REVIEW: Ribosome-Associated Quality Control in Bacteria
Maxim S. Svetlova
Center for Biomolecular Sciences, Department of Pharmaceutical Sciences, University of Illinois at Chicago, 60607 Chicago, Illinois, USA
Received May 14, 2021; Revised May 14, 2021; Accepted May 14, 2021
Translation of the genetic information into proteins, performed by the ribosome, is a key cellular process in all organisms. Translation usually proceeds smoothly, but, unfortunately, undesirable events can lead to stalling of translating ribosomes. To rescue these faulty arrested ribosomes, bacterial cells possess three well-characterized quality control systems, tmRNA, ArfA, and ArfB. Recently, an additional ribosome rescue mechanism has been discovered in Bacillus subtilis. In contrast to the “canonical” systems targeting the 70S bacterial ribosome, this latter mechanism operates by first splitting the ribosome into the small (30S) and large (50S) subunits to then clearing the resultant jammed large subunit from the incomplete nascent polypeptide. Here, I will discuss the recent microbiological, biochemical, and structural data regarding functioning of this novel rescue system.
KEY WORDS: translation, ribosome stalling, quality control, polyalanine-tailingDOI: 10.1134/S0006297921080058
Abbreviations: CAT tail, Carboxy-terminal Alanine and Threonine tail; RQC, Ribosome-associated Quality Control; tmRNA, transfer-messenger RNA.
CANONICAL RIBOSOME RESCUE MECHANISMS IN BACTERIA
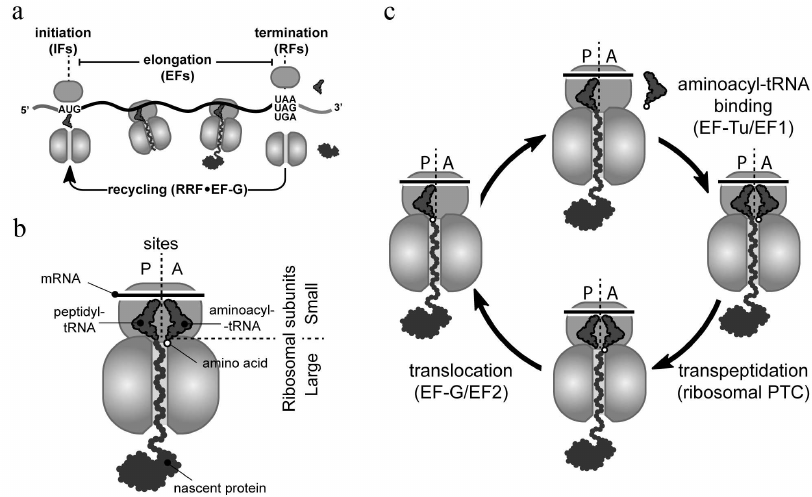
Ribosomes carry out protein synthesis in all living cells by the process known as translation (Fig. 1). Ribosomes start protein synthesis by recognizing the start site (usually the AUG codon and less commonly GUG or UUG) on a messenger RNA (mRNA) (Fig. 1a). This initiation step is followed by multiple elongation cycles during which ribosomes move through the mRNA open reading frame (the segment of mRNA encoding a specific protein), reading its nucleotide sequence and converting (translating) it into the amino acid sequence of the synthesized protein (Fig. 1c). Each elongation cycle begins with the delivery of the appropriate aminoacyl-tRNA to the ribosomal A site, followed by its reaction with the peptidyl-tRNA located in the P site, and ends with the translocation of the newly formed peptidyl-tRNA, from the A to the P site. The complete elongation cycle and its main participants are depicted in Fig. 1, a and c. Finally, ribosomes terminate translation and release mature protein, when one of the three stop codons (UAA, UAG, or UGA) is reached (Fig. 1a).
Fig. 1. Translation cycle in bacteria. a) With assistance of initiation factors (IFs), small (30S) and large (50S) ribosomal subunits join to form the full 70S ribosome at the mRNA start site (AUG, GUG, or UUG codons). Elongation factors (EFs) help the ribosome to polymerize amino acids into protein during the elongation stage. The full-length protein is released by release factors (RFs) from the ribosome that has reached a stop codon (UAA, UAG, or UGA). After termination, recycling factors (RRF and EF-G in bacteria) split the ribosome into subunits to initiate a new round of translation. b) Schematic representation of elongating ribosome at the pre-transpeptidation stage. This ribosome carries translated mRNA (located on the small subunit), aminoacyl-tRNA (in the ribosomal A site), and peptidyl-tRNA (in the P site). c) Overview of an elongation cycle. The cycle consists of three sequential steps: (i) EF-Tu-mediated binding of appropriate aminoacyl-tRNA to the ribosomal A site, (ii) reaction of the aminoacyl-tRNA with the P-site peptidyl-tRNA catalyzed by the ribosomal peptidyl transferase center, and (iii) EF-G-mediated translocation of the newly formed peptidyl-tRNA augmented by one amino acid residue from the A to the P site.
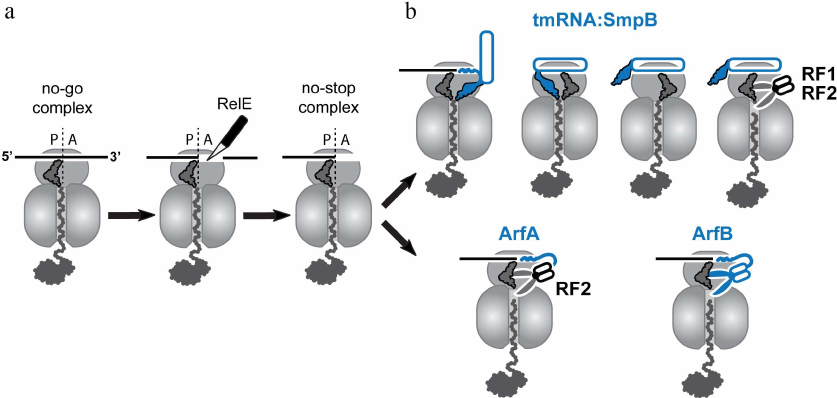
In actively growing bacteria, the ribosome-catalyzed polymerization of nascent peptides occurs at a rate of around 15 amino acids per second [1, 2]. Notably, this rate is not constant, and rapid progression of the ribosome along mRNA can be interrupted by pauses. In many cases, ribosomal pauses during translation are part of the cell physiology since they can play an essential role, for example, in co-translational folding, acquisition of chemical modification into the proteins, or sorting of nascent proteins to appropriate compartments [3, 4]. On the other hand, the ribosome pauses can be detrimental for the cell, when they are caused by translation of damaged mRNAs or polymerization of problematic amino acid segments in the nascent peptides. Cells utilize several translational accessory factors to relieve these unwanted pauses and eliminate their harmful effect. For instance, the bacterial elongation factor-P (EF-P) and its archaeal and eukaryotic homologs eIF5A, alleviate translational pauses caused by polyproline stretches in the nascent peptides [5, 6]. To this end, the factors stabilize peptidyl-tRNA in a productive conformation capable of reacting with Pro-tRNAPro that comes to the ribosomal A site [7]. Another translation elongation factor EF4 (LepA) has been suggested to contribute to relieve translational pauses in unfavorable conditions such as high magnesium concentrations or low temperature [8]. However, in some cases, translational pauses are hard or even impossible to relieve, and translating ribosomes may get stuck at a specific site on mRNAs. Ribosomal stalling is especially escalated during different stresses such as amino acid starvation [9], temperature shifts [10], and treatment with ribosome-targeting inhibitors [11]. Under these conditions, ribosomes can stall at the 3′-end of truncated mRNAs lacking their normal stop codon. Such no-stop mRNAs usually appear in a cell as a result of premature transcription termination or nuclease cleavage of full-length mRNA transcripts. Stalling can also occur on intact mRNAs leading to accumulation of the no-stop or no-go ribosomal complexes. The no-stop complexes (which are structurally identical to the complexes formed on the truncated mRNAs) are accumulated, when ribosomes erroneously read through the normal stop codons and then get stuck at the 3′-ends of the undamaged mRNAs. The absence of a codon in the ribosomal A-site is a distinguishable feature of these complexes recognized by the “canonical” bacterial rescue systems. In contrast to the no-stop complexes, the no-go ribosomes stall in the middle of translated mRNAs. The “canonical” systems cannot rescue these ribosomes due to the occupancy of their mRNA channels by mRNA. As a first step towards rescuing no-go complexes, the bacterial toxin RelE, an endonuclease activated by amino acid starvation, can bind to them and cut off the mRNA invading the ribosomal A site. As a result of the RelE cleavage, the no-go complexes are converted into no-stop stalled ribosomes that can be targeted by the “canonical” rescue mechanisms (Fig. 2a). These mechanisms, including SmpB•tmRNA-mediated trans-translation, ArfA, and ArfB, has been thoroughly reviewed [12-14], and I will only briefly describe them below.
Fig. 2. Rescue of stalled ribosomes by canonical bacterial systems. a) Ribosome stalling can result in formation of the no-stop or no-go complexes. Canonical rescue systems can directly recognize only the no-stop ribosomes with empty mRNA channels, whereas no-go ribosomes, in order to be recycled, need to be converted to no-stop ribosomes by endonuclease RelE. b) The resulting no-stop ribosomes are rescued by trans-translation mediated by the transfer-messenger RNA in complex with SmpB protein. Translation of the mRNA-like segment of the tmRNA is terminated by the canonical release factors RF1 or RF2. If activity of this quality control system is compromised, stalled peptidyl-tRNA can be hydrolyzed by the ArfA-recruited RF2 or by the stop-codon-independent release factor ArfB. All three canonical bacterial rescue systems, SmpB•tmRNA, ArfA, and ArfB (colored in blue), recognize an empty or partly vacant mRNA channel in the small subunit of the no-stop ribosome.
Trans-translation is the primary mechanism that rescues no-stop ribosomes in bacteria [15]. This mechanism relies on the operation of two cellular factors: a small protein SmpB and transfer-messenger RNA (tmRNA), a hybrid of tRNA charged with an alanine residue and mRNA encoding a special short degron tag (Fig. 2b). SmpB and tmRNA form a complex that is delivered to the no-stop ribosome by EF-Tu [16]. Being delivered to the ribosome, SmpB interacts with the small subunit and verifies that the mRNA channel and the A site are empty, while tmRNA mimics aminoacyl-tRNA and binds to the ribosome placing its CCA end aminoacylated with alanine into the ribosomal peptidyl-transferase center where it reacts with the peptidyl-tRNA. After the peptide transfer to tmRNA, the stalled ribosome translates the degron-coding segment of the tmRNA. Termination of tmRNA translation at the stop codon located at the end of the degron-coding sequence results in the recycling of the ribosomes through the standard mechanism, and the released C-terminally tagged peptide is targeted to proteolytic degradation.
Although components of trans-translation were identified in all sequenced bacterial species, disruption of this system usually is not lethal [17] due to the existence of two auxiliary rescue mechanisms, ArfA and ArfB, in some bacteria. These systems support bacterial growth, when the activity of SmpB•tmRNA complex is compromised or insufficient. ArfA is a small protein that binds to the A site in the small ribosomal subunit when it contains no mRNA codon and then recruits release factor 2 (RF2) to the stalled ribosome to hydrolyze the peptidyl-tRNA [18, 19] (Fig. 2b). Interestingly, such selectivity for RF2 is not uniform among bacteria; in Francisella tularensis, a small ArfA-analogous protein ArfT can recruit either RF1 or RF2 to no-stop ribosomes [20]. ArfB (previously known as YaeJ) is a homolog of the release factors (RFs) whose peptidyl-tRNA hydrolase activity is stop-codon-independent [21, 22] (Fig. 2b). Similar to ArfA, ArfB binds only to the ribosome positioned at the 3′-end of mRNA with no mRNA codons placed in the A site. Interestingly, both backup rescue systems discriminate between the stalled and active ribosomes in the same way as the SmpB•tmRNA complex; they bind to the empty mRNA entry channel on the small ribosomal subunit normally occupied by the translated mRNA to initiate the ribosome rescue. Given the existence of these backup rescue systems in many bacteria, the recent discovery of a completely different ribosome rescue mechanism in Bacillus subtilis became an unexpected and pleasant surprise. Even more striking is the fact that this rescue mechanism is reminiscent of the Ribosome-associated Quality Control (RQC) system that operates in eukaryotes. To understand similarities of the bacterial and eukaryotic RQC systems, I will briefly describe how the stalled ribosomes are rescued in eukaryotic cells.
RESCUE OF THE STALLED RIBOSOMES IN EUKARYOTES
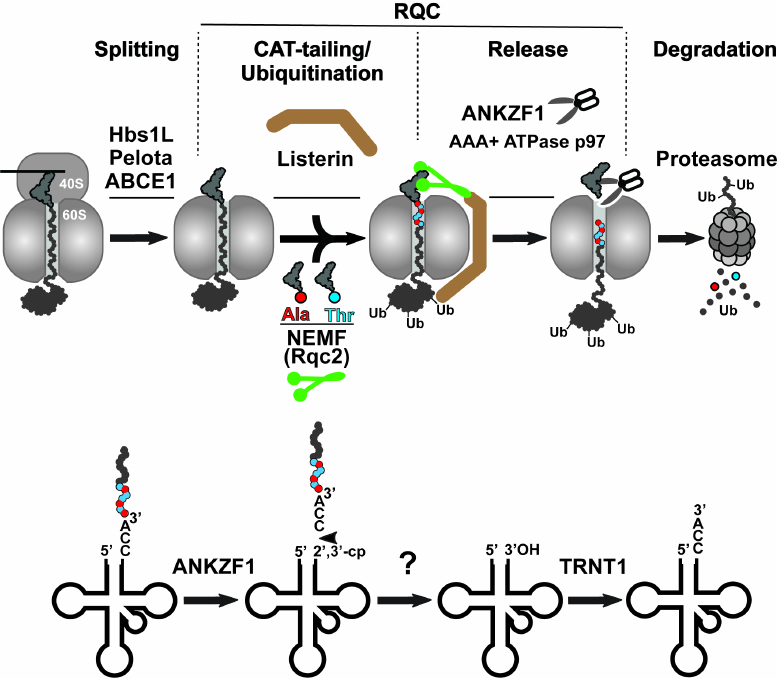
While the aforementioned bacterial rescue mechanisms target intact stalled ribosomes, the prolonged stalling of translation in eukaryotes induces splitting of the 80S ribosome into its constituent subunits (small 40S and large 60S), followed by their independent recycling for the next rounds of translation [13]. The model for the rescue of the stalled 80S ribosome is shown in Fig. 3 (top panel). Ribosome splitting requires concerted action of three factors: Hbs1L (yeast Hbs1p), which binds in the vicinity of the unoccupied mRNA entry channel, eRF1 paralog Pelota (Dom34p in yeast), which occupies the empty A site of the stalled ribosome, and ATPase ABCE1 (yeast Rli1p), which is recruited to the stalled ribosome by Pelota after Hbs1L dissociation to induce splitting of the stalled ribosome into subunits (Fig. 3, Splitting step). After ribosome splitting, mRNA is released from the 40S subunit to be degraded by the 5′-3′ exoribonuclease Xrn1 and the 3′-5′ exosome [23], and then the mRNA-free subunit is recycled by ABCE1 [24]. Simultaneously, the 60S subunit dissociates still carrying peptidyl-RNA because Pelota/Dom34p lacks peptidyl-tRNA hydrolase activity. Fate of the 60S•peptidyl-tRNA complex depends on the length of the nascent peptide conjugated with the tRNA. If the nascent peptide is relatively short, then the peptidyl-tRNA can simply dissociate from the large subunit [25]. In contrast, if the nascent peptide is long enough to establish stable interactions within the nascent peptide exit tunnel or fold outside the ribosome, the peptidyl-tRNA remains stuck in the 60S subunit. In this situation, the peptidyl-tRNA-jammed large ribosomal subunit is recognized and resolved by the proteins of the special rescue pathways named RQC (thoroughly reviewed in [26]) (Fig. 3). The RQC pathway is triggered by the selective binding of the nuclear export mediator factor (NEMF/yeast Rqc2) to the interface between the 60S subunit and the peptidyl-tRNA, where this protein plays at least three roles: (i) it prevents 40S from rejoining with the 60S•peptidyl-tRNA complex, (ii) it recruits E3 ubiquitin ligase Listerin (yeast Ltn1p) to ubiquitinate the stalled nascent peptide, and (iii) it recruits Ala-tRNAAla and Thr-tRNAThr and promotes the mRNA-independent addition of alanine and threonine residues to the C-terminus of the nascent peptide (the C-terminal extension called CAT tail) to expose the nascent chain-lysine residues that might be hidden inside the exit tunnel of the 60S subunit for the Listerin-mediated ubiquitination (Fig. 3, CAT-tailing/Ubiquitination step) [27]. The ubiquitinated nascent peptide is then cleaved from tRNA by the endonuclease ankyrin repeat and zinc finger domain-containing protein 1 (ANKZF1/yeast Vms1p) [28] (Fig. 3, Release step). Notably, in contrast to the peptide release during the regular translation termination, when the canonical RFs provoke ribosomal peptidyl-transferase center to hydrolyze the ester bond between the tRNA and peptide moieties in the peptidyl-tRNA, ANKZF1 selectively cleaves off its terminal 3′ CCA nucleotides [29]. The resulting CCA-nascent peptide conjugate is extracted from the RQC complexes with the assistance of AAA+ ATPase p97 (yeast Cdc48p) and its cofactors, UFD1 and NPLOC4 (yeast Ufd1p and Npl4p), and then is delivered by them to the proteasome for degradation (Fig. 3, Degradation step). Simultaneously, the ANKZF1-generated truncated tRNA dissociates from the 60S subunit and is recycled through a two-step process that includes removing 2′,3′-cycle phosphate by a mammalian homolog of RNaseZ, ELAC1 [30], followed by restoration of its 3′-CCA end by the CCA-adding enzyme TRNT1 [29] (Fig. 3, model for tRNA recycling shown on the bottom panel). Finally, the large ribosomal subunit free of tRNA and nascent peptide can be recruited for the new rounds of translation.
Fig. 3. Eukaryotic ribosome-associated quality control pathway. Top panel) the ribosome-associated quality control (RQC) is preceded by splitting of the stalled 80S ribosome with assistance of three proteins Pelota, Hbs1L, and ABCE1. Next, NEMF (green) and Listerin (brown) bind to the resulting 60S•peptidyl-tRNA complex and modify the nascent peptide. NEMF extends the C-terminus of the nascent peptide with alanine/threonine residues (CAT-tail) to expose its lysine residues, otherwise hidden inside the ribosomal exit tunnel, for Listerin-catalyzed ubiquitination. Finally, the nascent peptide is released from the 60S subunit by endonuclease ANKZF1 hydrolyzing peptidyl-tRNA and delivered by AAA+ ATPase p97 to the proteasome for degradation. Bottom panel) model for recycling of tRNA after ANKZF1-mediated nascent peptide release. ANKZF1 cuts off the 3′-CCA of the stalled peptidyl-tRNA leading to the release of the CCA-nascent peptide conjugate degraded by proteosome and generation of the truncated tRNA carrying a 2′,3′-cyclic phosphate. After removing the cyclic phosphate by ELAC1, the CCA-adding enzyme TRNT1 recycles the tRNA by adding back the 3′-CCA nucleotides to its 3′-end.
RQC-LIKE RIBOSOME RESCUE SYSTEM IN BACTERIA
Until recently, it had been thought that the RQC mechanism operates exclusively in eukaryotes (eRQC) whereas ribosome rescue in bacteria is solely achieved by the “canonical” bacteria-specific systems such as SmpB•tmRNA, ArfA/ArfT, and ArfB. This conclusion was based on several facts, including the following:
1.In many bacteria, combined deletion of the “canonical” rescue systems is synthetically lethal [31, 32], suggesting absence of any other backup rescue systems in the studied species.
2.There are no obvious bacterial homologs of the key eRQC factors, such as the splitting triad Pelota-Hbs1-ABCE1, the E3 ubiquitin ligase Ltn1, and 60S-dependent peptidyl-tRNA endonuclease ANKZF1/Vms1p.
3.Activity of the eRQC system is tightly coupled with ubiquitination of the 60S-bound stalled peptides, modification that is not common in bacteria.
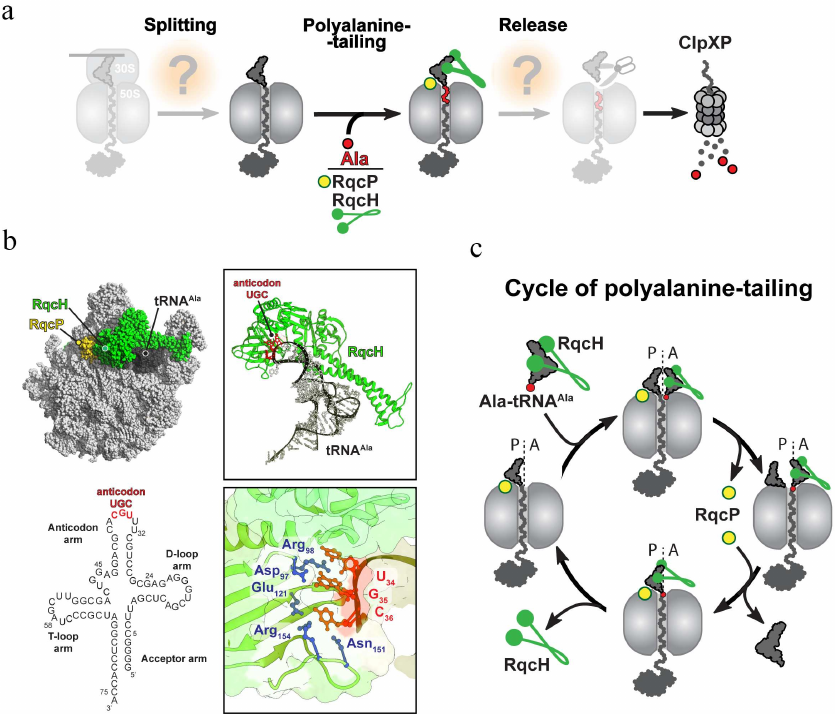
Nevertheless, while most of the RQC factors are eukaryote-specific, NEMF/Rqc2 homologs are found in archaea as well as in some bacterial species where they are known as members of the fibronectin-binding protein A (FbpA) family of virulence factors [33]. FbpA proteins produced by pathogenic bacteria were suggested to bind to fibronectin present in the extracellular matrix, thereby ensuring host adhesion [34]. However, Lytvynenko et al. [35] recently showed that a NEMF/Rqc2 homolog from Bacillus subtilis, RqcH, is directly involved in the ribosome rescue. It was found that RqcH can selectively bind and deliver Ala-tRNAAla(UGC) to the peptidyl-tRNA-occupied 50S ribosomal subunit (that becomes split from the 30S subunit by a still-unknown ribosome rescue factor) to facilitate the C-terminal tagging of the incomplete nascent peptide with a polyalanine tail. This nascent peptide modification is reminiscent of the CAT-tailing reaction mediated by NEMF/Rqc2 in eukaryotes. However, in contrast to the eRQC, where the CAT tail indirectly facilitates peptide degradation by promoting its ubiquitination, the C-terminal polyalanine tail in bacteria acts directly as a degron signal recognized by ClpXP protease (Fig. 4a).
Fig. 4. Bacterial RQC-like pathway. a) In general, the bacterial RQC-like pathway consists of the same basic steps as the analogous pathway in eukaryotes (see Fig. 3). However, the only well-studied step of this mechanism is addition of the polyalanine degron tail to the C-terminus of the 50S-bound nascent peptide. In B. subtilis, this step is assisted by the NEMF/Rqc2 homolog RqcH and the small protein RqcP. b) Cryo-EM structure of the bacterial RQC complex from B. subtilis (PDB: 7AS8). The RQC complex consists of the stalled 50S ribosome (light grey), tRNAAla (dark grey, the secondary structure of the tRNAAla is shown below the Cryo-EM structure), RqcH (green), and RqcP (yellow). In this complex, Asp97, Arg98, Glu121, Asn151, and Arg154 residues (shown in blue) of RqcH read G35 and C36 nucleotides (shown in red) of UGC anticodon in tRNAAla. c) Polyalanine tailing cycle in B. subtilis (see main text for details).
Structural studies shed more light on several aspects of the mechanism of polyalanine tailing in bacteria [36, 37] (Fig. 4b). The cryoelectron microscopy (Cryo-EM)-generated structures of the B. subtilis RQC complexes revealed that the incoming Ala-tRNA forms extensive interactions with the ribosome-bound RqcH. While the D- and T-loops of the alanyl-tRNAAla interact with the positively charged patch on the RqcH surface, two anticodon nucleotides G35 and C36 are engaged in the formation of a specific network of interactions with several residues of RqcH (Fig. 4b). Such RqcH-mediated tRNA decoding rationalizes high selectivity of the RQC complexes for tRNAAla. The structural, biochemical, and genetic evidence further identified the Hsp15 homolog YabO as an important RqcH partner for polyalanine tagging (Fig. 4, b and c). This small protein stabilizes an otherwise wobbly peptidyl-RNA in the P site of the 50S subunit (and therefore, YabO has been renamed as RqcP) in a productive conformation competent for reacting with the incoming Ala-tRNAAla recruited by RqcH to the ribosomal A site. Finally, the RQC structures captured at different stages of polyalanine tailing allowed to suggest a model of how this reaction proceeds on the ribosome (Fig. 4c). According to this model, following the ribosomal splitting of the stalled ribosome, the peptide moiety of the RqcP-stabilized peptidyl-tRNA in the P site is transferred to the RqcH-recruited Ala-tRNAAla in the A site, extending the nascent peptide with a C-terminal alanine. Dissociation of RqcP from the RQC complex allows for the deacylated P-site tRNA to relocate into the E site, and the newly formed peptidyl-tRNA adopts an A/P-like hybrid conformation. Subsequent spontaneous dissociation of deacylated tRNA from E site allows rebinding of RqcP to the complex, followed by the shift of the A/P-site peptidyl-alanyl-tRNAAla into the P site and completion of the elongation cycle. This elongation cycle of alanine addition is repeated several times to produce polyalanine-tagged peptides. Finally, the tagged peptide is released by another yet unknown factor to the cytosol, where it is degraded by ClpXP protease (Fig. 4a).
Discovery of the RqcH- and RqcP-mediated polyalanine tailing revealed existence of the RQC-like ribosome rescue system in at least some bacteria. Nevertheless, many aspects regarding functioning of this system remain unknown. It is unclear what cellular conditions initiate splitting of the stalled ribosomes in bacteria and what cellular factor(s) stimulates the splitting. It is also unknown how the peptidyl-tRNA-jammed large 50S subunits are rescued in the gamma-proteobacteria and actinobacteria, where the key factor of C-terminal polyalanine tailing, RqcH, is missing. It also remains to be established, what serves as a signal for termination of polyalanine tagging and what cellular factor hydrolyzes the resultant peptidyl-tRNA. Some of the important unanswered questions pertaining to the mechanisms of RQC-like ribosome rescue in bacteria are discussed below.
HOW ARE THE STALLED RIBOSOMES SPLIT IN BACTERIA?
Splitting of the stalled ribosome into subunits is the critical step that initiates the RQC-mediated rescue mechanism. Conditions that induce such splitting in bacteria and cellular factors involved in this process are not known yet. Interestingly, it has been shown that the stretches of acidic amino acids (aspartate and glutamate) incorporated into a nascent peptide can destabilize the translating ribosome and induce its dissociation into subunits, apparently without the involvement of other factors [38].
Even though this example illustrates a factor-independent dissociation of ribosomes, such spontaneous splitting events are apparently not widespread because they are induced by very specific – and therefore very rare – amino acid motifs in the nascent peptides, which seem to be under evolutionary pressure to be excluded from proteins [39]. Therefore, the existence of factor(s) specialized in recognizing and actively splitting stalled ribosomes in bacterial cells remains a distinct possibility. During normal bacterial translation, Ribosome Recycling Factor (RRF) and EF-G split the ribosome after termination and release of the mature protein. However, it is implausible that these proteins dissociate the stalled ribosomes carrying peptidyl-tRNAs, since in vitro experiments have clearly shown that the dissociation activity of the RRF•EF-G complex is limited exclusively to the post-termination ribosomes carrying deacylated tRNA [40].
It is worth noting that translational stalling increases during certain extreme conditions, such as heat shocks. It was found that the bacterial heat shock GTPase HflX can dissociate vacant and mRNA-associated ribosomes carrying deacylated P-site tRNA [41]. Based on this data, it was suggested that HflX could serve as an alternative splitting factor that recycles the post-termination ribosomes (generated via the regular or rescue system-induced peptide release) under conditions such as heat shock, when activity of the RRF•EF-G recycling complex is compromised. Although the dissociation activity of HflX on post-termination ribosomes is established, its ability to split stalled ribosomes carrying peptidyl-tRNA remains to be examined. The homologs of HflX, ribosome-dependent ATPase YchF and GTPase ObgE, could be other potential candidates for the ribosome-splitting activity in bacteria [42, 43].
HOW ARE THE JAMMED LARGE RIBOSOMAL SUBUNITS RESCUED IN BACTERIA
THAT LACK RqcH?
In B. subtilis, the C-terminal polyalanine tailing of the stalled nascent peptides is facilitated by the concerted action of RqcH and RqcP. Noteworthy, the ability of the RqcP homolog from E. coli, Hsp15, to bind the peptidyl-tRNA-jammed 50S subunits has been well documented [44]. Nevertheless, phylogenetic analysis has not been able to find the RqcH homologs in actinobacteria and gamma-proteobacteria (such as E. coli), raising the question of how the large ribosomal subunits occupied with peptidyl-tRNA are rescued in these bacterial species. It was speculated that E. coli Hsp15 can stabilize the 50S-bound peptidyl-tRNA in a P-site conformation competent for its hydrolysis by some release factor [45, 46]. If this is the case, then the ribosome rescue mechanism in actinobacteria and gamma-proteobacteria could, in principle, bypass the polyalanine-tailing step. Alternatively, polyalanine tailing in these species may be facilitated by the concerted action of Hsp15 and some RqcH-unrelated protein. It remains to be established which of these scenarios accounts for the resolution of the 50S•peptidyl-tRNA complexes in these species.
HOW IS THE NASCENT PEPTIDE RELEASED FROM THE 50S RIBOSOMAL
SUBUNIT?
Regardless of the polyalanine tailing, the ester bond of the large subunit-associated peptidyl-tRNA has to be hydrolyzed, and the nascent peptide needs to be released. The “canonical” bacterial rescue systems (SmpB•tmRNA, ArfA/ArfT, and ArfB) trigger hydrolytic activity of the ribosomal peptidyl-transferase center leading to peptidyl-tRNA hydrolysis. However, since these mechanisms operate exclusively upon the full 70S ribosomes, their involvement in the peptide release during the RQC-mediated rescue is unlikely. The peptidyl-tRNA Hydrolase (PTH), an essential protein that hydrolyzes ribosome-free peptidyl-tRNA [47, 48], and a mysterious RF-homolog PrfH, whose function in the cell remains enigmatic [49], could be candidates for such release activity. Lastly, existence of some tRNA-specific endonuclease with Vms1/ANKZF1-like activity also cannot be excluded.
MEDICAL SIGNIFICANCE OF THE BACTERIAL RQC SYSTEM
Infectious diseases caused by pathogenic bacteria are usually treated with antibiotics, small molecules that inhibit vital processes in bacterial cells, thereby preventing their proliferation [50]. Since the RQC-like rescue mechanism is not essential for viability of Bacillus subtilis [35], it may appear to be a poor target for antibiotics. However, RqcH homologs are essential for virulence of some pathogens, including S. pneumoniae, S. aureus, E. faecalis, and L. monocytogenes [51-54]. This fact makes bacterial RQC an attractive target for developing a fundamentally new class of medicines. In contrast to the traditional antibiotics that inhibit growth or kill bacteria, these drugs should keep pathogens at bay by neutralizing their virulence. In theory, this strategy could significantly reduce propensity of pathogens for developing drug resistance. Further studies of ribosome rescue systems in bacteria, including the RQC pathway, may reveal new cellular targets for the development of better anti-infectives. High potential of this strategy is confirmed by the recent discovery of acylaminooxadiazoles (including the so-called KKL-35 compound) as inhibitors of trans-translation with pronounced antibacterial properties [55, 56].
Acknowledgments. I want to express my deepest gratitude to my teacher Dr. Alexander Spirin and members of the Institute of Protein Research in Pushchino, where I gained priceless experience in the fields of protein biosynthesis and ribosomology. I am grateful to Alexander Richardson, Dr. Yury Polikanov, Dr. Nora Vázquez-Laslop, and Dr. Alexander Mankin for critical reading and fruitful discussion of the review.
Funding. This work was supported by the NIH grant R21-AI137584.
Ethics declarations. The author declares no conflicts of interest in financial or any other sphere. This article does not contain any studies involving human participants or animals performed by the author.
REFERENCES
1.Dai, X., Zhu, M., Warren, M., Balakrishnan, R.,
Patsalo, V., et al. (2016) Reduction of translating ribosomes enables
Escherichia coli to maintain elongation rates during slow
growth, Nat. Microbiol., 2, 16231.
2.Zhu, M., Dai, X., and Wang, Y.-P. (2016) Real time
determination of bacterial in vivo ribosome translation
elongation speed based on LacZɑ complementation system,
Nucleic Acid Res., 44, e155.
3.Jha, S., and Komar, A. A. (2011) Birth, life and
death of nascent polypeptide chains, Biotechnol. J., 6,
623-640.
4.Samatova, E., Daberger, J., Liutkute, M., and
Rodnina, M. V. (2021) Translational control by ribosome pausing in
bacteria: how a non-uniform pace of translation affects protein
production and folding, Front. Microbiol., 11,
619430.
5.Doerfel, L. K., Wohlgemuth, I., Kothe, C., Peske,
F., Urlaub, H., and Rodnina, M. V. (2013) EF-P is essential for rapid
synthesis of proteins containing consecutive proline residues,
Science, 339, 85-88.
6.Ude, S., Lassak, J., Starosta, A. L., Kraxenberger,
T., Wilson, D. N., and Jung, K. (2013) Translation elongation factor
EF-P alleviates ribosome stalling at polyproline stretches,
Science, 339, 82-85.
7.Huter, P., Arenz, S., Bock, L. V., Graf, M.,
Frister, J. O., et al. (2017) Structural basis for polyproline-mediated
ribosome stalling and rescue by the translation elongation factor EF-P,
Mol. Cell, 68, 515-527.
8.Pech, M., Karim, Z., Yamamoto, H., Kitakawa, M.,
Qin, Y., and Nierhaus, K. H. (2011) Elongation factor 4 (EF4/LepA)
accelerates protein synthesis at increased Mg2+
concentrations, Proc. Natl. Acad. Sci. USA, 108,
3199-3203.
9.Betney, R., de Silva, E., Krishnan, J., and
Stansfield, I. (2010) Autoregulatory systems controlling translation
factor expression: thermostat-like control of translational accuracy,
RNA, 16, 655-663.
10.Shalgi, R., Hurt, J. A., Krykbaeva, I., Taipale,
M., Lindquist, S., and Burge, C. B. (2013) Widespread regulation of
translation by elongation pausing in heat shock, Mol. Cell,
49, 439-452.
11.Vazquez-Laslop, N., and Mankin, A. S. (2018) How
macrolide antibiotics work, Trends Biochem. Sci., 43,
668-684.
12.Muller, C., Crowe-McAuliffe, C., and Wilson, D.
N. (2021) Ribosome rescue pathways in bacteria, Front.
Microbiol., 12, 652980.
13.Buskirk, A. R., and Green, R. (2017) Ribosome
pausing, arrest and rescue in bacteria and eukaryotes, Philos.
Trans. R. Soc. Lond. B. Biol. Sci., 372, 20160183.
14.Keiler, K. C. (2015) Mechanisms of ribosome
rescue in bacteria, Nat. Rev. Microbiol., 13,
285-297.
15.Keiler, K. C., Waller, P. R., and Sauer, R. T.
(1996) Role of a peptide tagging system in degradation of proteins
synthesized from damaged messenger RNA, Science, 271,
990-993.
16.Karzai, A. W., Susskind, M. M., and Sauer, R. T.
(1999) SmpB, a unique RNA-binding protein essential for the
peptide-tagging activity of SsrA (tmRNA), EMBO J., 18,
3793-3799.
17.Himeno, H., Nameki, N., Kurita, D., Muto, A., and
Abo, T. (2015) Ribosome rescue systems in bacteria, Biochimie,
114, 102-112.
18.Chadani, Y., Ito, K., Kutsukake, K., and Abo, T.
(2012) ArfA recruits release factor 2 to rescue stalled ribosomes by
peptidyl-tRNA hydrolysis in Escherichia coli, Mol.
Microbiol., 86, 37-50.
19.Shimizu, Y. (2012) ArfA recruits RF2 into stalled
ribosomes, J. Mol. Biol., 423, 624-631.
20.Goralski, T. D. P., Kirimanjeswara, G. S., and
Keiler, K. C. (2018) A new mechanism for ribosome rescue can recruit
RF1 or RF2 to nonstop ribosomes, mBio, 9, e02436-18.
21.Handa, Y., Inaho, N., and Nameki, N. (2011) YaeJ
is a novel ribosome-associated protein in Escherichia
coli that can hydrolyze peptidyl-tRNA on stalled ribosomes,
Nucleic Acids Res., 39, 1739-1748.
22.Gagnon, M. G., Seetharaman, S. V., Bulkley, D.,
and Steitz, T. A. (2012) Structural basis for the rescue of stalled
ribosomes: structure of YaeJ bound to the ribosome, Science,
335, 1370-1372.
23.Shoemaker, C. J., and Green, R. (2012)
Translation drives mRNA quality control, Nat. Struct. Mol.
Biol., 19, 594-601.
24.Pisarev, A. V., Skabkin, M. A., Pisareva, V. P.,
Skabkina, O. V., Rakotondrafara, A. M., et al. (2010) The role of ABCE1
in eukaryotic post-termination ribosomal recycling, Mol. Cell,
37, 196-210.
25.Shoemaker, C. J., Eyler, D. E., and Green, R.
(2010) Dom34:Hbs1 promotes subunit dissociation and peptidyl-tRNA
drop-off to initiate no-go decay, Science, 330,
369-372.
26.Joazeiro, C. A. P. (2019) Mechanisms and
functions of ribosome-associated protein quality control, Nat. Rev.
Mol. Cell Biol., 20, 368-383.
27.Shen, P. S., Park, J., Qin, Y., Li, X., Parsawar,
K., et al. (2015) Rqc2p and 60S ribosomal subunits mediate
mRNA-independent elongation of nascent chains, Science,
347, 75-78.
28.Verma, R., Reichermeier, K. M., Burroughs, A. M.,
Oania, R. S., Reitsma, J. M., et al. (2018) Vms1/ANKZF1 peptidyl-tRNA
hydrolases releases nascent chains from stalled ribosomes,
Nature, 557, 446-451.
29.Yip, M. C. J., Keszei, A. F. A., Feng, Q., Chu,
V., McKenna, M. J., and Shao, S. (2019) Mechanism for recycling tRNAs
on stalled ribosomes, Nat. Struct. Mol. Biol., 26,
343-349.
30.Yip, M. C. J., Savickas, S., Gygi, S. P., and
Shao, S. (2020) ELAC1 repairs tRNA cleaved during ribosome-associated
quality control, Cell Rep., 30, 2106-2114.
31.Chadani, Y., Ono, K., Kutsukake, K., and Abo, T.
(2011) Escherichia coli YaeJ protein mediates a novel ribosome-rescue
pathway distinct from SsrA- and ArfA-mediated pathways, Mol.
Microbiol., 80, 772-785.
32.Keiler, K. C., and Feaga, H. A. (2014) Resolving
nonstop translation complexes is a matter of life or death, J.
Bacteriol., 196, 2123-2130.
33.Burroughs, A. M., and Aravind, L. (2014) Analysis
of two domains with novel RNA-processing activities throws light on the
complex evolution of ribosomal RNA biogenesis, Front. Genet.,
5, 424.
34.Henderson, B., Nair, S., Pallas, J., and
Williams, M. A. (2011) Fibronectin: a multidomain host adhesin targeted
by bacterial fibronectin-binding proteins, FEMS Microbiol. Rev.,
35, 147-200.
35.Lytvynenko, I., Paternoga, H., Thrun, A., Balke,
A., Muller, T. A., et al. (2019) Alanine tails signal proteolysis in
bacterial ribosome-associated quality control, Cell, 178,
76-90.
36.Filbeck, S., Cerullo, F., Paternoga, H.,
Tsaprailis, G., Joazeiro, C. A. P., and Pfeffer, S. (2021) Mimicry of
canonical translation elongation underlies alanine tail synthesis in
RQC, Mol. Cell, 81, 104-114.
37.Crowe-McAuliffe, C., Takada, H., Murina, V.,
Polte, C., Kasvandik, S., et al. (2021) Structural basis for bacterial
ribosome-associated quality control by RqcH and RqcP, Mol. Cell,
81, 115-126.
38.Chadani, Y., Niwa, T., Izumi, T., Sugata, N.,
Nagao, A., et al. (2017) Intrinsic ribosome destabilization underlies
translation and provides an organism with a strategy of environmental
sensing, Mol. Cell, 68, 528-539.
39.Chadani, Y., Sugata, N., Niwa, T., Ito, Y.,
Iwasaki, S., and Taguchi, H. (2021) Nascent polypeptide within the exit
tunnel ensures continuous translation elongation by stabilizing the
translating ribosome, bioRxiv, doi:
10.1101/2021.02.02.429294.
40.Peske, F., Rodnina, M. V., and Wintermeyer, W.
(2005) Sequence of steps in ribosome recycling as defined by kinetic
analysis, Mol. Cell, 18, 403-412.
41.Zhang, Y., Mandava, C. S., Cao, W., Li, X.,
Zhang, D., et al. (2015) HflX is a ribosome-splitting factor rescuing
stalled ribosomes under stress conditions, Nat. Struct. Mol.
Biol., 22, 906-913.
42.Tomar, S. K., Kumar, P., and Prakash, B. (2011)
Deciphering the catalytic machinery in a universally conserved ribosome
binding ATPase YchF, Biochem. Biophys. Res. Commun., 408,
459-464.
43.Feng, B., Mandava, C. S., Guo, Q., Wang, J., Cao,
W., et al. (2014) Structural and functional insights into the mode of
action of a universally conserved Obg GTPase, PLos Biol.,
12, e1001866.
44.Korber, P., Stahl, J. M., Nierhaus, K. H., and
Bardwell, J. C. (2000) Hsp15: a ribosome-associated heat shock protein,
EMBO J., 19, 741-748.
45.Jiang, L., Schaffitzel, C., Bingel-Erlenmeyer,
R., Ban, N., Korber, P., et al. (2009) Recycling of aborted ribosomal
50S subunit-nascent chain-tRNA complexes by the heat shock protein
Hsp15, J. Mol. Biol., 386, 1357-1367.
46.Starosta, A. L., Lassak, J., Jung, K., and
Wilson, D. N. (2014) The bacterial translation stress response, FEMS
Microbiol. Rev., 38, 1172-1201.
47.Das, G., and Varshney, U. (2006) Peptidyl-tRNA
hydrolase and its critical role in protein biosynthesis,
Microbiology, 152, 2191-2195.
48.Sharma, S., Kaushik, S., Sinha, M., Kushwaha, G.
S., Singh, A., et al. (2014) Structural and functional insights into
peptidyl-tRNA hydrolase, Biochim. Biophys. Acta, 1844,
1279-1288.
49.Burroughs, A. M., and Aravind, L. (2019) The
origin and evolution of release factors: implications for translation
termination, ribosome rescue, and quality control pathways, Int. J.
Mol. Sci., 20, 1981.
50.Lewis, K. (2020) The science of antibiotic
discovery, Cell, 181, 29-45.
51.Holmes, A. R., McNab, R., Millsap, K. W., Rohde,
M., Hammerschmidt, S., et al. (2001) The pavA gene of
Streptococcus pneumoniae encodes a fibronectin-binding
protein that is essential for virulence, Mol. Microbiol.,
41, 1395-1408.
52.Pracht, D., Elm, C., Gerber, J., Bergmann, S.,
Rohde, M., et al. (2005) PavA of Streptococcus pneumoniae modulates
adherence, invasion, and meningeal inflammation, Infect. Immun.,
73, 2680-2689.
53.Singh, K. V., La Rossa, S. L., Somarajan, S. R.,
Roh, J. H., and Murray, B. E. (2015) The fibronectin-binding protein
EfbA contributes to pathogenesis and protects against infective
endocarditis caused by Enterococcus faecalis, Infect.
Immun., 83, 4487-4494.
54.Dramsi, S., Bourdichon, F., Cabanes, D., Lecuit,
M., Fsihi, H., and Cossart, P. (2004) FbpA, a novel multifunctional
Listeria monocytogenes virulence factor, Mol.
Microbiol., 53, 639-649.
55.Ramadoss, N. S., Alumasa, J. N., Cheng, L., Wang,
Y., Li, S., et al. (2013) Small molecule inhibitors of
trans-translation have broad-spectrum antibiotic activity, Proc.
Natl. Acad. Sci. USA, 110, 10282-10287.
56.Aron, Z. D., Mehrani, A., Hoffer, E. D.,
Connolly, K. L., Srinivas, P., et al. (2021) trans-Translation
inhibitors bind to a novel site on the ribosome and clear
Neisseria gonorrhoeae in vivo, Nat.
Commun., 12, 1799.