CD44-Associated Tn Antigen as a New Biomarker of Tumor Cells with Aberrant Glycosylation
M. L. Shuvalova1, A. T. Kopylov2, D. V. Mazurov1, A. V. Pichugin1, N. V. Bovin3, and A. V. Filatov1,4,a*
1Institute of Immunology National Research Center, Federal Medical and Biological Agency of Russia, 115522 Moscow, Russia2 Orekhovich Institute of Biomedical Chemistry, 119121 Moscow, Russia
3Shemyakin and Ovchinnikov Institute of Bioorganic Chemistry, Russian Academy of Sciences, 117997 Moscow, Russia
4Department of Immunology, Faculty of Biology, Lomonosov Moscow State University, 119234 Moscow, Russia
* To whom correspondence should be addressed.
Received June 30, 2020; Revised July 8, 2020; Accepted July 30, 2020
Tn antigen is a tumor-associated antigen that appears on cancer cells as a result of aberrant O-glycosylation. The most studied form of Tn antigen is found in mucins, in particular, in mucin 1 (MUC1). Antibodies against this form of Tn antigen are used to diagnose tumors, as well as to generate T-killers with a chimeric receptor. Some carcinomas do not carry MUC1 and antibodies of a different specificity are required to detect Tn antigen on these cells. In our work, we searched for anti-Tn antibodies without preliminary assumptions about the proteins that may be carriers of the Tn antigen. For this purpose, we obtained several pairs of isogenic cell lines with the wild type and knockout of the Cosmc gene, which is essential for correct protein O-glycosylation. Using the created lines as immunogens, we generated a monoclonal antibody AKC3, which reacted with the Cosmc-deficient A549 lung adenocarcinoma cells and did not bind to the wild-type cells. Using mass spectrometry, as well as co-immunoprecipitation, it was shown that the AKC3 antibody recognized the Tn antigen in the context of CD44 protein – a protein important for tumor growth. The AKC3 antibody can be used for tumor diagnosis, and to generate T cells with a chimeric receptor for treatment of tumors that do not express mucins.
KEY WORDS: Tn antigen, O-glycosylation, neoantigens, CD44, human lung adenocarcinomaDOI: 10.1134/S0006297920090060
Abbreviations: GalNAc, N-acetyl-D-galactosamine; LRG1, leucine rich alpha-2-glycoprotein 1; MUC1, mucin 1; MW, molecular weight; PAGE, polyacrylamide gel electrophoresis; PBS, phosphate-buffered saline; SDS, sodium dodecyl sulfate; WB, Western blotting.
INTRODUCTION
Tumor-associated and tumor-specific antigens are important in the diagnosis and immunotherapy of malignant neoplasms. The search for new biomarkers of tumor cells is carried out both at the proteomic and immunochemical levels. While the first method determines structure of the neoantigens, the second method facilitates creation of reagents for their detection. Aberrant glycosylation is one of the ways through which neoantigens are formed. In the process of protein O-glycosylation, the synthesis of a glycan chain starts with addition of N-acetylgalactosamine (GalNAc) to serine or threonine residues [1]. In the case of Cosmc chaperone deficiency further elongation of the glycan chain is stopped, and the terminal GalNAc, together with the adjacent amino acid residues, forms the so-called Tn antigen [2]. Mutations in the Cosmc gene that lead to formation of the Tn antigen have been observed in a number of tumor cells. More than 70% of carcinomas are positive for Tn antigen [3]. Tn antigen is absent in normal cells.
To date, a significant number of antibodies against Tn antigen have been generated [4, 5], which can be divided into two large groups: antibodies that are dependent and independent on the GalNAc peptide environment. Peptide-independent antibodies have a wide range of reactivity and bind to Tn antigen in a wide variety of proteins [6]. Unfortunately, these antibodies belong to the IgM class and have a low binding constant. Peptide-dependent antibodies are of more interest because they are characterized by high affinity, but they have a narrow spectrum of reactivity and react with Tn antigen only in specific proteins. Typically, anti-Tn antibodies were generated by immunizing with glycopeptides from mucin – the most well-known carrier protein of the Tn antigen. Very limited data are available on the aberrant glycosylation biomarkers presented on other proteins.
Previously we proposed a method using Cosmc isogenic cell lines to search for peptide-specific anti-Tn antibodies on unknown carrier proteins [7]. For immunization and screening, we used a pair of cell lines: Jurkat wt, which is constitutively defective in the Cosmc gene, and Jurkat cells, transduced with the wild-type Cosmc gene. This allowed us to generate antibodies against Tn antigen associated with CD45 and CD43 proteins, which were not previously known as possible carriers of Tn antigen [8].
The objective of this study is to search for new biomarkers of carcinomas that are formed as a result of aberrant glycosylation and that can later be used for diagnosis and therapy of tumors. To accomplish this, we set the following tasks: (i) to create isogenic cell lines by knocking out the Cosmc gene, which, in contrast to the wild-type cells, carry the Tn antigen; (ii) to generate peptide-specific monoclonal antibodies against the Tn antigen expressed on the lung carcinoma cells; (iii) to identify new biomarkers of the tumor cells with aberrant glycosylation.
MATERIALS AND METHODS
Cell culture, antibodies, and flow cytometry. Human A549, MCF-7 and RPMI 8226 cells were cultured in a DMEM/F12 medium supplemented with 10% fetal calf serum, 4 mM L-glutamine, and gentamicin (80 mg/liter) (Paneco, Russia) at 37°C, in a humidified atmosphere containing 5% CO2. Antibodies He7 (IgG2a, anti CD44), ER1 (IgG2b, anti CD99), JA1 (IgM, anti-Tn antigen) were produced previously in our laboratory [7]. To generate new monoclonal antibodies, BALB/c mice were immunized three times with target cells, after which the spleen cells from immunized mice were hybridized with Sp2/0 myeloma. The culture supernatants of hybrid cells after 10 days were tested using an immunofluorescence assay with target cells, and positive clones were selected [7].
For immunofluorescence staining, 1 × 105 cells were mixed with 50 µl of unlabeled primary antibodies and incubated at 4°C for 30 min. The cells were washed twice in a phosphate-buffered saline (PBS) by centrifugation at 300g for 5 min. Then the cells were incubated with FITC-labeled secondary antibodies against mouse IgG or IgM (Merck, USA) and washed. The stained cells were analyzed with a CytoFLEX S flow cytometer (Beckman-Coulter, USA). The cells treated only with the secondary antibodies were used as a negative control.
Generation of Cosmc gene knockout cell lines. Selection of the target sequence in the Cosmc gene (Gene ID: 29071) was performed using the online resource described in the publication by Hsu et al. [9]. The gRNA sequence was encoded into a pair of complementary DNA oligonucleotides for subsequent cloning into a vector: 5-gR-Cosmc: 5′-CACCGCAATGATAGCAAACGTAGTG-3′ and 3-gR-Cosmc: 5′-AAACCACTACGTTTGCTATCATTGC-3′. Along with this, the gRNA sequence reported by Stolfa et al. was used [10].
Oligonucleotides were annealed and cloned into the PKS-gRNA-BB vector using the BbsI restriction site [11]. One million cells were transfected using a Neon electroporation system (Thermo Fisher, USA). Five days after transfection, expression of the Tn antigen on the cells was evaluated using the JA1 antibody, the cells were cultivated and Tn positive population was sorted out using a FACSAria II device (Becton Dickinson Biosciences, USA).
Periodate oxidation. A549 cells were fixed in a 4% paraformaldehyde solution for 10 min and treated with 20 mM sodium periodate in PBS for 1 h. The cells were washed 2 times in PBS and the formed aldehyde groups were blocked for 30 min in a complete DMEM medium containing 10% of fetal calf serum [8].
Analysis of the carbohydrate specificity of antibodies with glycochip. Glycochips were produced by printing glycans on glass slides (Schott-Nexterion, Germany) activated with hydroxysuccinimide. On each slide, about 400 different ligands were immobilized, which were synthetic components of mammalian glycoproteins and glycolipids, as well as 200 bacterial polysaccharides. A complete list of ligands can be found in [12]. The glycochip was incubated with an antibody solution, washed in a PBS solution containing 0.1% Tween-20, stained with the secondary anti-mouse IgG antibodies labeled with Alexa555, and washed again. Fluorescence was recorded using an Innoscan 710 Fluorescence Scanner (Innopsys, France).
Fluorescent immunoprecipitation analysis. Twenty million cells were labeled with Cy3 aminoreactive fluorescent dye (10 mg/ml) (Lumiprobe, Russia) [13]. Cells were lysed for 30 min at 4°C in 1 ml of a 20 mM Tris HCl lysing buffer, pH 8.0, containing 1% Triton X-100, 150 mM NaCl, 5 mM EDTA, 1 mM phenylmethylsulfonyl fluoride (Merck). Immunoprecipitation was performed by adding 1 ml of the lysate to 20 µl of Protein A agarose pretreated with a complex of precipitating antibodies and subsequent incubation for 2 h at 4°C. Immunoprecipitates were washed three times, each time 800 µl of the lysing buffer was added. The bound fluorescently labeled proteins were eluted from the sorbent by heating with the sample buffer for electrophoresis for 5 min at 95°C.
Electrophoresis and Western blotting. Eluted proteins were separated by Laemmli’s sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) in 10% SDS-PAGE under reducing conditions [7]. Proteins in the gels were visualized with a fluorescent scanner Molecular Imager FX Pro (Bio-Rad, USA) recording fluorescence at 600 nm.
The separated proteins were transferred by a semi-dry method from the gel to a PVDF membrane and developed using primary and then secondary antibodies against mouse IgG labeled with horseradish peroxidase (GE Healthcare, USA). The blots were then visualized with a ChemiDoc XRS System (Bio-Rad) using chemiluminescence reagents (Merck).
Mass spectrometry analysis. Protein bands (~5 mm3) were cut from the gel, washed 3 times in deionized water, and trypsinolysis was performed as described previously [13]. A mixture of peptides in a volume of 1.5-2.0 µl was loaded on an Ultimeta 3000 Plus RSLC nano column (Thermo Fisher) and separated using reversed-phase chromatography using a linear gradient of 2→37% of the mobile phase (acetonitrile with 0.1% formic acid and 0.03% acetic acid). The signal was recorded using a Q-Exact HF-X mass spectrometer (Thermo Fisher) in the range of 425-1300 m/z. Identification of proteins was performed based on the tandem mass spectra (MS/MS) of tryptic peptides using the SearchGUI and Peptideakher programs [13].
RESULTS
Generation of cell lines deficient in the Cosmc gene. The CRISP/Cas9 gene editing technology was used to produce MCF-7, A549, and RPMI 8226 cell lines with aberrant O-glycosylation. The cells were transfected with components of the CRISPR/Cas9 system and after 5 days from 5 to 10% Tn positive cells were detected using a peptide-independent anti-Tn antibody JA1. After two rounds of sorting, cell lines were obtained in which at least 90% of the cells were Tn positive.
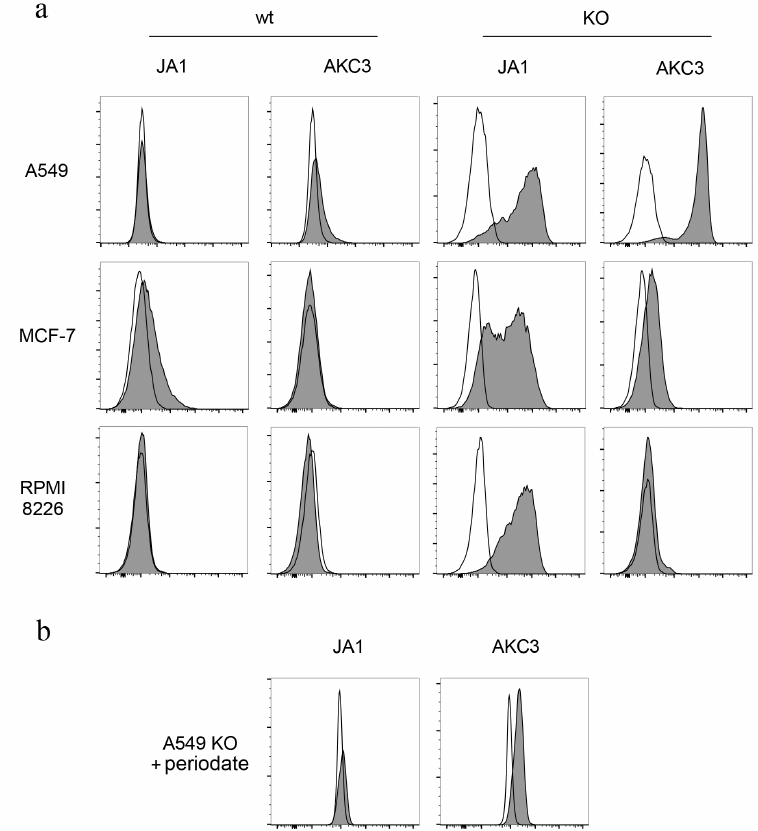
Generation of a monoclonal antibody against A549 cells with aberrant glycosylation. To generate new monoclonal antibodies against Tn-antigen, BALB/c mouse was immunized with the Cosmc knockout A549 cells (A549-Cosmc-KO). Mouse spleen cells were hybridized with Sp2/0 myeloma. At the first stage of screening, hybrids were selected that secrete antibodies reacting with the A549-Cosmic-KO cells. At the next stage of screening, clones that reacted with the wild-type A549 cells were excluded. As a result of selection, an AKC3 clone was generated, secreting an IgG1 antibody. This antibody stained A549-Cosmc-KO cells efficiently, and exhibited only minimal binding to the wild-type A549 cells (Fig. 1a). At the same time, the AKC3 antibody did not react with Tn positive MCF-7 and RPMI 8226 cells that were deficient in the Cosmc gene (Fig. 1a).
Fig. 1. Tn antigen expression on Cosmc gene knockout cells. a) A549 cells (top row), MCF-7 (middle row), and RPMI 8226 (bottom row) stained with JA1 or AKC3 antibody, then with a secondary FITC-labeled antibody against mouse Ig, and analyzed by flow cytometry (filled histograms). Cells stained only with secondary antibodies served as negative control (open plots). In the two left columns wild type cells (wt) are presented, in the two right columns – Cosmc knockout cells (KO). b) A549 Cosmc knockout cells treated with 20 mM sodium periodate and then stained with JA1 or AKC3 antibody.
Biochemical characterization of AKC3 antigen. In order to check association of the AKC3 antibody binding with the presence of carbohydrate-based determinants, mild oxidation of cells with periodate was conducted. A549-Cosmc-KO cells treated with sodium periodate, lost their ability to be stained with the AKC3 antibody (Fig. 1b). Testing of the AKC3 antibody on a glycochip containing more than 400 different glycans, including GalNAcα, GalNAcα-Ser and several dozen oligosaccharides with terminal or internal GalNAcα fragments, gave negative results [13]. Thus, the AKC3 binding was dependent on the presence of the carbohydrate chain, but was not completely determined by it and depended on the specific carrier proteins that were expressed by the A549 cells.
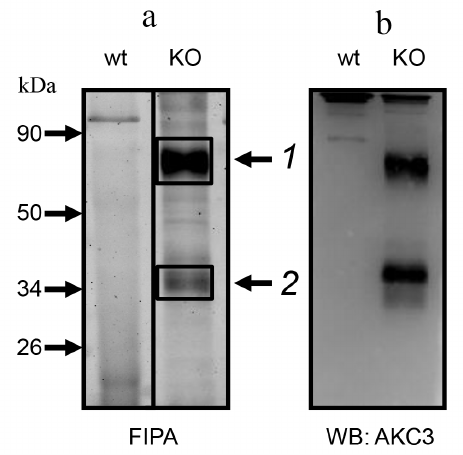
In order to characterize the proteins carrying the AKC3 antigen, we used the method of fluorescent immunoprecipitation (FIPA) [13]. AKC3 antigen purified with an immunosorbent, migrated in SDS-PAGE gel in the form of two bands (Fig. 2a). Major upper band matched a protein with molecular weight of 85 kDa. Lower band was less intense and often was observed as two closely located bands close to the position of the 35-kDa protein marker. The same bands were observed in Western blots (Fig. 2b). Position of the bands did not depend on whether the reducing or non-reducing conditions of electrophoresis were used. Thus, the AKC3 antigen is present on at least two carrier proteins, which are single-chain molecules.
Fig. 2. Immunoprecipitation of the antigen detected by the AKC3 antibody. a) Cells labeled with Cy3 dye, lysed in 1% (w/v) Triton X-100, and immunoprecipitated with AKC3 antibody. The isolated proteins were separated on 12% SDS-PAGE under reducing conditions and fluorescence of the gel was recorded (FIPA). Protein bands 1 and 2 were excised and subjected to mass spectrometry analysis. b) Proteins from the gel were transferred onto a PVDF membrane and developed using Western blotting with the AKC3 antibody. Wild-type A549 (wt) or Cosmc knockout (KO) A549 cells were used as target cells.
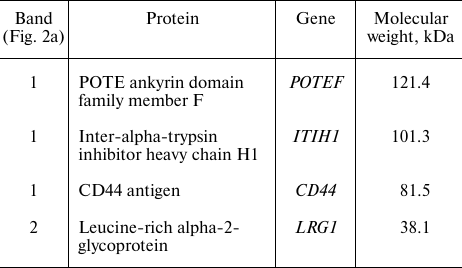
Mass spectrometry analysis of the AKC3 antigen. In order to identify proteins carrying the Tn antigen, two bands obtained by immunoprecipitation with the AKC3 antibody (Fig. 2a) were excised from the gel and treated with trypsin. The obtained peptides were first separated using liquid chromatography and next analyzed with tandem mass spectrometry with electrospray ionization. The most relevant identified proteins are presented in the Table. For band 1, the CD44 protein was of greatest interest, for which a more detailed study was carried out.
Mass spectrometry identification of proteins immunopurified using the
AKC3 antibody
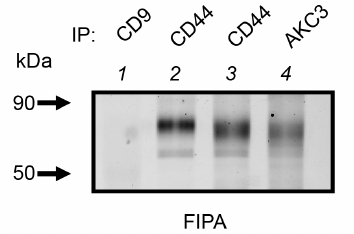
Co-immunoprecipitation analysis of the AKC3 antigen. It is interesting to note that the CD44 precipitation band obtained from the lysate of the A549-Cosmc-KO cells quite closely matches the upper band of the AKC3 antigen (Fig. 3, lanes 3 and 4). It can also be noted that CD44 isolated from the wild-type cells had a slightly lower motility than CD44 from the A549-Cosmc-KO cells (Fig. 3, lane 2). This observation is consistent with the fact that the degree of CD44 glycosylation should be higher in the wild-type cells than in the knockout cells.
Fig. 3. Comparison of electrophoretic mobility of CD44 protein isolated from the wild type A549 cells and the Cosmc knockout A549 cells. Immunoprecipitation was performed with antibodies against CD44, AKC3, or CD9 (negative control). Wild-type A549 cells (lane 2) or Cosmc knockout A549 cells (lanes 1, 3, and 4) were used as target cells.
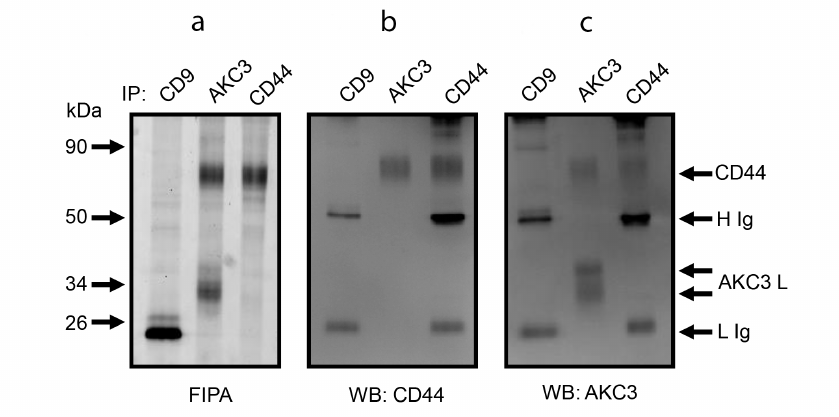
To validate the assumption that CD44 is a carrier protein of the Tn antigen, experiments on mutual co-immunoprecipitation of CD44 and AKC3 antigen were performed. For this purpose, CD44 and AKC3 antigens were first isolated using immunoaffinity sorbents based on the Protein A agarose (Fig. 4a). Then the obtained proteins were stained by the WB method using antibodies against CD44 (Fig. 4b) and AKC3 antigen (Fig. 4c). These experiments showed that the CD44 band and the upper band of the AKC3 antigen were equally developed by both anti-CD44 and AKC3 antibodies (Fig. 4, b and c). On the contrary, the lower band of the AKC3 antigen in WB was detected only by the AKC3 antibody, but not by the anti-CD44 antigen antibody. Anti-CD9 immunoprecipitation was used as a negative control in these experiments. The bands marked on the WB with symbols H and L appeared in both the test and control lanes. These bands emerge as a result of dissociation of the light and heavy chains of antibodies that were used for immunoprecipitation. With the exception of chains H and L, no other bands were detected in the negative control lane.
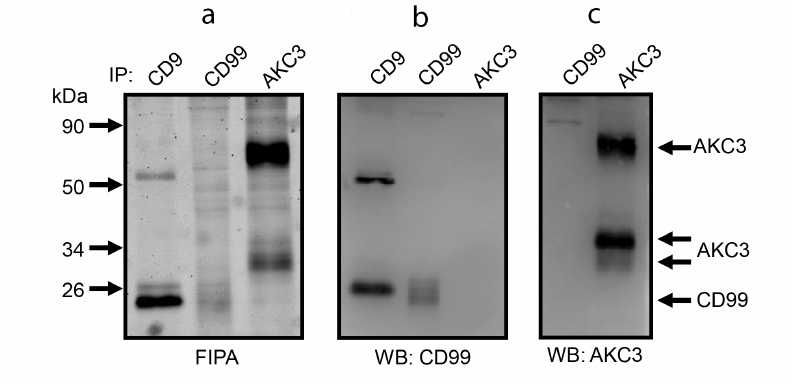
Fig. 4. Co-immunoprecipitation of CD44 protein and AKC3 antigen from the lysate of Cosmc knockout A549 cells. Immunoprecipitation was performed with antibodies against CD9, CD44, or AKC3. The isolated proteins were separated in 12% SDS-PAGE and fluorescence of the gel was recorded (a), next the proteins were transferred to a PVDF membrane and developed using WB with antibodies against CD44 (b) or AKC3 (c).
Molecular weight (MW) corresponding to the lower band of the AKC3 antigen is quite close to that of the CD99 protein. CD99 is known to have at least one O-glycosylation site, i.e., can theoretically carry a Tn antigen. In order to test possible association between CD99 and AKC3 co-immunoprecipitation experiments were performed for the pair of antibodies against CD99 and AKC3, similarly to the way as it was done for the pair of antibodies against CD44 and AKC3 (Fig. 5). However, no co-immunoprecipitation was observed for CD99 and AKC3. In addition, direct comparison of electrophoretic mobility of CD99 and AKC3 antigen showed that the latter had a markedly higher MW. It is obvious that the lower band of the AKC3 antigen is not associated with the CD99 protein.
Fig. 5. Co-immunoprecipitation of CD99 protein and AKC3 antigen from the lysate of Cosmc knockout A549 cells. Immunoprecipitation was performed on antibodies against CD9, CD99, or AKC3. a) Isolated proteins were separated in 12% SDS-PAGE and fluorescence of the gel was recorded, then the proteins were transferred onto a PVDF membrane and developed using Western blotting with antibodies against CD99 (b) or AKC3 (c).
DISCUSSION
By knocking out the Cosmc gene, we produced 3 pairs of isogenic cell lines that differed in the expression of the Tn antigen. Using isogenic A549 cells, we generated a monoclonal antibody AKC3 that reacted with A549-Cosmc-KO cells and did not bind to the A549 wt line. Oxidation of carbohydrate residues with sodium periodate eliminated binding of the AKC3 antibody to the cells. Thus, AKC3 is an anti-Tn antibody in terms of its specificity. A feature of our work was that AKC3 was generated without any preliminary assumptions regarding the proteins that might carry the Tn antigen.
Using the method of fluorescence immunoprecipitation, as well as WB, we found out that AKC3 reacted with two proteins with MW 85 and 34 kDa. Mass spectrometry analysis showed that peptides from the CD44 protein were found in the 85 kDa band. Similarity between the AKC3 antigen and the CD44 protein was confirmed by direct comparison of electrophoretic mobility of these molecules, as well as in experiments on co-immunoprecipitation. This is also consistent with the lack of reactivity of the AKC3 antibody toward MCF-7 and RPMI 8226 cell sublines, which are deficient in the Cosmc gene and carry the Tn antigen, but do not express the CD44 protein.
The standard form of human CD44 (CD44s) consists of 361 amino acid residues and has a calculated MW of 39.4 kDa. CD44 is a highly glycosylated protein causing it to migrate in the 85-90 kDa region during electrophoresis. CD44s has 6 potential N-glycosylation sites and 32 potential O-glycosylation sites [14]. O-glycosylation of the 4 sites in the Ser-Gly motifs, which are located in the extracellular part of CD44s directly adjacent to the transmembrane domain, is considered the most probable glycosylation scenario [15]. According to another estimate, CD44s is O-glycosylated at 7 sites [16]. It is worth mentioning that CD44 isolated from the knockout cells has a higher electrophoretic mobility than the one from the wild-type cells. This is in good agreement with the fact that O-glycosylation should be incomplete in the knockout cells. Thus, based on its characteristics, CD44s is indeed the antigen recognized by AKC3.
CD44 is of great functional importance. CD44 is a hyaluronic acid receptor and it also binds to other components of extracellular matrix. As a consequence, CD44 is involved in the processes of cell adhesion, migration, and cell differentiation. CD44 plays an essential role in tumor growth and metastasis [17]. Tumor growth is affected by both total expression of CD44 protein [18] and presence of its individual glycoforms. In particular, changes in the degree of CD44 glycosylation can regulate tumor growth [19]. Glycosylation affects hyaluronic acid binding and thereby changes adhesive properties of the CD44 molecule [15]. It was shown that the cells from the primary melanoma, as well as from metastases differed in the degree of O-glycosylation of the CD44 molecule [15].
Mass spectrometry data suggest that the carrier protein of the Tn antigen from the lower band (Fig. 2) may be the leucine-rich alpha-2-glycoprotein 1 (LRG1). LRG1 has MW of 38 kDa, which is quite close to the mass corresponding to the lower band. It possesses an O-glycosylation site. According to these criteria, LRG1 is suitable for the role of the target protein; however, LRG1 is expressed predominantly in neutrophils, has no membrane localization, and is secreted into plasma [20]. Thus, it is unlikely that the AKC3 antigen is associated with the LRG1 protein. Possibility for the AKC3 antibody to react with two different proteins also seems problematic, although this cannot be ruled out completely. For example, the anti-Tn antibody JA5 reacted simultaneously with the CD45 and CD43 proteins [7]. The highly O-glycosylated surface glycoprotein CD99 has MW of 32 kDa, which roughly corresponds to the mass of the AKC3 antigen from the lower band (Fig. 2). However, direct comparison of electrophoretic mobility of CD99 and the lower band of the AKC3 antigen showed that they are different proteins. Co-immunoprecipitation experiments also exclude association of these proteins.
It should be noted that in addition to the standard CD44 molecule (CD44s), there is the CD44 isoform with a shortened cytoplasmic domain (CD44 short-tail; CD44st). Based on the amino acid sequence, CD44st has a calculated MW of 32 kDa [14]. With minimal glycosylation, the CD44st MW corresponds to the lower band of the AKC3 antigen. Expression of the CD44st isoform was observed in Lovo, K562, and HL-60 cells [21].
An approach called SimpleCells was proposed previously to search for the tumor-associated proteins with aberrant glycosylation [22]. Using this strategy, an antibody against a tumor biomarker was obtained, which was a Tn antigen associated with the dysadherin protein [23]. As a result of a large-scale proteomic study on Cosmc knockout stomach cancer cells, 499 O-glycosylated proteins were identified, which carried more than 1000 different glycosites. Interestingly, the CD44 protein was also found among the proteins with aberrant glycosylation [22].
Taking into account that the Tn antigen is a tumor-associated antigen, it has been considered for a long time as an attractive target for the treatment of carcinomas. Antibodies against the Tn antigen can be used as a passive vaccine in cancer immunotherapy [24]. Antibodies against the Tn antigen are being used to develop T killers with a chimeric antigen receptor [25, 26]. Most of the known anti-Tn antibodies recognize this antigen in mucins. We generated a new anti-Tn antibody that reacted with the Tn antigen in the context of the CD44 protein. Sequencing the Ig genes of the AKC3 hybridoma could provide a good basis for generation of T cells with CAR receptor for treatment of CD44+ tumors that do not express mucins.
Funding. This work was financially supported by the Russian Foundation for Basic Research (project no. 18-29-07025 mk).
Ethics declarations. The authors declare no conflict of interest in financial or any other sphere. This article does not contain any studies with human participants performed by any of the authors. All applicable international, national, and/or institutional guidelines for the care and use of animals were followed.
REFERENCES
1.Ju, T., Otto, V. I., and Cummings, R. D. (2011) The
Tn antigen-structural simplicity and biological complexity, Angew.
Chem. Int. Ed., 50, 1770-1791,
doi: 10.1002/anie.201002313.
2.Ju, T., Lanneau, G. S., Gautam, T., Wang, Y., Xia,
B., Stowell, S. R., Willard, M. T., Wang, W., Xia, J. Y., Zuna, R. E.,
Laszik, Z., Benbrook, D. M., Hanigan, M. H., and Cummings, R. D. (2008)
Human tumor antigens Tn and sialyl Tn arise from mutations in
Cosmc, Cancer Res., 68, 1636-1646,
doi: 10.1158/0008-5472.CAN-07-2345.
3.Springer, G. F. (1997) Immunoreactive T and Tn
epitopes in cancer diagnosis, prognosis, and immunotherapy, J. Mol.
Med., 75, 594-602, doi: 10.1007/s001090050144.
4.Schietinger, A., Philip, M., Yoshida, B. A., Azadi,
P., Liu, H., Meredith, S. C., and Schreiber, H. (2006) A mutant
chaperone converts a wild-type protein into a tumor-specific antigen,
Science, 314, 304-308,
doi: 10.1126/science.1129200.
5.Tarp, M. A., and Clausen, H. (2008) Mucin-type
O-glycosylation and its potential use in drug and vaccine development,
Biochim. Biophys. Acta, 1780, 546-563,
doi: 10.1016/j.bbagen.2007.09.010.
6.Persson, N., Stuhr-Hansen, N., Risinger, C.,
Mereiter, S., Polónia, A., Polom, K., Kovács, A.,
Roviello, F., Reis, C. A., Welinder, C., Danielsson, L., Jansson, B.,
and Blixt, O. (2017) Epitope mapping of a new anti-Tn antibody
detecting gastric cancer cells, Glycobiology, 27,
635-645, doi: 10.1093/glycob/cwx033.
7.Blixt, O., Lavrova, O. I., Mazurov, D. V., Clo, E.,
Kracun, S. K., Bovin, N. V., and Filatov, A. V. (2012) Analysis of Tn
antigenicity with a panel of new IgM and IgG1 monoclonal antibodies
raised against leukemic cells, Glycobiology, 22, 529-542,
doi: 10.1093/glycob/cwr178.
8.Mazurov, D., Ilinskaya, A., Heidecker, G., and
Filatov, A. (2012) Role of O-glycosylation and expression of CD43 and
CD45 on the surfaces of effector T cells in human T cell leukemia virus
type 1 cell-to-cell infection, J. Virol., 86, 2447-2458,
doi: 10.1128/JVI.06993-11.
9.Hsu, P. D., Scott, D. A., Weinstein, J. A., Ran, F.
A., Konermann, S., Agarwala, V., Li, Y., Fine, E. J., Wu, X., Shalem,
O., Cradick, T. J., Marraffini, L. A., Bao, G., and Zhang, F. (2013)
DNA targeting specificity of RNA-guided Cas9 nucleases, Nat.
Biotechnol., 31, 827-832, doi: 10.1038/nbt.2647.
10.Stolfa, G., Mondal, N., Zhu, Y., Yu, X., Buffone,
A., and Neelamegham, S. (2016) Using CRISPR-Cas9 to quantify the
contributions of O-glycans, N-glycans and Glycosphingolipids to human
leukocyte-endothelium adhesion, Sci. Rep., 6, 30392,
doi: 10.1038/srep30392.
11.Tarasevich, A., Filatov, A., Pichugin, A., and
Mazurov, D. (2015) Monoclonal antibody profiling of cell surface
proteins associated with the viral biofilms on HTLV-1 transformed
cells, Acta Virol., 59, 247-256,
doi: 10.4149/av_2015_03_247.
12.Dobrochaeva, K., Khasbiullina, N., Shilova, N.,
Antipova, N., Obukhova, P., Ovchinnikova, T., Galanina, O., Blixt, O.,
Kunz, H., Filatov, A., Knirel, Y., LePendu, J., Khaidukov, S., and
Bovin, N. (2020) Specificity of human natural antibodies referred to as
anti-Tn, Mol. Immunol., 120, 74-82,
doi: 10.1016/j.molimm.2020.02.005.
13.Filatov, A. V., Krotov, G. I., Zgoda, V. G., and
Volkov, Y. (2007) Fluorescent immunoprecipitation analysis of cell
surface proteins: a methodology compatible with mass-spectrometry,
J. Immunol. Methods, 319, 21-33,
doi: 10.1016/j.jim.2006.09.014.
14.Azevedo, R., Gaiteiro, C., Peixoto, A.,
Relvas-Santos, M., Lima, L., Santos, L. L., and Ferreira, J. A. (2018)
CD44 glycoprotein in cancer: a molecular conundrum hampering clinical
applications, Clin. Proteomics, 15, 22,
doi: 10.1186/s12014-018-9198-9.
15.Gasbarri, A., Del Prete, F., Girnita, L.,
Martegani, M. P., Natali, P. G., and Bartolazzi, A. (2003) CD44s
adhesive function spontaneous and PMA-inducible CD44 cleavage are
regulated at post-translational level in cells of melanocytic lineage,
Melanoma Res., 13, 325-337,
doi: 10.1097/00008390-200308000-00001.
16.Ponta, H., Sherman, L., and Herrlich, P. A.
(2003) CD44: from adhesion molecules to signalling regulators, Nat.
Rev. Mol. Cell Biol., 4, 33-45,
doi: 10.1038/nrm1004.
17.Naor, D., Sionov, R. V., and Ish-Shalom, D.
(1997) CD44: structure, function and association with the malignant
process, Adv. Cancer Res., 71, 241-319,
doi: 10.1016/S0065-230X(08)60101-3.
18.Hu, B., Ma, Y., Yang, Y., Zhang, L., Han, H., and
Chen, J. (2018) CD44 promotes cell proliferation in non-small cell lung
cancer, Oncol. Lett., 15, 5627-5633
doi: 10.3892/ol.2018.8051.
19.Du, T., Jia, X., Dong, X., Ru, X., Li, L., Wang,
Y., Liu, J., Feng, G., and Wen, T. (2020) Cosmc disruption-mediated
aberrant O-glycosylation suppresses breast cancer cell growth via
impairment of CD44, Cancer Manag. Res., 12, 511-522,
doi: 10.2147/CMAR.S234735.
20.O’Donnell, L. C., Druhan, L. J., and
Avalos, B. R. (2002) Molecular characterization and expression analysis
of leucine-rich alpha2-glycoprotein, a novel marker of granulocytic
differentiation, J. Leukoc. Biol., 72, 478-485.
21.Fang, X. J., Jiang, H., Zhu, Y. Q., Zhang, L. Y.,
Fan, Q. H., and Tian, Y. (2014) Doxorubicin induces drug resistance and
expression of the novel CD44st via NF-κB in human breast cancer
MCF-7 cells, Oncol. Rep., 31, 2735-2742,
doi: 10.3892/or.2014.3131.
22.Campos, D., Freitas, D., Gomes, J.,
Magalhães, A., Steentoft, C., Gomes, C., Vester-Christensen, M.
B., Ferreira, J. A., Afonso, L. P., Santos, L. L., Pinto de Sousa, J.,
Mandel, U., Clausen, H., Vakhrushev, S. Y., and Reis, C. A. (2015)
Probing the O-glycoproteome of gastric cancer cell lines for biomarker
discovery, Mol. Cell. Proteomics, 14, 1616-1629,
doi: 10.1074/mcp.M114.046862.
23.Steentoft, C., Fuhrmann, M., Battisti, F., Van
Coillie, J., Madsen, T. D., Campos, D., Adnan Halim, A., Vakhrushev, S.
Y., Joshi, H. J., Schreiber, H., Mandel, U., and Narimatsu, Y. (2019) A
strategy for generating cancer-specific monoclonal antibodies to
aberrant O-glycoproteins: identification of a novel dysadherin-Tn
antibody, Glycobiology, 29, 307-319,
doi: 10.1093/glycob/cwz004.
24.Welinder, C., Baldetorp, B., Borrebaeck, C.,
Fredlund, B.-M., and Jansson, B. (2011) A new murine IgG1 anti-Tn
monoclonal antibody with in vivo anti-tumor activity,
Glycobiology, 21, 1097-1107,
doi: 10.1093/glycob/cwr048.
25.Posey, A. D., Schwab, R. D., Boesteanu, A. C.,
Steentoft, C., Mandel, U., Engels, B., Stone, J. D., Madsen, T. D.,
Schreiber, K., Haines, K. M., Cogdill, A. P., Chen, T. J., Song, D.,
Scholler, J., Kranz, D. M., Feldman, M. D., Young, R., Keith, B.,
Schreiber, H., Clausen, H., Johnson, L. A., and June, C. H. (2016)
Engineered CAR T cells targeting the cancer-associated Tn-glycoform of
the membrane mucin MUC1 control adenocarcinoma, Immunity,
44, 1444-1454, doi: 10.1016/j.immuni.2016.05.014.
26.Gorchakov, A. A., Kulemzin, S. V., Kochneva, G.
V., and Taranin, A. V. (2020) Challenges and prospects of chimeric
antigen receptor T-cell therapy for metastatic prostate cancer, Eur.
Urol., 77, 299-308,
doi: 10.1016/j.eururo.2019.08.014.