Studying the Structural Organization of Polyribosomes with Alexander S. Spirin
Bruno P. Klaholz1,2,3,4
1Centre for Integrative Biology (CBI), Department of Integrated Structural Biology, IGBMC (Institute of Genetics and of Molecular and Cellular Biology), 67404 Illkirch, France2Centre National de la Recherche Scientifique (CNRS) UMR 7104, 67404 Illkirch, France
3Institut National de la Santé et de la Recherche Médicale (INSERM) U964, 67404 Illkirch, France
4Université de Strasbourg, 67081 Strasbourg, France
Received May 20, 2021; Revised May 20, 2021; Accepted June 6, 2021
“Would it be possible to analyze molecular mechanisms and structural organisation of polyribosome assemblies using cryo electron tomography?” – we asked through a longstanding collaboration between my research group and that of Alexander S. Spirin. Indeed, it was: we found that double-row polyribosomes can have both circular and linear arrangements of their mRNA [Afonina, Z. A., et al. (2013) Biochemistry (Moscow)], we figured out how eukaryotic ribosomes assemble on an mRNA to form supramolecular left-handed helices [Myasnikov, A. G., et al. (2014) Nat. Commun.], that the circularization of polyribosomes is poly-A and cap-independent [Afonina, Z. A., et al. (2014) Nucleic Acids Res.], and that intermediary polyribosomes with open structures exist after a transition from a juvenile phase to strongly translating polysomes of medium size [Afonina, Z. A., et al. (2015) Nucleic Acids Res.] until they form densely packed helical structures with reduced activity. Our joint fruitful exchanges, hence, led to major advances in the field, which are reviewed here from a personal and historical perspective in memory of Alexander S. Spirin.
KEY WORDS: polyribosomes, polysomes, eukaryotic ribosomes, cryo electron microscopy, cryo electron tomographyDOI: 10.1134/S0006297921090030
Abbreviations: cryo-EM, cryo electron microcopy; cryo-ET, cryo electron tomography.
INTRODUCTION
“Studying and understanding the three-dimensional structural organization of eukaryotic polyribosome assemblies – would that ever become possible?” one could have thought, when we started doing so in collaboration with Alexander S. Spirin in 2007. Polyribosomes (also called polysomes) had been visualized in the mid 1960’s, in particular, in Warner’s early work [1, 2] or later as helical arrangements [3], but their three-dimensional (3D) organisation had remained an enigma.
I first met Alexander Sergeevich Spirin at the symposium in Pushchino in September 2001 on his 70th birthday anniversary, seemingly late but never too late (I was a postdoc at that time). It was a memorable meeting as it was scientifically excellent and also impressive with regards of seeing how his seminal scientific research had spread into many groups working worldwide – elucidating the molecular mechanisms underlying protein synthesis.
A few years later we met again on the occasion of a seminar he gave at the IGBMC, Strasbourg/Illkirch, France, in October 2007 (invited by Marat Yusupov). Fascinated by his latest work on the “Step-wise formation of eukaryotic double-row polyribosomes and circular translation of polysomal mRNA” (published shortly later in Nucleic Acids Res., 2008 [4], we started discussing together the issue of polyribosomes. Would it be possible to see the 3D organisation of eukaryotic polyribosomes and address some of the fundamental questions such as: how are individual ribosomes positioned on the mRNA, do they interact and how, do they cooperate, what is the global architecture of eukaryotic polysomes, what about mRNA circularization, specificity of eukaryotic regulation, localization of 5′- and 3′-ends, localization of translation factors, dynamics of the assembly process, etc.? We discussed together in depth and decided that we had everything needed to give it a trial: excellent biochemistry and the possibility of preparing translating wheat germ polyribosomes under various conditions, as well as technical tools to visualize and analyze these structures.
One key to provide “insights” into the ribosome function is to literately visualize ribosomes at work. At that time, the structural study of ribosomes in the field was concentrating on the analysis of isolated ribosomes using X-ray crystallography or single particle cryo electron microscopy (cryo-EM). But to address how a series of ribosomes act when bound to an mRNA molecule and form chains implied direct visualization and 3D reconstruction of a large macromolecular assembly comprising a chain of ribosomes. We thought of approaching the structural analysis of polysomes using cryo electron tomography (cryo-ET), a technique at the time being developed for cellular analysis (pioneered by the work of the W. Baumeister lab [5] and many other papers), but not applied yet to single particles or assemblies at that time (2007).
Indeed, we were in the process of installing the first high-resolution cryo electron microscope in France, the Polara (FEI company, The Netherlands), in a newly refurbished building of the Centre Européen de Biologie et Génomique Structurale (CEBGS), which included a new structural biology platform and gave rise shortly later to the creation of the Centre for Integrative Biology (CBI), Illkirch, France in 2014, that now hosts the FRISBI, Instruct-ERIC and iNEXT-Discovery infrastructures providing cutting-edge support for integrated structural biology. This high-resolution cryo electron microscope had the first transfer system for loading, selecting and analyzing several samples, a CCD camera that overcame the tedious usage of negative films and, hence, facilitated and in fact enabled automated tomography image data acquisition, and a stable goniometer that allowed tilt series acquisitions (much more stable than classical side-entry microscopes). Soon later, the Polara microscope was upgraded with the first-generation direct electron detection camera (Falcon 1), which had a significantly increased sensitivity being based on a CMOS device. In addition, it contained a specific sample holder system that allowed to do acquisitions in the modes of single- and dual-axis tomography (using a flip-flop device to rotate the sample in plane before acquiring the second tilt series). Image processing would then allow obtaining 3D reconstructions from the image tilt series acquired on a given region of the frozen-hydrated and, hence, preserved sample (this allowed overcoming limitations of negative staining EM).
Because thick cellular samples would be difficult to image in cryo-EM and gave low contrast we had to find ways to prepare adequate samples, ideally in a manner so that the thin samples can be obtained directly from the cryo-cooling (“freezing” without forming crystalline ice but instead forming amorphous, transparent ice) cryo-EM grids like for single particle cryo-EM. This became indeed possible by working with polysomes produced in a cell-free system developed by Alexander S. Spirin decades before [6]. Polysomes were obtained in a wheat germ translation system, the continuous exchange cell-free system (CECF) [4, 7], using a β-globin mRNA construct with GFP (green fluorescent protein) and a poly(A) tail or other constructs [8]. The polysomes were fractionated using gel chromatography on a Sephacryl column home-packed in a Stripette [9], which gave reasonable amounts for preparation of cryo-EM grids as compared to the sucrose gradients, which instead were used for sedimentation analysis and characterization; later, we managed to also work directly with the cell extracts, which further helped to keep the complexes intact and more homogenous. Our joint collaboration gave rise to a number of important breakthroughs and novel results (summarized in Figs. 1-4).
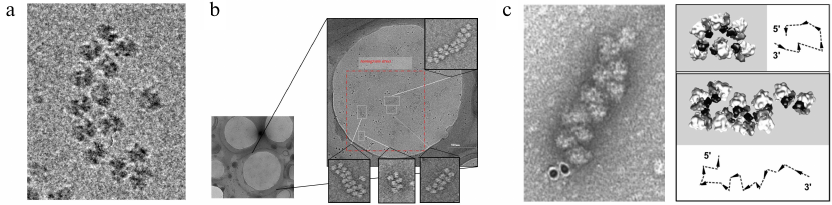
Fig. 1. a and b) Cryo-EM visualization of wheat germ polyribosomes under cryo conditions, i.e., in a preserved, non-stained state. The cryo-EM image shows a polysome on which a first attempt of helical annotation was done (Nov. 2008), shortly before the first cryo-tomograms were acquired (Dec. 2008). The small image shows a region of the cryo-EM grid with several holes, one of which was used to collect several tilt series on different regions (white boxed regions; dark dots are gold particles used as fiducial markers). The inset images show central sections of the 3D reconstruction. c) Double-row polyribosomes can have both circular and linear arrangements [Afonina, Z. A., et al. (2013) Biochemistry (Moscow)]. Negative staining EM image with nano-gold labelling of the 3′-end (left) and annotation of ribosomal particles (40S ribosomal subunit in dark and 60S subunit in light) based on 3D reconstruction obtained from cryo-ET. The annotation describes the mRNA path and its 5′-3′ directionality. Images presented with permission from Oxford University Press.
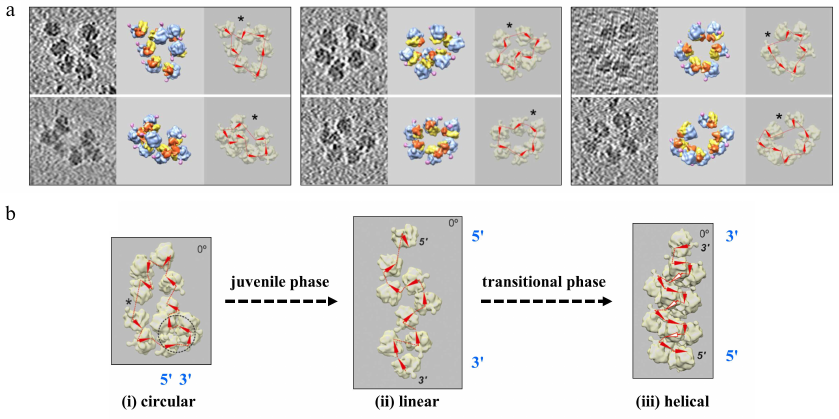
Fig. 2. a) Formation of circular polyribosomes on eukaryotic mRNA without cap-structure and poly(A)-tail, as evidenced by comparison of polyribosomes containing capped & polyadenylated mRNA (left), capped & non-polyadenylated mRNA (middle) or uncapped & non-polyadenylated mRNA (right), revealing that circularization of polyribosomes is poly-A and cap-independent [Afonina, Z. A., et al. (2014) Nucleic Acids Res.]. b) Conformation transitions of eukaryotic polyribosomes during multi-round translation. In an early, juvenile, phase polysomes form circular arrangements that undergo a transition to linearly arranged polysomes of medium size that are strongly translating, to finally form densely packed helical structures, which have reduced translation activity. This work showed that intermediary polyribosomes with open structures exist until they form more compact helical structures [Afonina, Z. A., et al. (2015) Nucleic Acids Res.]. Images presented with permission from Oxford University Press.
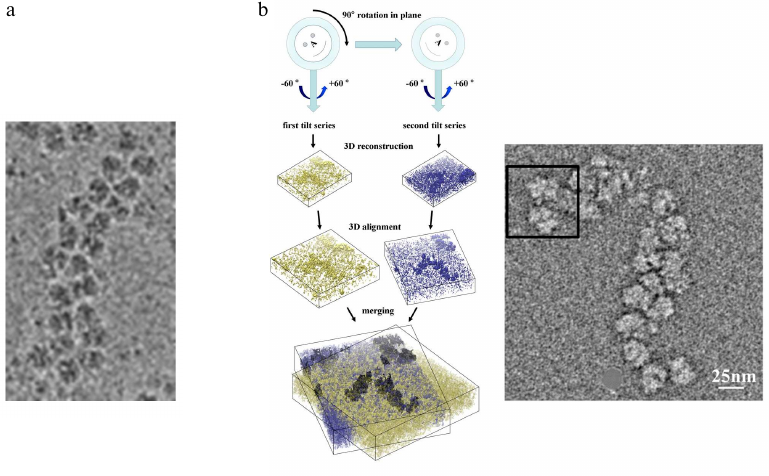
Fig. 3. a) Cryo-EM image example of a large polysome assembly used for cryo-ET. b) Concept of single tilt series acquisition and tomogram reconstruction (left), which can be combined with a second tilt series recorded after rotating the cryo-EM grid in plane by ~90°. 3D reconstructions from dual-axis tomography data provide a clearer picture with less distortions of the map and hence are easier to interpret (see text for details). This was implemented for polysomes and individual ribosomes [i.e., for the first time cryo-ET on single particles; Myasnikov, A. G., et al. (2013) Ultramicroscopy]. Images presented with permission from Elsevier.
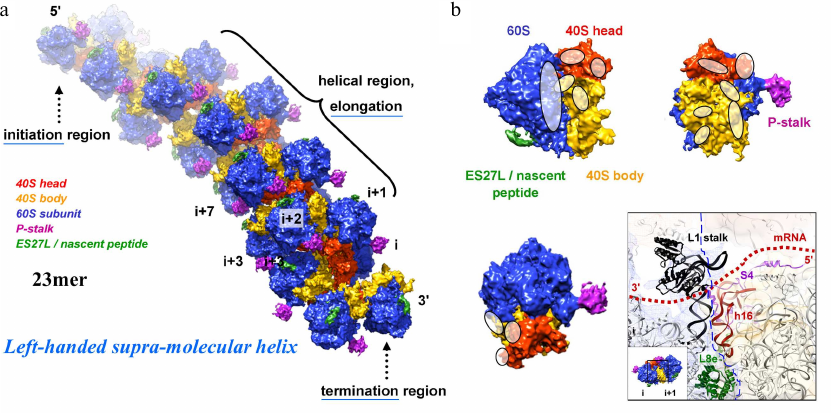
Fig. 4. a and b) 3D reconstruction of a polysome containing 23 ribosomes using cryo-ET and sub-tomogram averaging and atomic fitting into the maps. The averaged ribosome density is placed back into the tomogram to provide insights into the molecular assembly. The 60S ribosomal subunit is indicated in blue, the 40S ribosomal head and body parts in red and orange, the P-stalk and ES27L/nascent peptide in pink and green, respectively. These data revealed that helical polyribosomes form a left-handed supramolecular helix [Myasnikov, A. G., et al. (2014) Nat. Commun.]. The initiation, elongation and termination regions are annotated. Contact regions between the neighbouring ribosomes are indicated (white ovals; panel (b); a particularly interesting contact region (involving the L1 stalk and 18S rRNA helix h16) is the transition of the mRNA protruding from the exit of the mRNA channel (ribosome i) and entering the next ribosome (i+1) at the entry of its mRNA channel, hence being protected by the bound ribosomes against degradation.
In the first study, we jointly addressed the path of the mRNA within the eukaryotic polyribosomes by “marking” the 5′-end with added 40S ribosomal subunits and nano-gold labelling of the 3′-end. Visualization was done by cryo-EM and cryo-ET (using 150 kV acceleration voltage to increase the image contrast). Within the tomogram reconstructions, the individual ribosomes were cut out and aligned, i.e., processed by sub-tomogram averaging to improve the signal-to-noise ratio. The averaged density maps of each ribosome were placed back into the ribosome positions within the polysome (with correct rotation and translation parameters) and, thus, generated a 3D reconstruction of the entire polysome, which hence provided the mutual orientation of each ribosome revealing that the 40S ribosomal subunits were pointing inwards, the 60S subunits outwards – defining polarity (5′-3′ direction) of the mRNA path. This analysis showed that in the same sample double-row polyribosomes can have either a topologically circular or linear (zigzag-like) organization of their mRNA [8].
To optimize data collection strategies for cryo-ET we then compared single- and dual-axis tomography. The classical approach used in cryo-ET is to collect data from a single tilt series, but this causes artefacts in the 3D reconstructions due to systematically missing angular views (called the “missing wedge”). Furthermore, cryo-ET images suffer from particularly low signal-to-noise ratio due to dose limitations imposed by the risk of irradiation damage. Combination of a first data set with a second tilt series acquired after in plane rotation of the sample allows to reduce the missing wedge to a missing pyramid of data, providing less distortions in the 3D reconstruction and also improving the signal-to-noise ratio. Applied to polysomes and individual ribosomes (i.e., for the first time cryo-ET on single particles), this approach provided significantly improved 3D reconstructions and maps that were easier to interpret [9].
The really challenging part of the project should take several years to succeed: reconstructing the structure of a full, large polyribosome assembly. Through a series of cryo-ET studies we finally managed to address the supramolecular structure of eukaryotic polyribosomes. Cryo-ET (using nano-gold as fiducial marker for image alignment and 3D reconstruction) in combination with sub-tomogram averaging and molecular modelling allowed deriving the 3D structure of a polysome constituted of 23 ribosomes bound on the same mRNA molecule (selected as an example among many other polysomes). As this was among the largest asymmetric complexes ever solved (~110 MDa), it became a “special enterprise” to deposit the coordinates in the protein data bank (PDB). This structure revealed that helical polysomes form a left-handed supramolecular helix. It allowed visualization of the three functional parts of the polysome assembly: the central core region that forms a rather compact left-handed supramolecular helix, and the more open regions that harbor the initiation and termination sites at either end. The helical region forms a continuous mRNA channel where the mRNA strand bridges neighboring exit and entry sites of the ribosomes and prevents mRNA looping between ribosomes. This structure provided unprecedented insights into the protein- and RNA-mediated inter-ribosome contacts that involve conserved sites through 40S ribosomal subunits and long protruding RNA expansion segments. These findings shed new light on the molecular machinery of the ribosome and its mode of function [10]. We discussed these findings also in a review about multi-scale integration from the molecular to the cellular level from where this description is cited [11].
In parallel, we addressed the dependence of polysome circularization as a function of the 5′ cap structure of the mRNA and the 3′ poly(A) tail. We analyzed reference polysomes with GFP or β-globin mRNA containing a 5′ cap and a poly(A) tail and compared them to non-capped mRNA or to an mRNA lacking the 3′ poly(A) extension. Using cryo-ET and sub-tomogram averaging, we could trace the mRNA path connecting adjacent exit and entry sites at the ribosomes forming polysomes. To our surprise, it turned out that the polysome formation and circularization were not dependent on these elements that were though over decades to be key for circularization through cap-bound initiation factor eIF4F and poly(A)-binding protein, hence changing a paradigm in the field [12]. It remains, however, to be understood, which elements or associated protein factors of the polysome actually induce circularization.
Our last joint paper addressed the conformational transition of polysomes at different stages of polyribosome formation. Using time point analysis of the mRNA-induced polysomes in a multi-round cell-free translation system, sedimentation analysis, and cryo-ET reconstruction we were able to show that during a juvenile phase ring-shaped, circular polysomes with open structures and of medium size are formed, followed by the formation of intermediary, strongly translational linear polyribosomes with zigzag-like shapes, which eventually lead to densely packed helical structures that are less active. This conformational process is accompanied by decaying of the protein-synthesizing activity. This work illustrates dynamics of the eukaryotic polysomes and their conformational transitions as a function of their formation and transformation in sequential phases, i.e., an “ontogenesis” of polysomes [13].
Alexander S. Spirin presented the first obtained data at the 38th FEBS Congress in St. Petersburg, Russia, 2013. Other presentations of our joint results were given by Zhanna Afonina, Alexander Myasnikov and me at several international meetings, including at GTBio/French Crystallography Association, Paris, 2009; Ribosome Meeting, Orvieto, Italy 2010; EMBO Conference on Protein Synthesis and Translational Control, Heidelberg, 2011; J. Monod Conference, The translating ribosome: towards mature proteins, Roscoff, 2012; Gordon Research Conference on 3D-EM, Girona, Spain, 2014; Gordon Research Conference on 3D-EM, Colby Sawyer College, USA, 2015; EMBO Conference: Ribosome Structure and Function, Strasbourg, France, 2016.
The core team working on all these complexes included Zhanna Afonina from the Alexander S. Spirin’s and Vladimir Shirokov’s group, who came from Pushchino to Strasbourg/Illkirch for several visits during several months during her PhD to prepare polyribosomes under various conditions and with different mRNA constructs; Jean-François Ménétret in my group, who started with negative staining EM analysis and brought in his longstanding EM expertise; Alexander Myasnikov in my group who prepared cryo-EM samples, established single- and dual-axis data collection on the new Polara microscope in a rigorous manner and did all the image processing and tomogram analysis (sub-tomogram averaging, segmentation, atomic model fitting); Vladimir Shirokov, Alexander S. Spirin, and myself, who supervised and discussed experimental details with all team members. My gratefulness goes to all of them. The brainstorming moments and the great spirit of this “polysome team”, joined across two countries, will remain strongly in our minds, when thinking of the topology and arrangement of ribosomes in the polysome chain, how to prepare them, how to do tomography on these complex samples, discussing results and thinking of the next steps. Alexander S. Spirin passed away at the end of 2020 and Vladimir Shirokov in 2019. Life passes, but memories stay while time flows. Alexander S. Spirin has been of such great inspiration to everyone, an exceptional scientist – passionate and human at the same time.
As I wrote in an E-mail message to Alexander S. Spirin at the end of 2013, when our polysome supramolecular helix manuscript was getting ready for publication (published later in Nat. Commun., 2014 [10]) along with 2 papers in Nucleic Acids Res. [12, 13]: “Wherever [i.e., which journal] this work will be published, it represents the basis of understanding of a key aspect of protein synthesis. In this context, I’d like to express my gratitude to you, Alexander, when we met in autumn 2007 right after your seminar and started discussing about the possible 3D analysis of polysomes… Since then, it has been a major investment of each of us [meant were all people involved, i.e., Vladimir Shirokov, Zhanna Afonina, Alexander Myasnikov and Jean-François Ménétret) altogether to design the project, organize visits and exchanges, prepare adequate samples and start cryo electron tomography, which was not an obvious thing to do when we started. It was very exciting at the scientific level, and great at human level also, to work together on this.” He answered: “The successful collaboration with you and your group became a remarkable experience in scientific life of all my group and myself personally”; and in autumn 2014 when the paper in Nat. Commun. was about to come out he wrote: “In fact, we all can now state that our primary contact several years ago and the subsequent joint work proved to be very fruitful and led to novel and interesting results of fundamental significance.” Taken together, it was a great honor and pleasure to work together with Alexander Sergeevich Spirin over all these years.
Since this work in the period 2008-2015, during which other interesting studies on bacterial and eukaryotic polyribosomes appeared [14-16] (see also our reviews & book chapters [17, 18]) the field has been moving further, with multi-scale integration moving on [11, 17, 18] and high resolution cryo-EM analysis (3 Å resolution range and better) becoming feasible thanks to new direct electron detectors [11, 19] and image processing methods. In the future it might become possible to analyze the structure of polysomes at quasi atomic resolution, and also in situ directly in the cellular context, which would have been a dream to become true for Alexander S. Spirin. First studies in this direction using high-resolution cryo-ET of cellular ribosomes came out this year [20] and indicate a very promising future for the field.
Funding. I thank past and present members of my research group at CBI/IGBMC for dedicated research and members of the integrated structural biology platform at the CBI/IGBMC for support. I acknowledge support by Centre National pour la Recherche Scientifique (CNRS), Institut National pour la Recherche Médicale (Inserm), Association pour la Recherche sur le Cancer (ARC), Institut National du Cancer (INCa), the Fondation pour la Recherche Médicale (FRM), Ligue nationale contre le cancer (Ligue), Agence National pour la Recherche (ANR), and USIAS of the University of Strasbourg (USIAS-2018-012). The electron microscope facility was supported by the Région Grand Est and by the French Infrastructure for Integrated Structural Biology (FRISBI) and Instruct-ERIC.
Ethics declarations. The author declares no conflicts of interest in financial or any other sphere. This article does not contain any studies involving human participants or animals performed by the author.
REFERENCES
1.Warner, J. R., Rich, A., and Hall, C. E. (1962)
Electron microscope studies of ribosomal clusters synthesizing
hemoglobin, Science, 138, 1399-1403, doi:
10.1126/science.138.3548.1399.
2.Warner, J. R., Knopf, P. M., and Rich, A. (1963) A
multiple ribosomal structure in protein synthesis, Proc. Natl. Acad.
Sci. USA, 49, 122-129, doi: 10.1073/pnas.49.1.122.
3.Wooding, F. B. (1968) Ribosome helices in mature
cells, J. Ultrastruct. Res., 24, 157-164, doi:
10.1016/S0022-5320(68)80025-5.
4.Kopeina, G. S., Afonina, Zh. A., Gromova, K. V.,
Shirokov, V. A., Vasiliev, V. D., and Spirin, A. S. (2008) Step-wise
formation of eukaryotic double-row polyribosomes and circular
translation of polysomal mRNA, Nucleic Acids Res., 36,
2476-2488, doi: 10.1093/nar/gkm1177.
5.Medalia, O., Weber, I., Frangakis, A. S., Nicastro,
D., Gerisch, G., Baumeister, W. (2002) Macromolecular architecture in
eukaryotic cells visualized by cryoelectron tomography, Science,
298, 1209-1213, doi: 10.1126/science.1076184.
6.Spirin, A. S., Baranov, V. I., Ryabova, L. A.,
Ovodov, S., Alakhov, Y. B. (1988) A continuous cell-free translation
system capable of producing polypeptides in high yield, Science,
242, 1162-1164, doi: 10.1126/science.3055301.
7.Shirokov, V. A., Kommer, A. A., Kolb, V. A., and
Spirin, A. S. (2007) Continuous-exchange protein-synthesizing systems,
in Methods in Molecular Biology, In vitro Transcription and
Translation Protocols (Grandi, G., ed.) Humana Press Inc., Totowa,
NJ, 375, pp. 19-55, doi: 10.1007/978-1-59745-388-2_2.
8.Myasnikov, A. G., Afonina, Z., and Klaholz, B. P.
(2013) Single particle and molecular assembly analysis of polyribosomes
by single- and double-tilt cryo electron tomography,
Ultramicroscopy, 126, 33-39, doi:
10.1016/j.ultramic.2012.12.009.
9.Afonina, Z. A., Myasnikov, A. G., Khabibullina, N.
F., Belorusova, A. Yu., Ménétret, J.-F., et al. (2013)
Topology of mRNA chain in isolated eukaryotic double-row polyribosomes,
Biochemistry (Moscow), 78, 445-454, doi:
10.1134/S0006297913050027.
10.Myasnikov, A. G., Afonina, Z. A.,
Ménétret, J.-F., Shirokov, V. A., Spirin, A. S., and
Klaholz, B. P. (2014) The molecular structure of the left-handed
supra-molecular helix of eukaryotic polyribosomes, Nat. Commun.,
5, 5294, doi: 10.1038/ncomms6294.
11.Orlov, I., Myasnikov, A. G., Andronov, L.,
Natchiar, S. K., Khatter, H., et al. (2017) The integrative role of
cryo electron microscopy in molecular and cellular structural biology,
Biol. Cell, 109, 81-93, doi: 10.1111/boc.201600042.
12.Afonina, Z. A., Myasnikov, A. G., Shirokov, V.
A., Klaholz, B. P., Spirin, A. S. (2014) Formation of circular
polyribosomes on eukaryotic mRNA without cap-structure and
poly(A)-tail: a cryo electron tomography study, Nucleic Acids
Res., 42, 9461-9469, doi: 10.1093/nar/gku599.
13.Afonina, Z. A., Myasnikov, A. G., Shirokov, V.
A., Klaholz, B. P., and Spirin, A. S. (2015) Conformation transitions
of eukaryotic polyribosomes during multi-round translation, Nucleic
Acids Res., 43, 618-628, doi: 10.1093/nar/gku1270.
14.Ortiz, J. O., Förster, F., Kürner, J.,
Linaroudis, A. A., and Baumeister, W. (2006) Mapping 70S ribosomes in
intact cells by cryo electron tomography and pattern recognition, J.
Struct. Biol., 156, 334-341, doi:
10.1016/j.jsb.2006.04.014.
15.Brandt, F., Carlson, L. A., Hartl, F. U.,
Baumeister, W., Grünewald, K. (2010) The three-dimensional
organization of polyribosomes in intact human cells, Mol. Cell,
39, 560-569, doi: 10.1016/j.molcel.2010.08.003.
16.Brandt, F., Etchells, S. A., Ortiz, J. O.,
Elcock, A. H., Hartl, U., and Baumeister, W. (2009) The native 3D
organization of bacterial polysomes, Cell, 136, 261-271,
doi: 10.1016/j.cell.2008.11.016.
17.Simonetti, A., Marzi, S., Myasnikov, A. G.,
Ménétret, J.-F., and Klaholz, B. P. (2011) Insights into
translation initiation and termination complexes and into the polysome
architecture, in Ribosomes (Rodnina, M. V., Wintermeyer, W., and
Green, R., eds.) Springer, Vienna, doi:
10.1007/978-3-7091-0215-2_10.
18.Ménétret, J.-F., Khatter, H.,
Simonetti, A., Orlov, I., Myasnikov, A. G., et al. (2013) Integrative
structure–function analysis of large nucleoprotein complexes, in
RNA Structure and Folding (Klostermeier, D., and Hammann, C.,
eds.), doi: 10.1515/9783110284959.
19.Klaholz, B. P. (2019) Deriving and refining
atomic models in crystallography and cryo-EM: the latest Phenix tools
to facilitate structure analysis, Acta Cryst., D75,
878-881, doi: 10.1107/S2059798319013391.
20.Tegunov, D., Xue, L., Dienemann, C., Cramer, P.,
and Mahamid, J. (2021) Multi-particle cryo-EM refinement with M
visualizes ribosome-antibiotic complex at 3.5 Å in cells, Nat.
Methods, 18, 186-193, doi: 10.1038/s41592-020-01054-7.