10.1016/0005-2728(86)90115-5.
143.Dimroth, P. (1980) A new sodium-transport
system energized by the decarboxylation of oxaloacetate, FEBS
Lett., 122, 234-236, doi: 10.1016/0014-5793(80)80446-7.
144.Hilpert, W., and Dimroth, P. (1982) Conversion
of the chemical energy of methylmalonyl-CoA decarboxylation into a
Na+ gradient, Nature, 296, 584-585, doi:
10.1038/296584a0.
145.Hilpert, W., Schink, B., and Dimroth, P. (1984)
Life by a new decarboxylation-dependent energy conservation mechanism
with Na as coupling ion, EMBO J., 3, 1665-1670, doi:
10.1002/j.1460-2075.1984.tb02030.x.
146.Skulachev, V. P. (1991) Chemiosmotic systems in
bioenergetics: H+-cycles and Na+-cycles,
Biosci. Rep., 11, 387-441, doi: 10.1007/BF01130214.
147.Skulachev, V. P. (1989) The sodium cycle: a
novel type of bacterial energetics, J. Bioenerg. Biomemb.,
21, 635-647, doi: 10.1007/BF00762683.
148.Skulachev, V. P. (1989) Bacterial
Na+ energetics, FEBS Lett., 250, 106-114, doi:
10.1016/0014-5793(89)80693-3.
149.Mulkidjanian, A. Y., Makarova, K. S., Galperin,
M. Y., and Koonin, E. V. (2007) Inventing the dynamo machine: the
evolution of the F-type and V-type ATPases, Nat. Rev.Microbiol.,
5, 892-899, doi: 10.1038/nrmicro1767.
150.Mulkidjanian, A. Y., Galperin, M. Y., Makarova,
K. S., Wolf, Y. I., and Koonin, E. V. (2008) Evolutionary primacy of
sodium bioenergetics, Biol. Direct, 3, 13. doi:
10.1186/1745-6150-3-13.
151.Mulkidjanian, A. Y., Dibrov, P., and Galperin,
M. Y. (2008) The past and present of sodium energetics: may the
sodium-motive force be with you, Biochim. Biophys. Acta,
1777, 985-992, doi: 10.1016/j.bbabio.2008.04.028.
152.Mulkidjanian, A. Y., Galperin, M. Y., and
Koonin, E. V. (2009) Co-evolution of primordial membranes and membrane
proteins, Trends Biochem. Sci., 34, 206-215, doi:
10.1016/j.tibs.2009.01.005.
153.Mulkidjanian, A. Y., Bychkov, A. Y., Dibrova,
D. V., Galperin, M. Y., and Koonin, E. V. (2012) Origin of first cells
at terrestrial, anoxic geothermal fields, Proc. Natl. Acad. Sci.
USA, 109, E821-E830, doi: 10.1073/pnas.1117774109.
154.Kozlova, M. I., Bushmakin, I. M., Belyaeva, J.
D., Shalaeva D. N., Didrova, D. V., et al. (2020) Expansion of the
“sodium world” through evolutionary time and taxonomic
space, Biochemistry (Moscow), 85, 1518-1542, doi:
10.1134/S0006297920120056.
155.Gogarten, J. P., Kibak, H., Dittrich, P., Taiz,
L., Bowman, E. J., et al. (1989) Evolution of the vacuolar
H+-ATPase: implications for the origin of eukaryotes,
Proc. Natl. Acad. Sci. USA, 86, 6661-6665, doi:
10.1073/pnas.86.17.6661.
156.Weiss, M. C., Sousa, F. L., Mrnjavac, N.,
Neukirchen, S., Roettger, M., et al. (2016) The physiology and habitat
of the last universal common ancestor, Nat. Microbiol.,
1, 16116, doi: 10.1038/nmicrobiol.2016.116.
157.McInerney, J. O. (2016) Evolution: A four
billion year old metabolism, Nat. Microbiol., 1, 16139,
doi: 10.1038/nmicrobiol.2016.139.
158.Weiss, M. C., Preiner, M., Xavier, J. C.,
Zimorski, V., and Martin, W. F. (2018) The last universal common
ancestor between ancient Earth chemistry and the onset of genetics,
PLoS Genet., 14, e1007518, doi:
10.1371/journal.pgen.1007518.
159.Kagawa, Y., and Racker, E. (1971) Partial
resolution of the enzymes catalyzing oxidative phosphorylation. XXV.
Reconstitution of vesicles catalyzing
32Pi-adenosine triphosphate exchange, J. Biol.
Chem., 246, 5477-5487, doi:
10.1016/S0021-9258(18)61930-1.
160.Racker, E., and Stoeckenius, W. (1974)
Reconstitution of purple membrane vesicles catalyzing light-driven
proton uptake and adenosine triphosphate formation, J. Biol.
Chem., 249, 662-663, doi: 10.1016/S0021-9258(19)43080-9.
161.Mitchell, P. (2011) Chemiosmotic coupling in
oxidative and photosynthetic phosphorylation. 1966, Biochim.
Biophys. Acta, 1807, 1507-1538, doi:
10.1016/j.bbabio.2011.09.018.
162.Glagolev, A. N., and Skulachev, V. P. (1978)
The proton pump is a molecular engine of motile bacteria,
Nature, 272, 280-282, doi: 10.1038/272280a0.
163.Junge, W. (1999) ATP synthase and other motor
proteins, Proc. Natl. Acad. Sci. USA, 96, 4735-4737, doi:
10.1073/pnas.96.9.4735.
164.Dimroth, P., Wang, H., Grabe, M., and Oster, G.
(1999) Energy transduction in the sodium F-ATPase of Propionigenium
modestum, Proc. Natl. Acad. Sci. USA, 96,
4924-4929, doi: 10.1073/pnas.96.9.4924.
165.Murata, T., Yamato, I., Kakinuma, Y., Leslie,
A. G., and Walker, J. E. (2005) Structure of the rotor of the V-type
Na+-ATPase from Enterococcus hirae, Science,
308, 654-659, doi: 10.1126/science.1110064.
166.Meier, T., Polzer, P., Diederichs, K., Welte,
W., and Dimroth, P. (2005) Structure of the rotor ring of the F-type
Na+-ATPase from Llyobacter tartaricus,
Science, 308, 659-662, doi: 10.1126/science.1111199.
167.Wilkens, S. (2005) Rotary molecular motors,
Adv. Protein Chem., 71, 345-382, doi:
10.1016/S0065-3233(04)71009-8.
168.Kühlbrandt, W. (2014) The resolution
revolution, Science, 343, 1443-1444, doi:
10.1126/science.1251652.
169.Smith, M. T., and Rubinstein, J. L. (2014)
Structural biology. Beyond blob-ology, Science, 345,
617-619, doi: 10.1126/science.1256358.
170.Guo, H., and Rubinstein, J. R. (2018) Cryo-EM
of ATP synthases, Curr. Opin. Struct. Biol., 52, 71-79,
doi: 10.1016/j.sbi.2018.08.005.
171.Abbas, Y. M., Wu, D., Bueler, S. A., Robinson,
C. V., and Rubinstein, J. L. (2020) Structure of V-ATPase from the
mammalian brain, Science, 367, 1240-1246, doi:
10.1126/science.aaz2924.
172.Vasanthakumar, T., and Rubinstein, J. L. (2020)
Structure and roles of V-type ATPases, Trends. Biochem. Sci.,
45, 295-307, doi: 10.1016/j.tibs.2019.12.007.
173.Oot, R. A., Couoh-Cardel, S., Sharma, S., Stam,
N. J., and Wilkens, S. (2017) Breaking up and making up: the secret
life of the vacuolar H+-ATPase, Protein Sci.,
26, 896-909, doi: 10.1002/pro.3147.
174.Roh, S.-H., Stam, N. J., Hryc, C. F.,
Couoh-Cardel, S, Pintilie, G., et al. (2018) The 3.5 Å cryoEM
structure of nanodisc-reconstituted yeast vacuolar ATPase Vo
proton channel, Mol. Cell, 69, 993-1004, doi:
10.1016/j.molcel.2018.02.006.
175.Oot, R. A., Yao, Y., Manolson, M. F., and
Wilkens, S. (2021) Purification of active human vacuolar
H+-ATPase in native lipid-containing nanodiscs, J. Biol.
Chem., 297, 100964, doi: 10.1016/j.jbc.2021.100964.
176.Yasuda, R., Noji, H., Kinosita, K. Jr.,
Motojima, F., and Yoshida, M. (1997) Rotation of the gamma subunit in
F1-ATPase; evidence that ATP synthase is a rotary motor enzyme, J.
Bioenerg. Biomembr., 29, 207-209, doi:
10.1023/a:1022449708449.
177.Noji, H., Yasuda, R., Yoshida, M., and
Kinosita, K. Jr. (1997) Direct observation of the rotation of
F1-ATPase, Nature, 386, 299-302, doi:
10.1038/386299a0.
178.Noji, H., Häsler, K., Junge, W., Kinosita,
K. Jr., Yoshida, M., et al. (1999) Rotation of Escherichia coli
F(1)-ATPase, Biochem. Biophys. Res. Commun., 260,
597-599, doi: 10.1006/bbrc.1999.0885.
179.Omote, H., Sambonmatsu, N., Saito, K.,
Sambongi, Y., Iwamoto-Kihara, A., et al. (1999) The gamma-subunit
rotation and torque generation in F1-ATPase from wild-type
or uncoupled mutant Escherichia coli, Proc. Natl. Acad. Sci.
USA, 96, 7780-7784, doi: 10.1073/pnas.96.14.7780.
180.Sambongi, Y., Iko, Y., Tanabe, M., Omote, H.,
Iwamoto-Kihara, A., et al. (1999) Mechanical rotation of the c
subunit oligomer in ATP synthase (FoF1): direct observation,
Science, 286, 1722-1724, doi:
10.1126/science.286.5445.1722.
181.Futai, M., Omote, H., Sambongi, Y., and Wada,
Y. (2000) ATP synthase (H+ ATPase): coupling between
catalysis, mechanical work, and proton translocation, Biochim.
Biophys. Acta, 1458, 276-288, doi:
10.1016/s0005-2728(00)00080-3.
182.Tanabe, M., Nishio, K., Iko, Y., Sambongi, Y.,
Iwamoto-Kihara, A., et al. (2001) Rotation of a complex of the gamma
subunit and c ring of Escherichia coli ATP synthase. The
rotor and stator are interchangeable, J. Biol. Chem.,
276, 15269-15274, doi: 10.1074/jbc.M100289200.
183.Nishio, K., Iwamoto-Kihara, A., Yamamoto, A.,
Wada, Y., and Futai, M. (2002) Subunit rotation of ATP synthase
embedded in membranes: a or beta subunit rotation relative to the c
subunit ring, Proc. Natl. Acad. Sci. USA, 99,
13448-13452, doi: 10.1073/pnas.202149599.
184.Nakanishi-Matsui, M., Kashiwagi, S., Hosokawa,
H., Cipriano, D. J., Dunn, S. D., et al. (2006) Stochastic high-speed
rotation of Escherichia coli ATP synthase F1 sector: The epsilon
subunit-sensitive rotation, J. Biol. Chem., 281,
4126-4131, doi: 10.1074/jbc.M510090200.
185.Futai, M. (2006) Our research on proton pumping
ATPases over three decades: their biochemistry, molecular biology and
cell biology, Proc. Japan Acad. Sci., 82, 416-438, doi:
10.2183/pjab.82.416.
186.Nakanishi-Matsui, M., and Futai, M. (2006)
Stochastic proton pumping ATPases: from single molecules to diverse
physiological roles, IUBMB Life, 58, 318-322, doi:
10.1080/15216540600702255.
187.Nakanishi-Matsui, M., Sekiya, M., and Futai, M.
(2013) Rotating proton pumping ATPases: Subunit/subunit interactions
and thermodynamics, IUBMB Life, 65, 247-254, doi:
10.1002/iub.1134.
188.Hirata, T., Iwamoto-Kihara, A., Sun-Wada, G.
H., Okijama, T., Wada, Y., et al. (2003) Subunit rotation of
vacuolar-type proton pumping ATPase: relative rotation of the G and
c-subunits, J. Biol. Chem., 278, 23714-23719, doi:
10.1074/jbc.M302756200.
189.Sun-Wada, G.H., Wada, Y., and Futai, M. (2003)
Vacuolar H+ pumping ATPases in luminal acidic organelles and
extracellular compartments: common rotational mechanism and diverse
physiological roles, J. Bioenerg. Biomembr., 35, 347-358,
doi: 10.1023/a:1025780932403.
190.Futai, M., Sun-Wada, G. H., and Wada, Y. (2004)
Proton translocating ATPases: introducing unique enzymes coupling
catalysis and proton translocation through mechanical rotation, in
Handbook of ATPases: Biochemistry, Cell Biology, Pathophysiology
(Futai, M., Wada, Y., and Kaplan, J. H., eds) Wiley–VCH,
Weinheim, pp. 237-260.
191.Futai, M., Nakanishi-Matsui, M., Okamoto, H.,
Sekiya, M., and Nakamoto, R. K. (2012) Rotational catalysis in proton
pumping ATPases: from E. coli F-ATPase to mammalian V-ATPase,
Biochim. Biophys. Acta, 1817, 1711-1721, doi:
10.1016/j.bbabio.2012.03.015.
192.Futai, M., Sun-Wada, G.-H., Wada, Y.,
Matsumoto, N., and Nakanishi-Matsui, M. (2019) Vacuolar-type ATPase: a
proton pump to lysosomal trafficking, Proc. Jpn. Acad. Ser. B Phys.
Biol. Sci., 95, 261-277, doi: 10.2183/pjab.95.018.
193.Junge, W., Lill, H., and Engelbrecht, S. (1997)
ATP synthase: an electrochemical transducer with rotatory mechanics,
Trends Biochem. Sci., 22, 420-423, doi:
10.1016/s0968-0004(97)01129-8.
194.Yoshida, M., Muneyuki, E., and Hisabori, T.
(2001) ATP synthase – a marvellous rotary engine of the cell,
Nat. Rev. Mol. Cell Biol., 2, 669-677, doi:
10.1038/35089509.
195.Junge, W., and Nelson, N. (2005) Structural
biology. Nature’s rotary electromotors, Science,
308, 642-644, doi: 10.1126/science.1112617.
196.Hurtado-Lorenzo, A., El Annan, J., Bechoua, S.,
Futai, M., Bourgoin, S., et al. (2003) V-type ATPase as a possible
pH-sensing protein: a2-subunit of V-ATPase localizes in endosomes and
interacts with ARNO in kidney proximal tubules, Mol. Biol. Cell,
14, 237a-238a.
197.Hurtado-Lorenzo, A., El Annan, J., Futai, M.,
Bourgoin, S., Casanova, J., et al. (2004) The V-ATPase is a pH sensor:
novel role in vesicular trafficking, Mol. Biol. Cell, 15,
345.
198.Hurtado-Lorenzo, A., Skinner, M., El Annan, J.,
Futai, M., Sun-Wada, G. H., et al. (2006) V-ATPase interacts with ARNO
and Arf6 in early endosomes and regulates the protein degradative
pathway, Nat. Cell Biol., 8, 124-136, doi:
10.1038/ncb1348.
199.Recchi, C., and Chavrier, P. (2006) V-ATPase: a
potential pH sensor, Nat. Cell Biol., 8, 107-109, doi:
10.1038/ncb0206-107.
200.Hurtado-Lorenzo, A., Skinner, M., El Annan, J.,
Futai, M., Sun-Wada, G.-H., et al. (2006) Novel role of V-ATPase in
regulation of protein degradation pathway, Abstract: “20th
IUBMB International Congress of Biochemistry and Molecular Biology and
11th FAOBMB Congress” (Kyoto, Japan), 447.
201.Hosokawa, H., Dip, P. H., Merkulova, M.,
Bakulina, A., Zhuang, Z., et al. (2013) The N termini of a-subunit
isoforms are involved in signaling between vacuolar
H+-ATPase (V-ATPase) and cytohesin-2, J. Biol. Chem.,
288, 5896-5913, doi: 10.1074/jbc.M112.409169.
202.Marshansky, V., Hosokawa, H., Merkulova, M.,
Bakulina, A., Dip, P. V., et al. (2019) Structural model of a2-subunit
N-terminus and its binding interface for Arf-GEF CTH2: Implication for
regulation of V-ATPase, CTH2 funtion and rational drug design, Curr.
Top. Membr., 83, 77-106, doi:
10.1016/bs.ctm.2019.01.008.