REVIEW: MicroRNAs in Cancer: From Gene Expression Regulation to the Metastatic Niche Reprogramming
Ekaterina V. Semina1,2,a*, Karina D. Rysenkova1,2, Konstantin E. Troyanovskiy2, Anna A. Shmakova1, and Kseniya A. Rubina2
1National Cardiology Research Center, Ministry of Health of the Russian Federation, 121552 Moscow, Russia2Faculty of Fundamental Medicine, Lomonosov Moscow State University, 119192 Moscow, Russia
* To whom correspondence should be addressed.
Received March 11, 2021; Revised March 11, 2021; Accepted March 11, 2021
By 2003, the Human Genome project had been completed; however, it turned out that 97% of genome sequences did not encode proteins. The explanation came later when it was found the untranslated DNA contain sequences for short microRNAs (miRNAs) and long noncoding RNAs that did not produce any mRNAs or tRNAs, but instead were involved in the regulation of gene expression. Initially identified in the cytoplasm, miRNAs have been found in all cell compartments, where their functions are not limited to the degradation of target mRNAs. miRNAs that are secreted into the extracellular space as components of exosomes or as complexes with proteins, participate in morphogenesis, regeneration, oncogenesis, metastasis, and chemoresistance of tumor cells. miRNAs play a dual role in oncogenesis: on one hand, they act as oncogene suppressors; on the other hand, they function as oncogenes themselves and inactivate oncosuppressors, stimulate tumor neoangiogenesis, and mediate immunosuppressive processes in the tumors, The review presents current concepts of the miRNA biogenesis and their functions in the cytoplasm and nucleus with special focus on the noncanonical mechanisms of gene regulation by miRNAs and involvement of miRNAs in oncogenesis, as well as the authors’ opinion on the role of miRNAs in metastasis and formation of the premetastatic niche.
KEY WORDS: microRNA, RISC, Argonaute, microRNA biogenesis, microRNA functions in nucleus, exosomes, extracellular microRNA, oncogenesis, metastasis, metastatic nicheDOI: 10.1134/S0006297921070014
Abbreviations: Ago, Argonaute protein; EMT, epithelial-mesenchymal transition; MET, mesenchymal-epithelial transition; RISC, RNA-induced silencing complex; UTR, untranslated region.
INTRODUCTION
The first microRNA (miRNA) was identified in 1993 in the nematode Caenorhabditis elegans in the lin-4 gene locus [1]. Lee et al. found that a 22-nucleotide RNA transcript of this gene is complementary to the mRNA of another gene, lin-14, and suppresses translation of the lin-14 mRNA required for nematode transition from the first to the second larval stage [1]. In 2000, two independent research groups showed that a small 21-nuleotide RNA, let-7, plays an important role in the development of C. elegans larvae into adult organisms [2, 3]. Since then, miRNAs have been then reported in many evolutionary distant organisms, including humans [4]. At present, miRNAs are defined as small noncoding evolutionary conservative RNAs of 18-25 nucleotides that participate in the gene expression regulation. Thousands of miRNAs have been identified; their description can be found in numerous databases, including miRbase (http://www.mirbase.org/), miRDB (http://mirdb.org/), and miRTarBase (http://mirtarbase.cuhk.edu.cn/php/index.php).
The most studied function of miRNAs is the regulation of gene expression via binding to the 3′-untranslated regions (UTRs) of target mRNAs and inhibition of their translation. However, it has been found recently that miRNAs also interact with other targets, such as gene promoters, coding sequences, and 5′-UTRs [5]. A growing body of data indicates that miRNAs can shuttle between different cell compartments, where they regulate various processes, including transcription, translation, alternative splicing, and DNA repair. Moreover, miRNAs are secreted into the extracellular space and can serve as molecular markers of oncologic diseases, in the development of which they may play a key role. It was suggested that miRNAs act as both suppressors of tumor growth and oncogenes stimulating carcinogenesis [6]. This review describes different pathways of miRNA biosynthesis, their functions in cells and upon secretion into the extracellular space, and potential role in the formation of premetastatic niche.
BIOGENESIS OF miRNAs
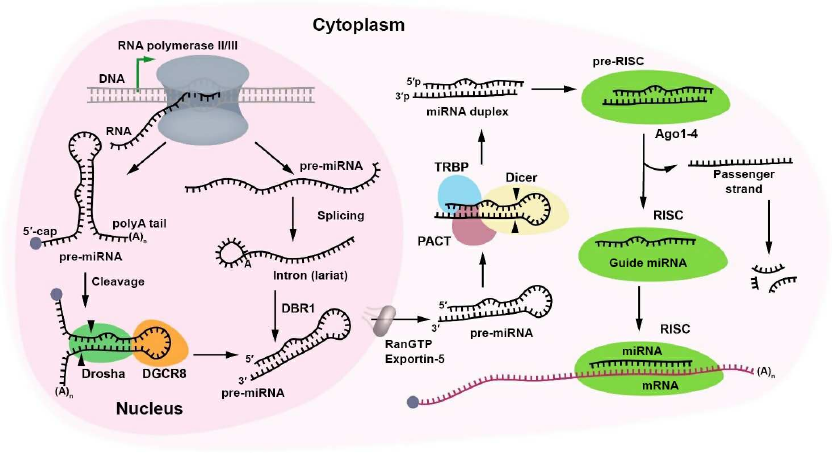
Biogenesis of miRNAs is a multistage process that starts with transcription of their genes. In the nucleus, miRNAs are transcribed by RNA polymerase II, either from their own promoters or from promoters of the host genes as long sequences called primary miRNAs (pri-miRNAs) [7]. The biogenesis of miRNAs can occur either through the canonical (Fig. 1) or noncanonical pathways, e.g., the mirtron pathway, in which miRNAs are produced in a Drosha-independent manner (see below).
Fig. 1. Biogenesis of miRNAs. Canonical biogenesis of miRNA starts with the synthesis of pri-miRNA transcript. A complex consisting of the ribonuclease III Drosha and DiGeorge syndrome critical region 8 (DGCR8) protein cleaves the primary pri-miRNA with the formation of miRNA precursor (pre-miRNA). Pri-miRNA is capped at the 5′-end and polyadenylated at the 3′-end. Pre-miRNA can also be produced by DBR1 (lariat debranching enzyme). After splicing, pre-miRNA is exported to the cytoplasm by the RanGTP/exportin-5 complex and cleaved by the ribonuclease Dicer in the presence of TRBP (transactivating response RNA-binding protein) and PACT (protein activator of interferon-induced protein kinase) cofactors with the generation of the miRNA duplex. The duplex binds the Ago1-4 proteins producing the RISC precursor. The passenger strand leaves the complex and is destroyed, whereas the guide miRNA remains in the RISC, ready for binding the mRNA target.
miRNA precursors can be encoded in short introns [8] that were named mirtrons. Such precursors have a hairpin structure similar to that of pre-miRNAs. Unlike classic pri-miRNAs, mirtrons are not processed by the Drosha–DGCR8 complex after their transcription (Fig. 1, right panel), but undergo splicing instead. The resulting product (similar to all other spliced introns) forms a lariat structure, in which the 5′-end of the intron is linked to the 2¢-OH group of adenosine residue. This 2¢-5′ phosphodiester bond is hydrolyzed by DBR1 (lariat debranching enzyme) [9]. The processed mirtron acquires the pre-miRNA structure and is exported to the cytoplasm, where it is cleaved by Dicer resulting in the generation of a 22-bp RNA duplex. RNA duplex is later transformed into the guide miRNA that regulates the expression of its target RNAs. In 2018, a program was developed for distinguishing mirtrons from precursors of canonical miRNAs based on the hairpin length and the GC-content, which might be for elucidating the mechanisms of mirtron processing [10].
At present, miRNA (miR)-451 is the only one known miRNA generated via a Dicer-independent pathway. It is processed by the enzyme Ago2, since Dicer cannot cleave it due to the short length of its hairpin (19 bp) [11]. Translation initiation eukaryotic factor 1A (EIF1A), which is a RISC component, plays an important role in the processing of pre-miR-451 by interacting with Ago2 and promoting its activation. miR-451 is then incorporated into the RISC in the cytoplasm and performs its well-known function, i.e., post-transcriptional regulation of gene expression [12].
FUNCTIONS OF miRNAs IN THE CYTOPLASM
The majority of miRNAs studies address the well-known activity of these molecules, such as binding to the 3′-UTRs of target mRNAs and their translational repression [13]. However, recently published papers report interactions between miRNA and the 5′-UTRs of mRNAs, leading to the opposite effect – activation of translation [14].
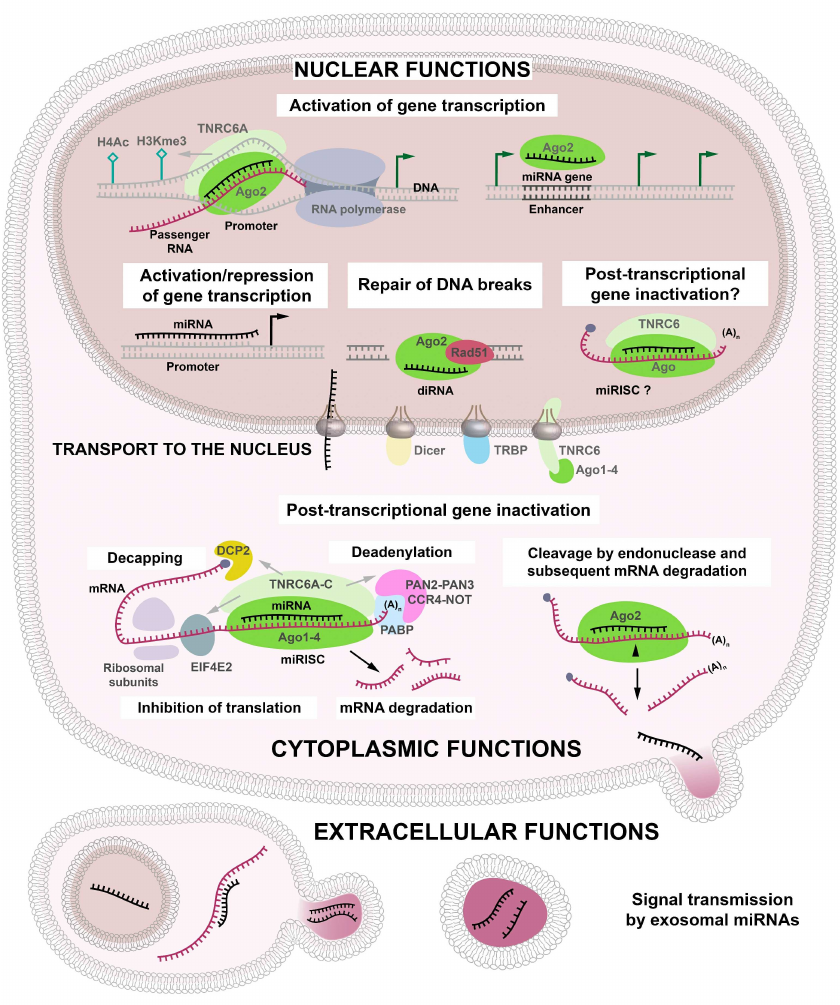
Ago proteins and guide miRNAs are the major components of the miRNA/RISC (miRISC) complex [15]. To initiate the endonuclease activity of Ago2 towards mRNA, it is necessary and sufficient to provide a complementary interaction between the seed sequence (nucleotides 2 to 8) of the miRNA and its target [16]. Seed sequences are commonly used for predicting the targets of miRNAs; however, it was shown that the complementarity of the seed sequence is not sufficient for the miRNA activity and that pairing between the 3′-end sequence of miRNA and the target RNA is required for the RISC functioning [5]. Moreover, interactions between miRNA and its target can be controlled by RNA-binding proteins. Some of these interactions are cell type specific and determine specific features of miRNA functioning in different cells [17]. The importance of Ago proteins in the miRISC formation has been confirmed by their involvement in the recruitment of TNRC6A-C proteins [18] (Fig. 2).
Fig. 2. Cytoplasmic, nuclear, and extracellular functions of miRNAs. In addition to their canonical role in the initiation of mRNA degradation in the cytoplasm, miRNAs can penetrate into the nucleus, where they participate in the activation/repression of gene transcription, repair of double-strand DNA breaks, and, presumably, post-transcriptional gene inactivation (PTGI). miRNAs incorporated into the exosomes can perform these functions in the recipient cells, either immediately in the zone of exosome release or after being transported to different organs and tissues with the blood flow.
Until recently, the general statement was that miRNAs suppress gene expression exclusively through degradation of the target mRNAs. However, in recent years, a large body of data has been accumulated indicating that miRNAs could act as both inhibitors and coactivators of translation. In particular, it was found that miRNA-15a interacts with the translation initiation factor eIF4E and inhibits its expression [19]. eIF4E recognizes the 5′-cap of the mRNA and recruits other factors, such as eIF4G, eIF2, eIF3, and RNA helicase eIF4A, as well as 40S small ribosomal subunit [20]. Other studies have shown that miRNAs can act as translation activators. Thus, a complex of let-7 with Ago2 and FXR1 (fragile-X-mental retardation-related) protein can activate translation in HeLa cells upon cell cycle arrest [21].
In the above examples, miRNAs were found to regulate gene expression in the cytoplasm: either directly, by degrading the target mRNAs, or indirectly, through modulating the translational complex activity, with these two mechanisms complementing each other. However, further studies are needed to find out which of the mechanisms predominates.
Recently, a new function of miR-1254 has been reported. In association with Ago2/miRISC, miR-1254 can interact with the 5′-UTR of the CCAR1 (cell cycle and apoptosis protein regulator) mRNA and upregulate CCAR1 expression. The 5′-UTR of the CCAR1 mRNA acts a natural stabilizer of miR-1254 (similarly to miRancers, which are artificially synthesized molecules that stabilize associated miRNAs). This interaction re-sensitized tamoxifen-resistant breast cancer cells to tamoxifen [22].
Unexpectedly, it has been discovered that several miRNAs can operate as either suppressors or activators of translation, depending on its pairing to the target (both processes required the presence of the Ago protein). Such miRNA was detected in the protozoan Giardia lamblia and named miR-3. miR-3 can repress translation of histone H2A mRNA in case of incomplete and imperfect pairing, but can enhance translation when the target mRNA is fully complementary [23]. It has been shown that a mismatch at the position 10 or 11 in the miRNA/target mRNA duplex makes the target RNA resistant to cleavage by Thermus thermophilus Argonaute protein [24]. Other examples of the dual role of miRNAs in various organisms also exist. miRNAs can influence the levels of mRNAs, micronucleoproteins, and ribonucleoproteins both directly and indirectly, through the regulation of their target promoters. Human miR-369-3 can activate translation of the of TNF-α mRNA upon the cell cycle arrest, but suppresses it in proliferating cell [21]. All these data indicate that miRNAs perform multiple functions in the cytoplasm that extend beyond their canonical role as the negative regulators of translation.
FUNCTIONS OF miRNAs IN THE NUCLEUS
It has been commonly accepted that miRNAs remain in the cell cytoplasm after their biogenesis, however, in 2004 Meister et al. detected microRNA-21 at high concentration in the nuclei of HeLa cells [25]. Later, the presence of mature miRNAs in the nuclear fraction has been confirmed in several studies. Hundreds of miRNAs have been identified by RNA sequencing in the nuclei of different cells. Nuclear localization of some of these miRNAs was corroborated by Northern blotting, RT-qPCR, RT-PCR, as well as in situ hybridization (ISH) – the methods which allow for excluding any signal detection from the miRNA precursors [26-29]. Three possible mechanisms ensuring nuclear localization of miRNAs are under consideration: (i) the presence of the nuclear localization signal in the miRNA sequence; (ii) independent nuclear biogenesis of miRNAs; (iii) shuttling of miRNAs between the cytoplasm and the nucleus via RISC [30] (Fig. 2).
Mechanisms of miRNA transport into the nucleus. The presence of nuclear localization signals has been demonstrated for miR-29b and miRNAs of the let-7 family [30]. miR-29b belongs to the miR-29 family and is one of the most extensively studied nuclear miRNAs. It differs from the other members of the family due to the presence of uridine at position 10 and the AGUGUU motif (positions 18-23) at the 3′-end. This motif is responsible for miR-29 translocation to the nucleus [31]. Two other miRNAs of the same family were detected in the nuclei of embryonic C166 cells [32]. Unlike miR-29b, miRs-29a and miR-29c lack the AGUGGU motif, but are significantly enriched in the nucleus, suggesting the existence of an alternative mechanisms for miRNA transport to the nucleus.
Moreover, miRNAs with the 5′-UUGCAUAGU-3′ and 5′-AGGUUGKSUG-3′ motifs (K = U/G) has been also detected in the nuclei of C166 cells. These motifs are present mainly in miRNAs of the let-7 family [32]. About a third of nuclear miRNAs has the ASUS consensus sequence (S = G/C). Therefore, it has been commonly accepted that some miRNAs contain nucleotide sequences that ensure their transport to the nucleus with the help of RNA-binding proteins, but the specific mechanisms and the proteins involved in the process, remain unresolved.
RISC components that have been initially identified in the cytoplasm (Ago1-4, TRBP, and TNRC6A), has also been found in the nuclei of mammalian cells [33-39]. The presence of these factors per se does not guarantee the loading of the nuclear miRNAs on the RISC, since the formation of the functional complex requires cytoplasmic HSP90, TRAX, and Translin proteins [34]. Apparently, some nuclear miRNAs, which are not functional can undergo degradation after their processing in the nucleus by Drosha and Dicer [30].
It has been reported that RISC formation may involve miRNA loading with assistance of some non-canonical proteins. One of the identified assistant proteins, AUF1, binds to miRNAs of the let-7 family and shuttles between the nucleus and the cytoplasm [40]. Although it remains still obscure, whether miRNAs are transported into the nucleus as a part of RISC, it was shown that the components of RISC can move between the nucleus and the cytoplasm operating as protein shuttles. Thus, proteins from the karyopherin family, such as exportin-1, exportin-5, importin-8, and karyopherins mediate shuttling of the proteins with classical nuclear localization and nuclear export signals through the nuclear pore complex [41].
Regulation of miRNA gene expression in the nucleus. As of today, miRNAs are commonly known to regulate gene expression by either transcriptional activating or repression. One of the mechanisms for gene transcription regulation is miRNA binding to gene promoters. In 2008 Place et al. demonstrated that miR-373 can activate gene expression of CSDC2 (cold shock domain-containing protein C2) and CDH1 (E-cadherin) proteins by interacting with their promoters in prostate carcinoma PC-3 cells, but fail to upregulate the synthesis of these proteins in another prostate carcinoma cell line LNCaP. In colorectal cancer HCT-116 cells only CSDC2 expression was induced [42], suggesting that miR-373 can differentially activate its target genes in various cell lines. It is also possible that gene activation by small RNAs depends on the epigenetic genome status. Thus, a small double-stranded (ds) RNA targeting the E-cadherin gene promoter induced E-cadherin expression in the prostate cancer PC-3 and DU-145 cell lines, but not in HeLa cells, because of the aberrant methylation of the E-cadherin gene in HeLa cells that prevented dsRNA binding. When used in combination with a demethylating agent, the same dsRNA was able to induce E-cadherin expression in HeLa cells [43]. Moreover, miR-373 interacted with sequences in the CDH1, CSCD2, and PDE4D genes in the breast carcinoma MCF-7 cells, but not in HeLa cells [44]. miR-552 inhibited expression of human cytochrome P450 2E1 (CYP2E1) via binding to the CYP2E1 gene promoter [45]. Taken together, these data confirm that the direct interaction of miRNAs with their target gene sequences exists as an additional mechanism of gene expression modulation besides the canonical binding of target mRNAs followed by the induction of RNA interference in the cytoplasm.
It has become clear that RISC components can localize both to the cytoplasm and to the nucleus, and therefore, can be involved in the miRNA-mediated post-translational inactivation of genes in the nucleus. It is also possible that precursors of miRNAs and other nuclear noncoding RNAs are undergo degradation in the nucleus. In 2013, Matsui et al. found that miR-589 inside the Ago2- and TNRC6A-containing complex can interact with the promoter-associated RNA of cyclooxygenase-2 (COX-2) gene, resulting in the activation of enzyme expression, although the exact mechanism of this regulation remains obscure. However, such interaction was accompanied by the increase in the chromatin marks associated with gene activation (H3K4me3) and histone H4 acetylation (H4Ac) [46]. MALAT1 (metastasis-associated lung adenocarcinoma transcript; >200 nucleotides) affiliated with the class of long noncoding RNAs (lncRNAs), was also shown to be controlled by post-transcriptional gene silencing: its binding to miR-9 resulted in Ago2-mediated MALAT1 cleavage [47]. Another example of this type of control is MACC1-AS1, which operates as an anti-sense lncRNA of the sixth intron of the MACC gene. The main function of this lncRNA is the modulation of cell proliferation and tumor progression in breast cancer. However, MACC1-AS1 binding to the tumor suppressors miR-384 and miR-145 promotes cell proliferation due to the increase in the expression of mRNA of pleiotrophin (PTN) and c-Myc mRNAs [48].
It has been shown that miRNAs-encoding sequences can be located in the enhancer loci of protein-coding genes. In a recent study [49], miR-26a-1, miR-339, miR-3179, miR-24-1, and miR-24-2 were found to induce expression of the neighboring genes. Thus, the miR-26a-1 gene is surrounded by the protein-coding ITGA9, CTDSPL, VILL, and PLCD1 genes. Overexpression of this miRNA in the kidney epithelium HEK293T cells causes transcriptional activation of two of these genes, ITGA9 and VILL, whereas miR-24-1 increases expression of the neighboring FBP1 and FANC genes [49]. Moreover, some miRNAs are indispensable for activation of gene transcription by their enhancers, since transcription was abolished when the enhancer sequence was deleted. Transcriptional activation of the neighboring genes also requires the presence of Ago2, indicating that miRNA alone cannot activate transcription. Taken together, these data suggest an existence of a new specific mechanism of gene expression regulation [49].
Interestingly, miRNAs can also participate in the regulation of alternative splicing factors. It was shown [50] that miRNAs are involved in alternative splicing coordination in the postnatal heart development through binding with the CELF (ELAV-like) proteins, which regulate mRNA stability and bind to the introns in pre-mRNA precursors, acting as alternative mediators of splicing. Two miRNAs, miR-23a and miR-23b, were found to target mRNAs for two CELF paralogs, CUG-binding proteins 1 and 2 (CUGBP1 and CUGBP2). Since these proteins have control over one half of all splice isoforms during heart embryogenesis, the authors suggested a hierarchy, in which rapid postnatal increase in specific miRs may control the expression of alternative splicing regulators and their downstream targets [50]. Another example of this regulation is the effect of miRNAs on the mRNA processing via participation of the splicing factors asd-2, hrp-2, and smu-2 that contain functional miRNA regulatory elements in their 3′-UTR sequences. It has been also discovered that the alternative splicing patterns of the respective downstream targets of these splicing factors (unc-60, unc-52, lin-10, and ret-1, main regulators of C. elegans development) was disrupted when the miRNA pathway was inhibited [51].
Mounting evidence of the last decade studies suggest that miRNAs participate in DNA repair. Double-strand DNA breaks (DSBs) initiate formation of a class of miRNAs [52] that are produced from the sequences in the vicinity of DSB sites and are processes by Dicer. Such miRNAs were termed diRNAs (DSB-induced small RNAs). A complex of diRNA with Ago2 can be transported to the DSB site, where Ago2 recruits RAD51, a key eukaryotic factor of DNA repair [52]. Different DNA modifications, such as oxidation, methylation, alkylation, and thymine replacement by uracil, can lead to spontaneous DNA damage. To protect DNA, cells utilize the base excision repair (BER) mechanism. BER enzymes specifically recognize and repair the damaged DNA. Some miRNAs are involved in the regulation of BER components. Thus, miR-16, miR-34c, and miR-199a can bind to the 3′-UTR of the mRNA that encodes the uracil-DNA glycosylase (UNG2), which is an enzyme responsible for elimination of uracil residues from DNA in tumor cells [53]. DNA polymerase β (Polβ) is regulated by miR-499, that binds to the 3′-UTR of the Polβ mRNA and activates its degradation [54].
In 2011, a group of Chinese researchers revealed another function of miRNAs, such as binding with pri-miRNAs in the nucleus. It was shown that miR-709 can bind to pri-miR-15a and pri-miR-16-1 preventing their further processing, thus suggesting a new, extremely unique role of miRNAs in the regulation of gene expression [55].
EXTRACELLULAR miRNAs AND THEIR FUNCTIONS AS COMPONENTS OF
EXOSOMES
Numerous studies indicate that miRNAs can be released into the extracellular medium and serve as biomarkers in various pathologies [56]. Extracellular miRNAs are extremely stable and withstand degradation for a prolonged period of time (days); for example, circulating miRNAs remain intact even when stored at room temperature or under unfavorable conditions, e.g., upon rapid change of temperature or in high or low pH medium [57]. Extracellular miRNAs exist as integral components of vesicles (exosomes, microvesicles, apoptotic bodies) or in a soluble form as protein-containing complexes (e.g., Ago2) [58]. High-density lipoproteins (HDLs) [59] and nucleophosmin 1 (NPM1) [60] can also bind extracellular miRNAs. These complexes ensure the miRNA stability outside the cells and in the blood.
Exosomes are extracellular vesicles with a diameter of 30-150 nm that are secreted by different types of cells. Exosomes contain a variety of biological molecules, such as proteins and nucleic acids [61] (see the ExoCarta database for is comprehensive description of the exosome composition; http://www.exocarta.org/). The loading of miRNAs into exosomes is controlled by hnRNPA2B1 and hnRNPA1 proteins that recognize specific sequence motifs [62]. The presence of extracellular complexes of miRNAs with Ago2 has been also described [63]. Exosomal miRNAs can be exported outside the cells via a ceramide-dependent mechanism, as the inhibition of neutral sphingomyelinase 2 (nSMase2), an enzyme involved in ceramide biosynthesis, downregulates secretion of exosomes and release of exosomal miRNAs into the extracellular space [64]. The above-described mechanisms of miRNA sorting into the exosomes suggest that exocytosis of miRNAs is a tightly controlled process. However, upon certain conditions, high miRNAs concentration in the exosomes can appear as a byproduct of other cellular processes, such as cell damage or death [65, 66].
At present, the mechanisms for the exosomal miRNAs uptake by cells are poorly understood, although several different pathways of miRNA penetration into the cells have been proposed. One of the possible mechanisms of exosomal miRNAs entry into the cells is via endocytosis, phagocytosis, micropinocytosis, the other is direct fusion of exosomes with the plasma membrane [67, 68]. Exosome-free miRNAs can also penetrate the cells through specific receptors [69].
Recently accumulated data indicate that extracellular miRNAs can act as signaling molecules and perform various functions in the recipient cells in both normal and pathological conditions (Table 1). Of special interest is the miRNA function as parts of extracellular vesicles and their role in the incipient and progressing malignancies. miRNAs-carrying exosomes released by tumor cells can be absorbed by the recipient cells. miRNAs that enter the cells can have an impact on tumor growth and stimulate/inhibit cell invasion, metastasis, and tumor neoangiogenesis. Moreover, exosomal miRNAs can also affect tumor microenvironment by modulating the extracellular matrix or through recruitment and activation of immune cells [70-74].
Table 1. Functions of some miRNAs in
physiological conditions
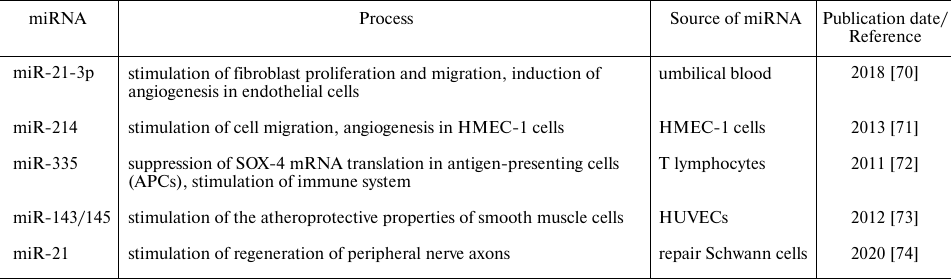
Recent studies have demonstrated that mesenchymal stromal cells (MSCs) actively produce miRNAs as a part of their secretome. These miRNAs operate as the anti-inflammatory and antifibrotic mediators and stimulate the growth of blood vessels and nerves during tissue regeneration [75]. However, the discovery of the anti-angiogenic miR-92a in the MSC secretome [76, 77] underlined the heterogeneity of the MSC population, as well as the dual role of this miRNA [78]. Thus, it was demonstrated that the effect of miR-92a is dependent on the specific cell line, experimental conditions, and cellular microenvironment [78]. The heterogeneity of the MSC population in terms of miR-29c and miR-21 production has been demonstrated by our colleagues who addressed the role of extracellular vesicles and incorporated miRs in association with fibrosis [79], despite of the well-known suppressing role of the MSC secretome in fibrosis [80].
To infer, numerous research groups in this field focus on miRNAs identification and studying their key functional features. At the same time, the mechanisms of miRNA delivery and uptake by recipient cells remain to be ascertained. We believe that further research in this area will offer a perspective on the context of potential practical application of exosomal miRNAs.
miRNAs AS ONCOSUPPRESSORS IN CANCER PROGRESSION AND
METASTASIS
Most of the papers in the field of miRNAs analyze the miRNA expression profiles in aberrant and normal cells. This comparative approach is also used in the studies addressing already known and yet to be identified miRNA and their functions (tumor suppressor or oncogenes) in cancer [6]. Oncosuppressor miRNAs apparently prevent tumor growth via suppression of oncogenes and/or genes controlling cell differentiation and apoptosis. Below, we discuss molecular mechanisms that underlie the oncosuppressor properties of several well-studied miRNAs and their potential effects on cancer cell invasion, metastasis, EMT, their proliferation and differentiation.
The studies on the role of miR-532-3p in prostate cancer progression in vitro revealed that overexpression of this miRNA suppressed invasion and migration of PC-3 prostate cancer cells due to the downregulation of the TRAF1/2/4 transcription factor expression and the decrease in NF-κB transcription factor activity. In vivo overexpression of miR-532-3p abrogated the formation of bone metastases by PC-3 cells [81]. A similar mechanism has been described for miR-3664-5P and miR-145-5p that suppress tumor progression through binding and inactivating the mediators of NF-κB signaling, thus decelerating invasion, migration, and metastasis of tumor cell of various origin [82, 83].
miRNAs can also control tumor growth and metastasis by suppressing the EMT, one of the essential processes in carcinogenesis that triggers metastasis and supports the chemoresistance of tumor cells. Downregulation of miR-2392 expression revealed that this miRNA suppresses invasion, migration, and metastasis of gastric cancer cells in vitro and in vivo. miR-2392 targets the MAML3 and WHSC1 genes and their downstream targets Slug and Twist1, respectively, which are transcriptional repressors of the CDH1 gene. Expression of the CDH1 gene supports the epithelial phenotype of tumor cells and prevents the onset of mesenchymal markers expression, typical for the migrating cells [84]. These features of miR-2392 and its ability to inhibit the EMT makes it a promising target in the therapy of highly metastatic gastric cancer. Another likely oncosuppressor miRNA that inhibits the EMT is miR-143-5p. It has been revealed that miR-143-5p exerts its inhibitory effects through downregulation of HIF-1α expression, which further decreases Twist1 and suppresses the EMT in the gall bladder cancer cells [85].
Disruption of the Wnt/β-catenin/TCF signaling in tumor cells can cause their hyperproliferation, dedifferentiation, and chemoresistance [86]. Recently, miR-148a was shown to suppress the Wnt/β-catenin-mediated proliferation and invasion of the PANC-1 pancreatic cancer cells in vitro and in vivo and to trigger apoptosis by upregulating the oncosuppressor MEG-3 (maternally expressed gene-3) [87]. miR-506 is another miRNA that controls the Wnt/β-catenin signaling pathway and EMT. The overexpression of this oncosuppressor miRNA inhibited expression of the homeobox protein LHX2 and transcription factor TCF4, as well as downregulated Wnt/β-catenin and Twist activity in a xenograft model in vivo, thus triggering apoptosis and suppressing metastasis of nasopharyngeal carcinoma cells in the lymph nodes. These experimental results are now being tested using clinical specimens [88].
Although the oncosuppressor properties of miRNAs attract a considerable interest of oncologists due to their potential use in cancer therapy, they require further detailed investigation. For example, in prostate cancer miR-532-3p acts as an oncosuppressor [81]; whereas in lung adenocarcinoma its expression is upregulated and contributes significantly to cancer progression thus functioning in this case as an oncogene [89].
miRNAs AS ONCOGENES IN CANCER PROGRESSION AND
METASTASIZING
miRNAs which are upregulated in tumors may be considered as oncogenes promoting tumor progression and metastasis acting via inhibition of tumor suppressor genes and/or genes involved in the control of cell differentiation and apoptosis. In 2009, the term metastamiR was proposed for metastasis-associated miRNAs [90], and since then, the role of such miRNAs has been actively studied. Metastasis is a stage of tumor progression associated with the negative course of oncologic diseases. It is a complex multistage process that includes dissemination of tumor cells from the primary tumor site into the surrounding stroma (invasion), blood or lymph, and then to the distant organs and tissues. Table 2 summarizes the functions of the most well-known miRNAs involved in the tumor progression.
Table 2. miRNAs involved in oncogenesis and
metastasis in various types of cancer
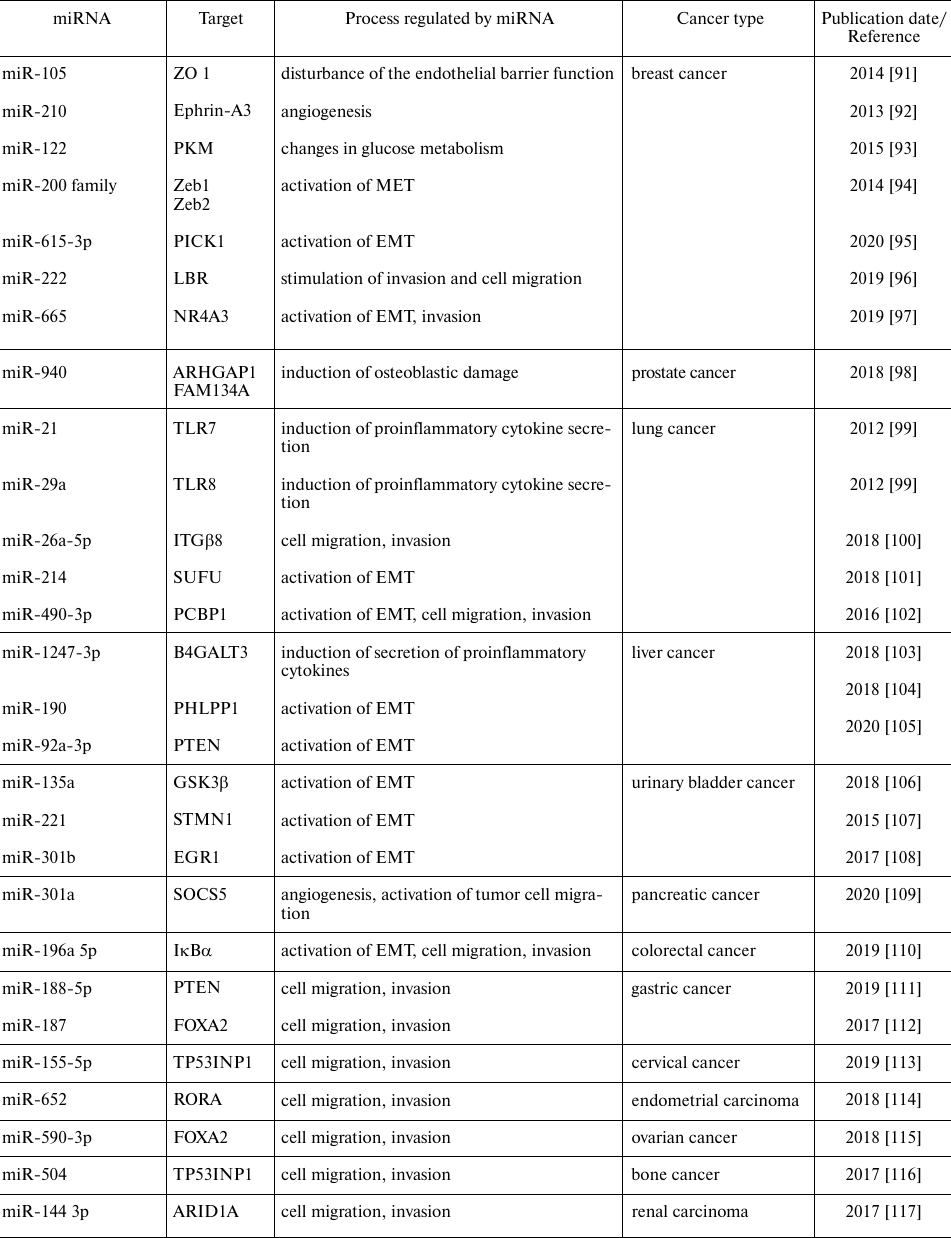
It is well known that tumor cells actively produce miRNAs-containing extracellular vesicles, which are used for information exchange between cancer cells and the surrounding stroma [118]. Invasion is preceded by the remodeling of the cancer cell niche and demolition of the structural proteins and cell-cell contacts, which facilitates the entry of tumor cells into the circulation. It has been shown that the highly metastatic breast cancer cells release the exosomal miR-105 that downregulates expression of the tight junction protein ZO-1 in the endothelial cells, thus disturbing the endothelial barrier function and facilitating migration of tumor cell across the vascular wall [91]. Stimulation of tumor neoangiogenesis is directly associated with the tumor growth and metastasis. It was shown that nSMase2 in the content of exosomes released by HUVECs (human umbilical vein endothelial cells) regulates expression of miR-210 in the breast cancer cells. In turn, exosomal miR-210 stimulates neoangiogenesis and metastasis by affecting expression of ephrin-A3 [92].
Another specific feature of tumor progression is reprograming of glucose metabolism not only in cancer cells per se, but also in the surrounding cells in the metastatic niche. Fong et al. demonstrated that exosomal miR-122 released by the breast cancer cell exerts their effects on normal pulmonary fibroblasts and astrocytes, i.e., on cells which are typically affected upon metastatic spreading in breast cancer. In particular, the authors revealed that tumor exosomes downregulated expression of the glycolytic enzyme pyruvate kinase in normal cells in vitro and in vivo [93]. Such reprogramming of systemic metabolism can lead to the increased availability of glucose for the tumor cells and promote tumor progression and formation of favorable environment in the metastatic niche.
Prostate cancer is characterized by bone metastasis and destruction of osteoblasts; however, the mechanism of this targeted and selective damage of osteoblasts is unknown. Exosomal miR-940 released by the tumor was shown to stimulate the osteogenic differentiation of human MSCs in vitro and induced large osteoblastic and osteolytic metastases formation in vivo in mice [98]. The fact that MSCs can reside in the bone tissue and differentiate in the osteogenic direction provides evidence that tumor miRNAs can reprogram the metastatic niche and generate the environment favorable for the development of secondary tumors.
Secretion of proinflammatory cytokines by tumor cells can change the tumor microenvironment and stimulate the growth and invasion of tumor cells. miR-21 and miR-29a in the exosomes released by A-549 and SK-MES lung cancer cells can bind Toll-like receptors 8 and 7 (TLR8 and TLR7) on the immune cells resulting in their activation. This interaction resulted in the NF-κB activation that triggered the cytokine secretion ultimately leading to further activation of the tumor microenvironment and resulting in invasion of tumor cells, metastasis, and secondary tumors [99]. In another study, cells of a the highly metastatic hepatocellular carcinoma secreted exosomal miR-1247-3p, which directly targeted the B4GALT3 gene, leading to the activation of the β1-integrin/NF-κB signaling pathway in the fibroblasts. As a result, the fibroblasts reprogrammed into cancer-associated fibroblasts (CAFs) that contribute to the tumor progression via secretion of the proinflammatory cytokines IL-6 and IL-8 and the formation of the tumor microenvironment [103].
High metastatic potential of cancer cells is a negative predictor of the clinical outcome, while the invasive status of the primary tumor cells serving as a diagnostic marker. miR-188-5p has been recognized to provoke metastasis of AGS and MGC803 gastric cancer cells by promoting their invasion with subsequent increase in the number of metastases to the lungs [111].
It is known that the level of miRNAs regulating the EMT process are elevated in blood serum of patients with metastatic cancers [119]. The miR-200 family comprises miR-200a, miR-200b, miR-200c, miR-429, and miR-141. The major function of these miRNAs is EMT suppression and triggering of the reverse process – the mesenchymal-epithelial transition (MET). MET is induced by direct binding of miR-200 with the Zeb1/2 transcriptional factors, followed by their subsequent inhibition and further repression of a various mesenchymal genes [119]. Lee et al. showed that the exosomal miR-200 produced by the highly metastatic 4T1 breast cancer cell line was absorbed by cells of the non-metastatic mammary gland tumor line, resulting in their altered gene expression and MET activation [94]. We demonstrated [120] that uPAR knock-down in Neuro2a neuroblastoma cells resulted in upregulated expression of miR-34c-5p and downregulated expression of miR-141-3p, miR-28a-5p, miR-291-3p, and miR-295-5p. Using bioinformatics, we have identified target gene clusters for these miRNAs and classified them into several functional group, such as genes involved in the EMT (Snai1, Zeb2), apoptosis (Bcl6, p21), proliferation (Atf1), cell adhesion and migration (CD93, ITGAV), exosome biogenesis (TSPAN2, TSPAN11, Rab11b, Rab21) [120].
Taken together, the above data point to an important role of the secreted extracellular vesicles containing miRNAs, which can act as humoral mediators of cell–cell communication, remodel the extracellular matrix, reprogram the premetastatic niche, and finally enable tumor cells to form metastasis and secondary tumors.
CONCLUSIONS
It is common knowledge that upon cancer progression and metastasis tumor cells can exert their effects far beyond their immediate microenvironment via paracrine signaling through the production of extracellular vesicles containing proteins, miRNAs, receptors, and signaling molecules. miRNAs of extracellular vesicles can be considered as markers of tumor chemoresistance and active fibrosis in the tumor surrounding tissues. In this connection, miRNAs application for diagnostics or as a potential therapeutic target will significantly expand an arsenal of current approaches in oncology, especially taking into consideration the fact that miRNA-containing exosomes lack the side effects and limitations of cellular technologies.
It should be emphasized that the bioinformatic search for miRNA targets is based on the in silico identification of the complementary sequences in the 3′-UTR of target mRNAs. The approaches based on the analysis of direct interactions of miRNAs with their target gene sequences or target proteins are rare. High expectations reside in the identification of miRNA receptors, which supposedly mediate endocytosis of miRNAs and determine the specificity of their effects on target cells. As demonstrated for the well-known let-7i, miRNAs not only perform their “classical” functions, such as regulation of gene expression and suppression of tumor cell invasion, but also act as ligands for Toll-like receptors [121] and activate the TLR-dependent signaling. Taking into consideration an important role of these receptors in antitumor immunity, the application of let-7i-based exosomes might become a new efficient method in the antitumor therapy.
To summarize, numerous data on the expression and functioning of exosomal miRNAs indicate that these molecules can be considered as humoral mediators in oncogenesis. miRNAs are not only the byproducts of the primary tumors, but can actively reprogram the cells in the metastatic niche. Therefore, the suppression of oncogenic miRNAs biosynthesis or the application of the antisense sequences might be a promising strategy in oncotherapy of the primary tumors and invasion/metastasis.
Funding. This work was supported by the Russian Foundation for Basic Research (project no. 20-015-00186).
Ethics declarations. The authors declare no conflict of interests. This article does not describe any research involving humans or animals conducted by any of the authors.
Open access. This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution, and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
REFERENCES
1.Lee, R. C., Feinbaum, R. L., and Ambros, V. (1993)
The C. elegans heterochronic gene lin-4 encodes small RNAs with
antisense complementarity to lin-14, Cell, 75, 843-854,
doi: 10.1016/0092-8674(93)90529-Y.
2.Slack, F. J., Basson, M., Liu, Z., Ambros, V.,
Horvitz, H. R., et al. (2000) The lin-41 RBCC gene acts in the C.
elegans heterochronic pathway between the let-7 regulatory RNA and the
LIN-29 transcription factor, Mol. Cell, 5, 659-669, doi:
10.1016/S1097-2765(00)80245-2.
3.Reinhart, B. J., Slack, F. J., Basson, M.,
Pasquinelli, A. E., Bettlnger, J. C., et al. (2000) The 21-nucleotide
let-7 RNA regulates developmental timing in Caenorhabditis
elegans, Nature, 403, 901-906, doi:
10.1038/35002607.
4.Pasquinelli, A. E., Reinhart, B. J., Slack, F.,
Martindale, M. Q., Kuroda, M. I., et al. (2000) Conservation of the
sequence and temporal expression of let-7 heterochronic regulatory RNA,
Nature, 408, 86-89, doi: 10.1038/35040556.
5.Broughton, J. P., Lovci, M. T., Huang, J. L., Yeo,
G. W., and Pasquinelli, A. E. (2016) Pairing beyond the seed supports
microRNA targeting specificity, Mol. Cell, 64, 320-333,
doi: 10.1016/j.molcel.2016.09.004.
6.Fasoulakis, Z., Daskalakis, G., Diakosavvas, M.,
Papapanagiotou, I., Theodora, M., et al. (2019) MicroRNAs determining
carcinogenesis by regulating oncogenes and tumor suppressor genes
during cell cycle, MicroRNA, 9, 82-92, doi:
10.2174/2211536608666190919161849.
7.Lee, Y., Kim, M., Han, J., Yeom, K. H., Lee, S., et
al. (2004) MicroRNA genes are transcribed by RNA polymerase II, EMBO
J., 23, 4051-4060, doi: 10.1038/sj.emboj.7600385.
8.Berezikov, E., Chung, W. J., Willis, J., Cuppen,
E., and Lai, E. C. (2007) Mammalian mirtron genes, Mol. Cell,
28, 328-336, doi: 10.1016/j.molcel.2007.09.028.
9.Ruby, J. G., Jan, C. H., and Bartel, D. P. (2007)
Intronic microRNA precursors that bypass Drosha processing,
Nature, 448, 83-86, doi: 10.1038/nature05983.
10.Rorbach, G., Unold, O., and Konopka, B. M. (2018)
Distinguishing mirtrons from canonical miRNAs with data exploration and
machine learning methods, Sci. Rep., 8, 7560, doi:
10.1038/s41598-018-25578-3.
11.Cifuentes, D., Xue, H., Taylor, D. W., Patnode,
H., Mishima, Y., et al. (2010) A novel miRNA processing pathway
independent of dicer requires argonaute2 catalytic activity,
Science, 328, 1694-1698, doi:
10.1126/science.1190809.
12.Yi, T., Arthanari, H., Akabayov, B., Song, H.,
Papadopoulos, E., et al. (2015) EIF1A augments Ago2-mediated
dicer-independent miRNA biogenesis and RNA interference, Nat.
Commun., 6, 7194, doi: 10.1038/ncomms8194.
13.Huntzinger, E., and Izaurralde, E. (2011) Gene
silencing by microRNAs: Contributions of translational repression and
mRNA decay, Nat. Rev. Genet., 12, 99-110, doi:
10.1038/nrg2936.
14.Kehl, T., Backes, C., Kern, F., Fehlmann, T.,
Ludwig, N., et al. (2017) About miRNAs, miRNA seeds, target genes and
target pathways, Oncotarget, 8, 107167-107175, doi:
10.18632/oncotarget.22363.
15.Kawamata, T., and Tomari, Y. (2010) Making RISC,
Trends Biochem. Sci., 35, 368-376, doi:
10.1016/j.tibs.2010.03.009.
16.Lewis, B. P., Burge, C. B., and Bartel, D. P.
(2005) Conserved seed pairing, often flanked by adenosines, indicates
that thousands of human genes are microRNA targets, Cell,
120, 15-20, doi: 10.1016/j.cell.2004.12.035.
17.Nussbacher, J. K., and Yeo, G. W. (2018)
Systematic discovery of RNA binding proteins that regulate microRNA
levels, Mol. Cell, 69, 1005-1016.e7, doi:
10.1016/j.molcel.2018.02.012.
18.Behm-Ansmant, I., Rehwinkel, J., Doerks, T.,
Stark, A., Bork, P., et al. (2006) mRNA degradation by miRNAs and GW182
requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes,
Genes Dev., 20, 1885-1898, doi: 10.1101/gad.1424106.
19.Li, G., Chong, T., Xiang, X., Yang, J., and Li,
H. (2017) Downregulation of microRNA-15a suppresses the proliferation
and invasion of renal cell carcinoma via direct targeting of eIF4E,
Oncol. Rep., 38, 1995-2002, doi:
10.3892/or.2017.5901.
20.Gingras, A. C., Raught, B., and Sonenberg, N.
(1999) eIF4 initiation factors: Effectors of mRNA recruitment to
ribosomes and regulators of translation, Annu. Rev. Biochem.,
68, 913-963, doi: 10.1146/annurev.biochem.68.1.913.
21.Vasudevan, S., Tong, Y., and Steitz, J. A. (2007)
Switching from repression to activation: MicroRNAs can up-regulate
translation, Science, 318, 1931-1934, doi:
10.1126/science.1149460.
22.Li, G., Wu, X., Qian, W., Cai, H., Sun, X., et
al. (2016) CCAR1 5′-UTR as a natural miRancer of miR-1254
overrides tamoxifen resistance, Cell Res., 26, 655-673,
doi: 10.1038/cr.2016.32.
23.Saraiya, A. A., Li, W., and Wang, C. C. (2013)
Correction: transition of a microRNA from repressing to activating
translation depending on the Extent of base pairing with the target,
PLoS One, 8, doi:
10.1371/annotation/cb23f7bd-0d8c-4fa2-8ce8-1d641c03f561.
24.Wang, Y., Juranek, S., Li, H., Sheng, G., Tuschl,
T., et al. (2008) Structure of an argonaute silencing complex with a
seed-containing guide DNA and target RNA duplex, Nature,
456, 921-926, doi: 10.1038/nature07666.
25.Meister, G., Landthaler, M., Patkaniowska, A.,
Dorsett, Y., Teng, G., et al. (2004) Human Argonaute2 mediates RNA
cleavage targeted by miRNAs and siRNAs, Mol. Cell, 15,
185-197, doi: 10.1016/j.molcel.2004.07.007.
26.Wong, J. J. L., Ritchie, W., Gao, D., Lau, K. A.,
Gonzalez, M., et al. (2014) Identification of nuclear-enriched miRNAs
during mouse granulopoiesis, J. Hematol. Oncol., 7, 42,
doi: 10.1186/1756-8722-7-42.
27.Ritland Politz, J. C., Hogan, E. M., and
Pederson, T. (2009) MicroRNAs with a nucleolar location, RNA,
15, 1705-1715, doi: 10.1261/rna.1470409.
28.Park, C. W., Zeng, Y., Zhang, X., Subramanian,
S., and Steer, C. J. (2010) Mature microRNAs identified in highly
purified nuclei from HCT116 colon cancer cells, RNA Biol.,
7, 606-614, doi: 10.4161/rna.7.5.13215.
29.Khudayberdiev, S. A., Zampa, F., Rajman, M., and
Schratt, G. (2013) A comprehensive characterization of the nuclear
microRNA repertoire of post-mitotic neurons, Front. Mol.
Neurosci., 6, 43, doi: 10.3389/fnmol.2013.00043.
30.Stavast, C. J., and Erkeland, S. J. (2019) The
non-canonical aspects of microRNAs: many roads to gene regulation,
Cells, 8, 1465, doi: 10.3390/cells8111465.
31.Hwang, H. W., Wentzel, E. A., and Mendell, J. T.
(2007) A hexanucleotide element directs microRNA nuclear import,
Science, 315, 97-100, doi: 10.1126/science.1136235.
32.Turunen, T. A., Roberts, T. C., Laitinen, P.,
Väänänen, M. A., Korhonen, P., et al. (2019) Changes in
nuclear and cytoplasmic microRNA distribution in response to hypoxic
stress, Sci. Rep., 9, 10332, doi:
10.1038/s41598-019-46841-1.
33.Sarshad, A. A., Juan, A. H., Muler, A. I. C.,
Anastasakis, D. G., Wang, X., et al. (2018) Argonaute-miRNA complexes
silence target mRNAs in the nucleus of mammalian stem cells, Mol.
Cell, 71, 1040-1050.e8, doi:
10.1016/j.molcel.2018.07.020.
34.Gagnon, K. T., Li, L., Chu, Y., Janowski, B. A.,
and Corey, D. R. (2014) RNAi factors are present and active in human
cell nuclei, Cell Rep., 6, 211-221, doi:
10.1016/j.celrep.2013.12.013.
35.Robb, G. B., Brown, K. M., Khurana, J., and Rana,
T. M. (2005) Specific and potent RNAi in the nucleus of human cells,
Nat. Struct. Mol. Biol., 12, 133-137, doi:
10.1038/nsmb886.
36.Rüdel, S., Flatley, A., Weinmann, L.,
Kremmer, E., and Meister, G. (2008) A multifunctional human
Argonaute2-specific monoclonal antibody, RNA, 14,
1244-1253, doi: 10.1261/rna.973808.
37.Nishi, K., Nishi, A., Nagasawa, T., and Ui-Tei,
K. (2013) Human TNRC6A is an Argonaute-navigator protein for
microRNA-mediated gene silencing in the nucleus, RNA, 19,
17-35, doi: 10.1261/rna.034769.112.
38.Roya, K., Jessica, A. H., Liande, L. I., Keith,
T. G., Viswanadham, S., et al. (2016) Stable association of RNAi
machinery is conserved between the cytoplasm and nucleus of human
cells, RNA, 22, 1085-1098, doi:
10.1261/rna.056499.116.
39.Ohrt, T., Mütze, J., Staroske, W., Weinmann,
L., Höck, J., et al. (2008) Fluorescence correlation spectroscopy
and fluorescence cross-correlation spectroscopy reveal the cytoplasmic
origination of loaded nuclear RISC in vivo in human cells,
Nucleic Acids Res., 36, 6439-6449, doi:
10.1093/nar/gkn693.
40.Yoon, J. H., Jo, M. H., White, E. J. F., De, S.,
Hafner, M., et al. (2015) AUF1 promotes let-7b loading on argonaute 2,
Genes Dev., 29, 1599-1604, doi:
10.1101/gad.263749.115.
41.Schraivogel, D., Schindler, S. G., Danner, J.,
Kremmer, E., Pfaff, J., et al. (2015) Importin-b facilitates nuclear
import of human GW proteins and balances cytoplasmic gene silencing
protein levels, Nucleic Acids Res., 43, 7447-7461, doi:
10.1093/nar/gkv705.
42.Place, R. F., Li, L. C., Pookot, D., Noonan, E.
J., and Dahiya, R. (2008) MicroRNA-373 induces expression of genes with
complementary promoter sequences, Proc. Natl. Acad. Sci. USA,
105, 1608-1613, doi: 10.1073/pnas.0707594105.
43.Li, L. C., Okino, S. T., Zhao, H., Pookot, D.,
Place, R. F., et al. (2006) Small dsRNAs induce transcriptional
activation in human cells, Proc. Natl. Acad. Sci. USA,
103, 17337-17342, doi: 10.1073/pnas.0607015103.
44.Xun, Y., Tang, Y., Hu, L., Xiao, H., Long, S., et
al. (2019) Purification and identification of miRNA target sites in
genome using DNA affinity precipitation, Front. Genet.,
10, 778, doi: 10.3389/fgene.2019.00778.
45.Miao, L., Yao, H., Li, C., Pu, M., Yao, X., et
al. (2016) A dual inhibition: MicroRNA-552 suppresses both
transcription and translation of cytochrome P450 2E1, Biochim.
Biophys. Acta Gene Regul. Mech., 1859, 650-662, doi:
10.1016/j.bbagrm.2016.02.016.
46.Matsui, M., Chu, Y., Zhang, H., Gagnon, K. T.,
Shaikh, S., et al. (2013) Promoter RNA links transcriptional regulation
of inflammatory pathway genes, Nucleic Acids Res., 41,
10086-10109, doi: 10.1093/nar/gkt777.
47.Leucci, E., Patella, F., Waage, J.,
Holmstrøm, K., Lindow, M., et al. (2013) MicroRNA-9 targets the
long non-coding RNA MALAT1 for degradation in the nucleus, Sci.
Rep., 3, 2535, doi: 10.1038/srep02535.
48.Zhang, X., Zhou, Y., Chen, S., Li, W., Chen, W.,
et al. (2019) LncRNA MACC1-AS1 sponges multiple miRNAs and RNA-binding
protein PTBP1, Oncogenesis, 8, 73, doi:
10.1038/s41389-019-0182-7.
49.Xiao, M., Li, J., Li, W., Wang, Y., Wu, F., et
al. (2017) MicroRNAs activate gene transcription epigenetically as an
enhancer trigger, RNA Biol., 14, 1326-1334, doi:
10.1080/15476286.2015.1112487.
50.Kalsotra, A., Wang, K., Li, P. F., and Cooper, T.
A. (2010) MicroRNAs coordinate an alternative splicing network during
mouse postnatal heart development, Genes Dev., 24,
653-658, doi: 10.1101/gad.1894310.
51.Kotagama, K., Schorr, A. L., Steber, H. S., and
Mangone, M. (2018) miRNA activity contributes to accurate RNA splicing
in C. elegans intestine and body muscle tissues, bioRxiv,
479832, doi: 10.1101/479832.
52.Gao, M., Wei, W., Li, M. M., Wu, Y. S., Ba, Z.,
et al. (2014) Ago2 facilitates Rad51 recruitment and DNA double-strand
break repair by homologous recombination, Cell Res., 24,
532-541, doi: 10.1038/cr.2014.36.
53.Hegre, S. A., Sætrom, P., Aas, P. A.,
Pettersen, H. S., Otterlei, M., et al. (2013) Multiple microRNAs may
regulate the DNA repair enzyme uracil-DNA glycosylase, DNA Repair
(Amst.), 12, 80-86, doi: 10.1016/j.dnarep.2012.10.007.
54.Wang, Y., Feng, J., Zang, W., Du, Y., Chen, X.,
et al. (2015) MIR-499 Enhances the cisplatin sensitivity of esophageal
carcinoma cell lines by targeting DNA polymerase b, Cell. Physiol.
Biochem., 36, 1587-1596, doi: 10.1159/000430321.
55.Tang, R., Li, L., Zhu, D., Hou, D., Cao, T., et
al. (2012) Mouse miRNA-709 directly regulates miRNA-15a/16-1 biogenesis
at the posttranscriptional level in the nucleus: evidence for a
microRNA hierarchy system, Cell Res., 22, 504-515, doi:
10.1038/cr.2011.137.
56.Pereira-da-Silva, T., Coutinho Cruz, M.,
Carrusca, C., Cruz Ferreira, R., Napoleão, P., et al. (2018)
Circulating microRNA profiles in different arterial territories of
stable atherosclerotic disease: a systematic review, Am. J.
Cardiovasc. Dis., 8, 1-13.
57.Turchinovich, A., Weiz, L., Langheinz, A., and
Burwinkel, B. (2011) Characterization of extracellular circulating
microRNA, Nucleic Acids Res., 39, 7223-7233, doi:
10.1093/nar/gkr254.
58.Turchinovich, A., and Burwinkel, B. (2012)
Distinct AGO1 and AGO2 associated miRNA profiles in human cells and
blood plasma, RNA Biol., 9, 1066-1075, doi:
10.4161/rna.21083.
59.Canfrán-Duque, A., Lin, C. S., Goedeke,
L., Suárez, Y., and Fernández-Hernando, C. (2016)
Micro-RNAs and high-density lipoprotein metabolism, Arterioscler.
Thromb. Vasc. Biol., 36, 1076-1084, doi:
10.1161/ATVBAHA.116.307028.
60.Hasan, S., Gadewal, N., Aher, S., Kumar, R.,
Varma, A., et al. (2018) Identification of miRNA-mRNA network in NPM1
mutated acute myeloid leukemia, Clin. Lymphoma Myeloma Leuk.,
18, S193, doi: 10.1016/j.clml.2018.07.035.
61.Li, M., Zeringer, E., Barta, T., Schageman, J.,
Cheng, A., et al. (2014) Analysis of the RNA content of the exosomes
derived from blood serum and urine and its potential as biomarkers,
Philos. Trans. R. Soc. B Biol. Sci., 369, 20130502, doi:
10.1098/rstb.2013.0502.
62.Villarroya-Beltri, C.,
Gutiérrez-Vázquez, C., Sánchez-Cabo, F.,
Pérez-Hernández, D., Vázquez, J., et al. (2013)
Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes
through binding to specific motifs, Nat. Commun., 4,
1-10, doi: 10.1038/ncomms3980.
63.Li, L., Zhu, D., Huang, L., Zhang, J., Bian, Z.,
et al. (2012) Argonaute 2 complexes selectively protect the circulating
microRNAs in cell-secreted microvesicles, PLoS One, 7,
e46957, doi: 10.1371/journal.pone.0046957.
64.Kubota, S., Chiba, M., Watanabe, M., Sakamoto,
M., and Watanabe, N. (2015) Secretion of small/microRNAs including
miR-638 into extracellular spaces by sphingomyelin phosphodiesterase 3,
Oncol. Rep., 33, 67-73, doi: 10.3892/or.2014.3605.
65.Makarova, J., Turchinovich, A., Shkurnikov, M.,
and Tonevitsky, A. (2021) Extracellular miRNAs and cell–cell
communication: problems and prospects, Trends Biochem. Sci.,
doi: 10.1016/j.tibs.2021.01.007.
66.Kakarla, R., Hur, J., Kim, Y. J., Kim, J., and
Chwae, Y. J. (2020) Apoptotic cell-derived exosomes: messages from
dying cells, Exp. Mol. Med., 52, 1-6, doi:
10.1038/s12276-019-0362-8.
67.Tian, T., Zhu, Y. L., Zhou, Y. Y., Liang, G. F.,
Wang, Y. Y., et al. (2014) Exosome uptake through clathrin-mediated
endocytosis and macropinocytosis and mediating miR-21 delivery, J.
Biol. Chem., 289, 22258-22267, doi:
10.1074/jbc.M114.588046.
68.Makarova, J. A., Shkurnikov, M. U., Wicklein, D.,
Lange, T., Samatov, T. R., et al. (2016) Intracellular and
extracellular microRNA: an update on localization and biological role,
Prog. Histochem. Cytochem., 51, 33-49, doi:
10.1016/j.proghi.2016.06.001.
69.Prud’homme, G. J., Glinka, Y., Lichner, Z.,
and Yousef, G. M. (2016) Neuropilin-1 is a receptor for extracellular
miRNA and AGO2/miRNA complexes and mediates the internalization of
miRNAs that modulate cell function, Oncotarget, 7,
68057-68071, doi: 10.18632/ONCOTARGET.10929.
70.Hu, Y., Rao, S. S., Wang, Z. X., Cao, J., Tan, Y.
J., et al. (2018) Exosomes from human umbilical cord blood accelerate
cutaneous wound healing through miR-21-3p-mediated promotion of
angiogenesis and fibroblast function, Theranostics, 8,
169-184, doi: 10.7150/thno.21234.
71.Van Balkom, B. W. M., Jong, O. G. D., Smits, M.,
Brummelman, J., den Ouden, K., et al. (2013) Endothelial cells require
miR-214 to secrete exosomes that suppress senescence and induce
angiogenesis in human and mouse endothelial cells, Blood,
121, 3997-4006, doi: 10.1182/blood-2013-02-478925.
72.Mittelbrunn, M., Gutiérrez-Vázquez,
C., Villarroya-Beltri, C., González, S., Sánchez-Cabo,
F., et al. (2011) Unidirectional transfer of microRNA-loaded exosomes
from T cells to antigen-presenting cells, Nat. Commun.,
2, 282, doi: 10.1038/ncomms1285.
73.Hergenreider, E., Heydt, S., Tréguer, K.,
Boettger, T., Horrevoets, A. J. G., et al. (2012) Atheroprotective
communication between endothelial cells and smooth muscle cells through
miRNAs, Nat. Cell Biol., 14, 249-256, doi:
10.1038/ncb2441.
74.López-Leal, R.,
Díaz-Viraqué, F., Catalán, R. J., Saquel, C.,
Enright, A., et al. (2020) Schwann cell reprogramming into repair cells
increases miRNA-21 expression in exosomes promoting axonal growth,
J. Cell Sci., 133, jcs239004, doi:
10.1242/jcs.239004.
75.Dong, R., Liu, Y., Yang, Y., Wang, H., Xu, Y., et
al. (2019) MSC-derived exosomes-based therapy for peripheral nerve
injury: a novel therapeutic strategy, Biomed Res. Int.,
2019, 6458237, doi: 10.1155/2019/6458237.
76.Bonauer, A., Carmona, G., Iwasaki, M., Mione, M.,
Koyanagi, M., et al. (2009) MicroRNA-92a controls angiogenesis and
functional recovery of ischemic tissues in mice, Science,
324, 1710-1713, doi: 10.1126/science.1174381.
77.Efimenko, A., Sagaradze, G., Akopyan, Z.,
Lopatina, T., and Kalinina, N. (2016) Data supporting that miR-92a
suppresses angiogenic activity of adipose-derived mesenchymal stromal
cells by down-regulating hepatocyte growth factor, Data Br.,
6, 295-310, doi: 10.1016/j.dib.2015.12.021.
78.Zhang, L., Zhou, M., Qin, G., Weintraub, N. L.,
and Tang, Y. (2014) MiR-92a regulates viability and angiogenesis of
endothelial cells under oxidative stress, Biochem. Biophys. Res.
Commun., 446, 952-958, doi: 10.1016/j.bbrc.2014.03.035.
79.Basalova, N., Sagaradze, G., Arbatskiy, M.,
Evtushenko, E., Kulebyakin, K., et al. (2020) Secretome of mesenchymal
stromal cells prevents myofibroblasts differentiation by transferring
fibrosis-associated microRNAs within extracellular vesicles,
Cells, 9, 1272, doi: 10.3390/cells9051272.
80.Chuang, H. M., Shih, T. E., Lu, K. Y., Tsai, S.
F., Harn, H. J., et al. (2018) Mesenchymal stem cell therapy of
pulmonary fibrosis: improvement with target combination, Cell
Transplant., 27, 1581-1587, doi:
10.1177/0963689718787501.
81.Wa, Q., Zou, C., Lin, Z., Huang, S., Peng, X., et
al. (2020) Ectopic expression of miR-532-3p suppresses bone metastasis
of prostate cancer cells via inactivating NF-kB signaling, Mol.
Ther. Oncolytics, 17, 267-277, doi:
10.1016/j.omto.2020.03.024.
82.Jin, C., Wang, A., Liu, L., Wang, G., Li, G., et
al. (2019) miR-145-5p inhibits tumor occurrence and metastasis through
the NF-kB signaling pathway by targeting TLR4 in malignant melanoma,
J. Cell. Biochem., 120, 11115-11126, doi:
10.1002/jcb.28388.
83.Jiao, Y., Yang, H., Qian, J., Gong, Y., Liu, H.,
et al. (2019) MiR-3664-5P suppresses the proliferation and metastasis
of gastric cancer by attenuating the NF-kB signaling pathway through
targeting MTDH, Int. J. Oncol., 54, 845-858, doi:
10.3892/ijo.2019.4680.
84.Li, J., Li, T., Lu, Y., Shen, G., Guo, H., et al.
(2017) MiR-2392 suppresses metastasis and epithelial-mesenchymal
transition by targeting MAML3 and WHSC1 in gastric cancer, FASEB
J., 31, 3774-3786, doi: 10.1096/fj.201601140RR.
85.He, M., Zhan, M., Chen, W., Xu, S., Long, M., et
al. (2017) MiR-143-5p Deficiency triggers EMT and metastasis by
targeting HIF-1a in gallbladder cancer, Cell. Physiol. Biochem.,
42, 2078-2092, doi: 10.1159/000479903.
86.Zhang, Y., and Wang, X. (2020) Targeting the
Wnt/b-catenin signaling pathway in cancer, J. Hematol. Oncol.,
13, 165, doi: 10.1186/s13045-020-00990-3.
87.Sun, Y., Zhu, Q., Zhou, M., Yang, W., Shi, H., et
al. (2019) Restoration of miRNA-148a in pancreatic cancer reduces
invasion and metastasis by inhibiting the Wnt/b-catenin signaling
pathway via downregulating maternally expressed gene-3, Exp. Ther.
Med., 17, 639-648, doi: 10.3892/etm.2018.7026.
88.Liang, T. S., Zheng, Y. J., Wang, J., Zhao, J.
Y., Yang, D. K., et al. (2019) MicroRNA-506 inhibits tumor growth and
metastasis in nasopharyngeal carcinoma through the inactivation of the
Wnt/b-catenin signaling pathway by down-regulating LHX2, J. Exp.
Clin. Cancer Res., 38, 97, doi:
10.1186/s13046-019-1023-4.
89.Subat, S., Inamura, K., Ninomiya, H., Nagano, H.,
Okumura, S., et al. (2018) Unique microRNA and mRNA interactions in
EGFR-mutated lung adenocarcinoma, J. Clin. Med., 7, 419,
doi: 10.3390/jcm7110419.
90.Edmonds, M. D., Hurst, D. R., and Welch, D. R.
(2009) Linking metastasis suppression with metastamiR regulation,
Cell Cycle, 8, 2673-2675, doi: 10.4161/cc.8.17.9303.
91.Zhou, W., Fong, M. Y., Min, Y., Somlo, G., Liu,
L., et al. (2014) Cancer-Secreted miR-105 destroys vascular endothelial
barriers to promote metastasis, Cancer Cell, 25, 501-515,
doi: 10.1016/j.ccr.2014.03.007.
92.Kosaka, N., Iguchi, H., Hagiwara, K., Yoshioka,
Y., Takeshita, F., et al. (2013) Neutral sphingomyelinase 2
(nSMase2)-dependent exosomal transfer of angiogenic micrornas regulate
cancer cell metastasis, J. Biol. Chem., 288, 10849-10859,
doi: 10.1074/jbc.M112.446831.
93.Fong, M. Y., Zhou, W., Liu, L., Alontaga, A. Y.,
Chandra, M., et al. (2015) Breast-cancer-secreted miR-122 reprograms
glucose metabolism in premetastatic niche to promote metastasis,
Nat. Cell Biol., 17, 183-194, doi: 10.1038/ncb3094.
94.Le, M. T. N., Hamar, P., Guo, C., Basar, E.,
Perdigão-Henriques, R., et al. (2014) MiR-200-containing
extracellular vesicles promote breast cancer cell metastasis, J.
Clin. Invest., 124, 5109-5128, doi: 10.1172/JCI75695.
95.Lei, B., Wang, D., Zhang, M., Deng, Y., Jiang,
H., et al. (2020) MiR-615-3p promotes the epithelial-mesenchymal
transition and metastasis of breast cancer by targeting PICK1/TGFBRI
axis, J. Exp. Clin. Cancer Res., 39, 71, doi:
10.1186/s13046-020-01571-5.
96.Chatterjee, A., Jana, S., Chatterjee, S.,
Wastall, L. M., Mandal, G., et al. (2019) MicroRNA-222 reprogrammed
cancer-associated fibroblasts enhance growth and metastasis of breast
cancer, Br. J. Cancer, 121, 679-689, doi:
10.1038/s41416-019-0566-7.
97.Zhao, X. G., Hu, J. Y., Tang, J., Yi, W., Zhang,
M. Y., et al. (2019) miR-665 expression predicts poor survival and
promotes tumor metastasis by targeting NR4A3 in breast cancer, Cell
Death Dis., 10, 479, doi: 10.1038/s41419-019-1705-z.
98.Hashimoto, K., Ochi, H., Sunamura, S., Kosaka,
N., Mabuchi, Y., et al. (2018) Cancer-secreted hsa-miR-940 induces an
osteoblastic phenotype in the bone metastatic microenvironment via
targeting ARHGAP1 and FAM134A, Proc. Natl. Acad. Sci. USA,
115, 2204-2209, doi: 10.1073/pnas.1717363115.
99.Fabbri, M., Paone, A., Calore, F., Galli, R.,
Gaudio, E., et al. (2012) MicroRNAs bind to Toll-like receptors to
induce prometastatic inflammatory response, Proc. Natl. Acad. Sci.
USA, 109, E2110-E2116, doi: 10.1073/pnas.1209414109.
100.Song, Q., Liu, B., Li, X., Zhang, Q., Cao, L.,
et al. (2018) MiR-26a-5p potentiates metastasis of human lung cancer
cells by regulating ITGb8- JAK2/STAT3 axis, Biochem. Biophys. Res.
Commun., 501, 494-500, doi: 10.1016/j.bbrc.2018.05.020.
101.Liu, C., Luo, J., Zhao, Y. T., Wang, Z. Y.,
Zhou, J., et al. (2018) TWIST1 upregulates miR-214 to promote
epithelial-to-mesenchymal transition and metastasis in lung
adenocarcinoma, Int. J. Mol. Med., 42, 461-470, doi:
10.3892/ijmm.2018.3630.
102.Li, J., Feng, Q., Wei, X., and Yu, Y. (2016)
MicroRNA-490 regulates lung cancer metastasis by targeting poly
r(C)-binding protein 1, Tumor Biol., 37, 15221-15228,
doi: 10.1007/s13277-016-5347-9.
103.Fang, T., Lv, H., Lv, G., Li, T., Wang, C., et
al. (2018) Tumor-derived exosomal miR-1247-3p induces cancer-associated
fibroblast activation to foster lung metastasis of liver cancer,
Nat. Commun., 9, 191, doi:
10.1038/s41467-017-02583-0.
104.Xiong, Y., Wu, S., Yu, H., Wu, J., Wang, Y., et
al. (2018) miR-190 promotes HCC proliferation and metastasis by
targeting PHLPP1, Exp. Cell Res., 371, 185-195, doi:
10.1016/j.yexcr.2018.08.008.
105.Yang, B., Feng, X., Liu, H., Tong, R., Wu, J.,
et al. (2020) High-metastatic cancer cells derived exosomal miR92a-3p
promotes epithelial-mesenchymal transition and metastasis of
low-metastatic cancer cells by regulating PTEN/Akt pathway in
hepatocellular carcinoma, Oncogene, 39, 6529-6543, doi:
10.1038/s41388-020-01450-5.
106.Mao, X. W., Xiao, J. Q., Li, Z. Y., Zheng, Y.
C., and Zhang, N. (2018) Effects of microRNA-135a on the
epithelial–mesenchymal transition, migration and invasion of
bladder cancer cells by targeting GSK3b through the wnt/b-catenin
signaling pathway, Exp. Mol. Med., 50, e429, doi:
10.1038/emm.2017.239.
107.Liu, J., Cao, J., and Zhao, X. (2015) MiR-221
facilitates the TGFbeta1-induced epithelial-mesenchymal transition in
human bladder cancer cells by targeting STMN1 urological oncology,
BMC Urol., 15, 36, doi: 10.1186/s12894-015-0028-3.
108.Yan, L., Wang, Y., Liang, J., Liu, Z., Sun, X.,
et al. (2017) MiR-301b promotes the proliferation, mobility, and
epithelial-to-mesenchymal transition of bladder cancer cells by
targeting EGR1, Biochem. Cell Biol., 95, 571-577, doi:
10.1139/bcb-2016-0232.
109.Hu, H., Zhang, Q., Chen, W., Wu, T., Liu, S.,
et al. (2020) MicroRNA-301a promotes pancreatic cancer invasion and
metastasis through the JAK/STAT3 signaling pathway by targeting SOCS5,
Carcinogenesis, 41, 502-514, doi:
10.1093/carcin/bgz121.
110.Xin, H., Wang, C., and Liu, Z. (2019)
MiR-196a-5p promotes metastasis of colorectal cancer via targeting
IkBa, BMC Cancer, 19, 30, doi:
10.1186/s12885-018-5245-1.
111.Li, Y., Yan, X., Shi, J., He, Y., Xu, J., et
al. (2019) Aberrantly expressed miR-188-5p promotes gastric cancer
metastasis by activating Wnt/b-catenin signaling, BMC Cancer,
19, 1-15, doi: 10.1186/s12885-019-5731-0.
112.Li, C., Lu, S., and Shi, Y. (2017) MicroRNA-187
promotes growth and metastasis of gastric cancer by inhibiting FOXA2,
Oncol. Rep., 37, 1747-1755, doi:
10.3892/or.2017.5370.
113.Li, N., Cui, T., Guo, W., Wang, D., and Mao, L.
(2019) MiR-155-5p accelerates the metastasis of cervical cancer cell
via targeting TP53INP1, Onco. Targets. Ther., 12,
3181-3196, doi: 10.2147/ott.s193097.
114.Sun, X., Dongol, S., Qiu, C., Xu, Y., Sun, C.,
et al. (2018) MiR-652 promotes tumor proliferation and metastasis by
targeting RORA in endometrial cancer, Mol. Cancer Res.,
16, 1927-1939, doi: 10.1158/1541-7786.MCR-18-0267.
115.Salem, M., O’Brien, J. A., Bernaudo, S.,
Shawer, H., Ye, G., et al. (2018) MiR-590-3p promotes ovarian cancer
growth and metastasis via a novel FOXA2-versican pathway, Cancer
Res., 78, 4175-4190, doi: 10.1158/0008-5472.CAN-17-3014.
116.Cai, Q., Zeng, S., Dai, X., Wu, J., and Ma, W.
(2017) MiR-504 promotes tumour growth and metastasis in human
osteosarcoma by targeting TP53INP1, Oncol. Rep., 38,
2993-3000, doi: 10.3892/or.2017.5983.
117.Xiao, W., Lou, N., Ruan, H., Bao, L., Xiong,
Z., et al. (2017) Mir-144-3p promotes cell proliferation, metastasis,
sunitinib resistance in clear cell renal cell carcinoma by
downregulating ARID1A, Cell. Physiol. Biochem., 43,
2420-2433, doi: 10.1159/000484395.
118.Vu, L. T., Gong, J., Pham, T. T., Kim, Y., and
Le, M. T. N. (2020) microRNA exchange via extracellular vesicles in
cancer, Cell Prolif., 53, e12877, doi:
10.1111/cpr.12877.
119.Madhavan, D., Zucknick, M., Wallwiener, M.,
Cuk, K., Modugno, C., et al. (2012) Circulating miRNAs as surrogate
markers for circulating tumor cells and prognostic markers in
metastatic breast cancer, Clin. Cancer Res., 18,
5972-5982, doi: 10.1158/1078-0432.CCR-12-1407.
120.Rysenkova, K. D., Rubina, K. A., Ivanova, K.
A., Karagyaur, M. N., and Semina, E. V. (2019) The role of the
urokinase system in carcinogenesis and metastasis of tumor cells with
the participation of microRNA, Genes Cells, 14,
200-200.
121.Bayraktar, R., Bertilaccio, M. T. S., and
Calin, G. A. (2019) The interaction between two worlds: microRNAs and
Toll-like receptors, Front. Immunol., 10, 1053, doi:
10.3389/fimmu.2019.01053.