In vitro Reconstitution of the S. aureus 30S Ribosomal Subunit and RbfA Factor Complex for Structural Studies
A. G. Bikmullin1,a*, L. I. Nurullina1, N. S. Garaeva1, E. A. Klochkova1, D. S. Blokhin1, A. A. Golubev1,2, Sh. Z. Validov1, I. Sh. Khusainov1,2,3, K. S. Usachev1,b*, and M. M. Yusupov1,2,c*
1Kazan Federal University, 420008 Kazan, Russia2Institute of Genetics and Molecular and Cellular Biology (IGBMC), 67400 Illkirch-Graffenstaden, France
3Max Planck Institute for Biophysics, 60438 Frankfurt-am-Main, Germany
* To whom correspondence should be addressed.
Received December 5, 2019; Revised February 17, 2020; Accepted March 14, 2020
Ribosome-binding factor A (RbfA) from Staphylococcus aureus is a cold adaptation protein that is required for the growth of pathogenic cells at low temperatures (10-15°C). RbfA is involved in the processing of 16S rRNA, as well as in the assembly and stabilization of the small 30S ribosomal subunit. Structural studies of the 30S–RbfA complex will help to better understand their interaction, the mechanism of such complexes, and the fundamental process such as 30S subunit assembly that determines and controls the overall level of protein biosynthesis. This article describes protocols for preparation of RbfA and the small 30S ribosomal subunits and reconstitution and optimization of the 30S–RbfA complex to obtain samples suitable for cryo-electron microscopy studies.
KEY WORDS: Staphylococcus aureus, ribosome, cryo-electron microscopy, RbfA, translation factor, 30S subunit assemblyDOI: 10.1134/S000629792005003X
INTRODUCTION
Staphylococcus aureus is currently one of the most common and dangerous pathogens. These gram-positive bacteria cause many hospital- and community-acquired diseases of the respiratory tract and skin in humans. The main method of combating this pathogen is antibiotic therapy, but its effectiveness is constantly reduced due to the progressive resistance of staphylococci to the antibiotics used against them. There are cases of S. aureus strains that have multiple resistance to different classes of antibiotics, for example, methicillin-resistant S. aureus (MRSA) [1-5].
A key element of all living cells that control their activity is the ribosome. The number of ribosomes and their functionality determine the overall level of protein synthesis at each stage of normal cell growth and under stressful conditions [6]. The ribosome is a macromolecular RNA–protein complex, its assembly is a complicated process that includes associated steps of biosynthesis of rRNA and 30S and 50S proteins, their modifications, and the correct folding and binding to each other. This process is strictly conservative and is tightly regulated at the level of rRNA molecules and depends on their abundance in the cell. The formation of ribosomal subunits also depends on the cell growth rate. Errors at the assembly stages reduce the overall metabolic rate and can even be fatal for microorganisms [7]. The assembly of the ribosome and its activity at various stages of life are regulated by special protein factors, which can interrupt the function of the protein biosynthesis, turning it on and off. Some factors are constantly active, whereas others are activated only under stress [8]. One of such stress-activated factors is ribosome-binding factor A (RbfA). It is a cold adaptation protein necessary for cell growth at low temperatures (10-15°C). RbfA has a specific affinity to 30S (not to 70S) and is needed for its maturation: a factor is required for efficient processing of 16S rRNA (contributing to the correct conformation of the h1 helix). It is also assumed that RbfA is involved in stabilization of the small subunit even after maturation of 16S rRNA, interacting with its 5′-end. RbfA-deficient Escherichia coli strain demonstrate a decrease of active 70S ribosomes (so an increase in the number of individual subunits), as well as an increase in the number of precursor 16S rRNA molecules (17S rRNA). Proteins of the RbfA family are found in most eubacteria and archaebacteria, as well as in plant chloroplasts and mitochondria of higher organisms including humans. RbfA from S. aureus is a small protein consisting of about 115 amino acid residues with molecular weight ~14 kDa [9-13].
Biogenesis of ribosomes is one of the most fundamental processes in all living organisms. The study of particular details of the assembly of ribosomes and their subunits in the human pathogen S. aureus will provide more information about this complex process as a whole, and it will also teach more about how the ribosome itself functions. The study of the structure of the S. aureus 30S–RbfA complex will demonstrate the localization of the factor on the subunit and describe the mechanism of its binding, i.e. will allow better understanding the stages of maturation of the small subunit and the formation of the translation initiation complex.
Cryo-electron microscopy today, along with nuclear magnetic resonance spectroscopy and X-ray diffraction analysis, is one of the most informative methods in molecular and structural biology. It is possible to obtain resolution above 2 Å by this method. The main feature of this type of microscopy is that macromolecules are fixed in their native shape due to instant freezing in liquid ethane or a mixture of ethane and propane [14-17].
With the development of cryo-electron microscopy, the spectrum of ribosomal complex structural studies, including that of bacterial ribosomes, has expanded significantly [18-21]. These studies have shown that, despite the structural homology of the ribosomes of various bacteria, their optimum ionic conditions, and the concentration of Mg2+ ions to maintain proper in vitro conformation are different. This is especially true for individual isolated ribosomal subunits, which in the absence of stabilizing contacts between subunits (intersubunit bridges) are the most susceptible to structural distortions, especially at the interface. At the same time, the small subunit is the most fragile, and it is often deformed in the neck that supports the orientation of the subunit head relative to the body. Such mobility of the subunit introduces not only distortions into the final structure, but also significantly increases the heterogeneity of the sample. Thus, the selection of the optimal conditions for purification and stabilization of the 30S subunits and the 30S–RbfA complex is a necessary and important requirement for preparing samples for structural analysis.
In this article, we present an optimized method for the reconstitution and purification of the complex of S. aureus RbfA factor and 30S ribosomal subunit for further analysis by cryo-electron microscope. Samples of high purity S. aureus RbfA and small subunits were obtained, then we induced their binding and their additional purification by NiNTA pull-down assay to increase the proportion of the 30S subunits associated with RbfA in the final sample. This technique can be further applied to a variety of proteins that bind to the small subunit of the S. aureus ribosome.
MATERIALS AND METHODS
Materials. In this work we used isopropyl-β-D-1-thiogalactopyranoside (IPTG), spermidine, kanamycin, chloramphenicol (Euromidex, France); LB, LB agar (Invitrogen, USA); a cocktail of protease inhibitors (Roche, Switzerland); Superflow NiNTA resin (QIAGEN, Germany); and bovine serum albumin (BSA) (Helicon, Russia). Other chemical reagents were classified as analytical grade, reagent grade, or special purity grade.
Preparing S. aureus RbfA. The S. aureus RbfA gene was amplified from chromosomal DNA with the insertion of restriction sites NdeI and HindIII. The following specific forward (5′-TTTTTTCATATGAGCAGTATGAGAGCAGAGCG-3′) and reverse (5′-TTTTTTAAGCTTATCTATCTTGTTTGTGTAAATCTTGAATCA-3′) primers (Eurogen, Russia) were used. The temperature mode of the polymerase chain reaction (PCR), as well as the composition of the reaction mixture was selected in accordance with the melting temperature of the primers (59°C) and according to the protocol of the commercial PCR enzyme Phusion Green High-Fidelity DNA Polymerase. Restriction of PCR fragments and pET28a plasmid with appropriate enzymes and purification of DNA components and their ligation were performed according to the protocols of commercial enzymes NdeI and HindIII, and the GeneJET Gel Extraction Kit and the T4 DNA Ligase Kit. Then the ligase mixture was transformed into E. coli strain DH5α and the vector was purified using a commercial GeneJET Plasmid MiniPrep kit. As a result, we obtained the vector RbfA_Sa :: pET28a carrying the rbfA gene with a six-histidine tag at the N-terminus (His-tag) under the control of the LacIq promoter. At all stages of cloning, enzymes and commercial kits manufactured by Thermo Fisher Scientific were used.
Protein expression was performed in E. coli BL21 strain (DE3, pLysS) on selective rich nutrient LB medium containing kanamycin and chloramphenicol. The cells were grown at 37°C with shaking at the rate of 180 rpm using an Inforse HT shaker incubator (Inforse, Germany). At optical density OD600 = 0.6, the expression of the RbfA was induced by adding IPTG to a final concentration of 1 mM. Expression was carried out for 6 h at the temperature of 30°C and shaking rate of 180 rpm. Then cells were collected by centrifugation (Beckman, USA), the pellet was frozen and stored at –20°C.
The cells were disrupted in basic lysis buffer 1 (see composition below) by endogenous T7 lysozyme (pLysS) and sonication with a HD2070 ultrasonic homogenizer (Bandelin, Germany) in the presence of protease inhibitor cocktail, including metalloproteases. Cell debris was pelleted by centrifugation at 25,000g for 30 minutes and 100,000g for 45 minutes using an Avanti JXN-26 (JA-25.50 rotor) and an Optima XPN (45Ti rotor) centrifuges (Beckman Coulter, USA), respectively.
RbfA purification. Purification of RbfA from resulting supernatant was performed sequentially by metal chelate affinity chromatography and gel filtration. Metal chelate chromatography on NiNTA resin was carried out in buffer 1 (20 mM Tris-HCl, pH 7.6, 0.5 M NH4Cl, 1 mM DTT), including intermediate steps of salt wash in buffer 2 (20 mM Tris-HCl (pH 7.6), 1 M NH4Cl, 1 mM DTT), low imidazole wash in buffer 3 (20 mm Tris-HCl (pH 7.6), 0.5 M NH4Cl, 20 mM imidazole, 1 mM DTT) followed by elution in buffer 4 (20 mM Tris-HCl (pH 7. 6), 0.5 M NH4Cl, 0.3 M imidazole, 1 mM DTT). Then, protein was precipitated by ammonium sulfate (80%, w/v) [22].
Gel filtration was performed using an NGC Discover chromatographic system (BioRad, USA) in buffer containing 50 mM sodium phosphate (pH 6.8) and 0.25 M NH4Cl. The purity of the sample was evaluated by polyacrylamide gel electrophoresis under denaturing conditions (SDS-PAGE) in pH 8.3 Tris-glycine buffer solution (25 mM Tris basic, 250 mM glycine, 0.1% sodium dodecyl sulfate (w/v)) at 20°C and operating voltage 140 V.
Purification of 70S ribosomes. Ribosomes of S. aureus were isolated according to a previously published protocol [23]. 30S subunits were obtained by dissociation of 70S ribosomes in a sucrose gradient (0-30%) in buffer D (30 mM NH4Cl, 1 mM MgOAc, 10 mM Hepes-K (pH 7.5), 1 mM DTT) on an Optima XPN-80 ultracentrifuge (Beckman Coulter) with SW28 rotor at 20,550 rpm for 16 h at 4°C. Gradient fractions containing small ribosome subunits were pooled and concentrated using Amicon Ultra spin centricons (Merck Millipore, Ireland; pore size 100 kDa) with buffer exchange to buffer G (10 mM NH4Cl, 10 mM MgOAc, 10 mM Hepes-K (pH 7.5), 50 mM KCl, 1 mM DTT, 2.5 mM spermidine). The purity of ribosomes and subunits was also evaluated by SDS-PAGE (conditions were similar to analysis of samples after gel filtration) and ethidium bromide agarose gel electrophoresis (0.8% agarose gel (w/v), TBE buffer (pH 8.0) with 89 mM Tris basic, 89 mM boric acid, 2 mM EDTA, 0.5 μg/ml ethidium bromide (w/v), at 20°C with operating voltage 180 V.
Reconstitution and purification of 30S–RbfA complex. Ten microliters (10 μl) of different concentrations of RbfA in phosphate buffer after gel filtration were added to 90 μl of a mixture of 30S subunits in buffer G so that the 30S/RbfA ratio was 1/1, 1/5, and 1/10. The reaction mixture was incubated for 40 minutes at 37°C.
To purify the 30S–RbfA complex from unbound 30S subunits, the mixture was subjected to NiNTA chromatography in the reconstitution buffer. The volume of the NiNTA resin was equal to the volume of the reaction mixture (100 μl). After sample loading, the NiNTA resin was washed with 10 column volumes of buffer G. The column was eluted step-wise with 3 column volumes of each elution buffer containing 100, 150, and 200 mM imidazole. As a control, we used free 30S subunits. Washing and elution stages were similar to the experiment with the complex.
The first five, the 10th, and the 20th washing fractions and the first six elution fractions were selected for further analysis. The fraction volume was 50 μl. The absorption of the selected fractions was analyzed at wavelengths of 260 nm and 280 nm (absorption maxima of nucleic acids and proteins, respectively). Also, these fractions were investigated using protein SDS-PAGE and nucleic acid electrophoresis in an agarose gel in the presence of ethidium bromide. The conditions of electrophoretic studies were similar to those described previously in the analysis of protein samples after gel filtration and subunits after dissociation of ribosomes. Fractions containing the complex were immediately frozen in liquid nitrogen and stored at –80°C. Samples should be transported at low temperatures in dry ice. The samples should be defrosted immediately before the cryo-microscopic studies.
Western blot. For additional qualitative identification of RbfA in a sample of the complex after NiNTA purification, we used Western-blot analysis of the elution fraction with the maximum optical absorption value using a commercial set of secondary chemiluminescent antibodies to the His-tag (HisProbeTM-HRP; Thermo Scientific, USA) containing reagent and substrate solutions for the chemiluminescent enzymatic reaction. Western blot was performed according to the protocol of the commercial kit at room temperature. SDS-PAGE analysis of the elution fractions as well as of free 30S subunits and purified RbfA was performed as described above. Proteins were transferred from a 15% PAGE gel to a polyvinylidene fluoride membrane (PVDF, 0.45 µm, BioRad) in transfer buffer (25 mM Tris-HCl (pH 8.3), 190 mM glycine, and 20% methanol (v/v)) on a Trans-blot SEMI-DRY transfer blot system device (BioRad) at 17 V for 40 min. Nonspecific binding was blocked in a 20% solution of BSA (w/v) in TBST buffer (25 mM Tris-HCl (pH 7.2), 150 mM NaCl, and 0.05% (w/v) Tween 20) upon swinging for 1 h. The membrane was washed twice in TBST buffer, incubated with antibodies with agitation for 1 h, and washed once more in TBST. Then the protein-bound antibodies were activated by mixing the substrate and reagent of the chemiluminescent reaction (1/1). The luminescence of antibodies bound to the recombinant RbfA His-tag was detected using an MP ChemiDoc gel documentation system (BioRad).
Evaluation of the quality of the 30S–RbfA complex sample. The quality of the 30S–RbfA complex sample was evaluated using a Titan Krios electron microscope (Thermo Fisher) at the Institute of Genetics, Molecular and Cellular Biology (Strasbourg, France). After thawing on ice, the sample was applied onto a carbon-coated grid using a Vitrobot device (FEI Company/Thermo Fisher, USA) at 4°C and 100% relative humidity. After soaking for 30 s, the grid was frozen in liquid ethane. A Falcon 3 detector (FEI Company/Thermo Fisher) was used at 75,000 magnification (corresponding to a physical pixel size of 0.85 Å/pix) with defocus of 3 µm and total dose 60 e/Å2.
Binding of RbfA to the large ribosome subunit. To confirm the specificity of binding of RbfA to the small ribosomal subunit, we performed binding of RbfA to the large subunit. The conditions for this binding (50S/RbfA = 1/10), as well as the analysis of samples after NiNTA purification, were identical to binding RbfA to 30S. Elution was performed at 100 mM imidazole concentration. As a control, the free 50S subunits were loaded on NiNTA resin. The washing and elution were carried out identically to the experiment with the complex. Samples obtained after elution were analyzed by SDS-PAGE.
RESULT AND DISCUSSIONS
To prepare the 30S–RbfA complex we used the following strategy: a mixture of 30S subunits and purified RbfA was incubated in vitro at physiologically normal temperature for S. aureus (37°C) in buffer G, which has the optimal composition for ribosomes. The molar ratios of 30S subunits to RbfA molecules were 1/1, 1/5, and 1/10.
The total volume of the reaction mixture was 100 μl. The stability of the small ribosome subunit depends strongly on the buffer conditions, namely, the concentration of Mg2+ in the solution. When RbfA in phosphate buffer is added to 30S subunits, the buffer G in which they are located is diluted; accordingly, the concentration of Mg2+ in the volume decreases. Dilution of the total volume of the reaction mixture by 10% or less is not critical for the stability of the ribosome subunit [24]. Therefore, we added 10 μl of a solution of RbfA in phosphate buffer to 90 μl of 30S subunits in buffer G.
According to published data [9], RbfA binds to the 30S subunit in the region of the decoding center, and the C-terminal end of the protein molecule binds to the core of the subunit. The His-tag binding to the NiNTA resin is located at the N-terminus of the factor. Thus, in theory, RbfA should bind to the NiNTA resin with its N-terminus, while retaining the C-terminus to bind the ribosome subunit. This makes it possible to purify the sample of the 30S–RbfA complex from subunits that did not bind to the factor using the NiNTA resin. As a control experiment, free 30S subunits were loaded on the resin, and they were washed and eluted identically to the experiment with the complex.
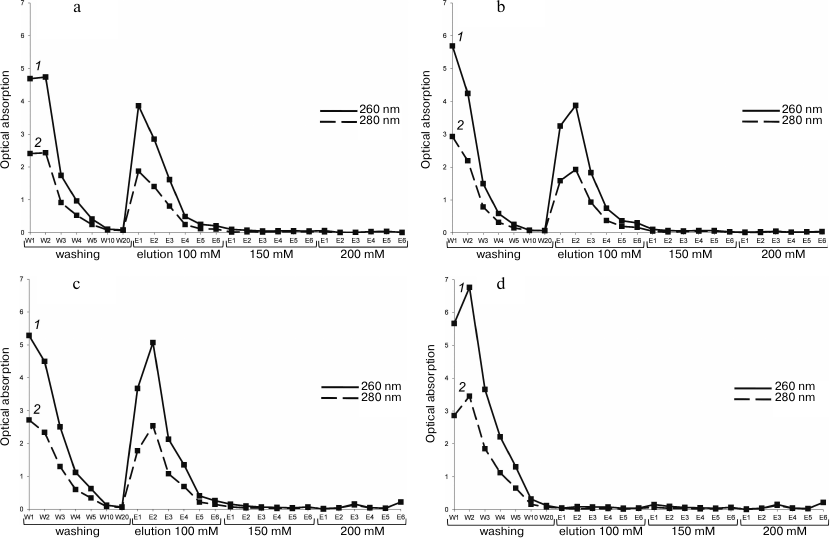
The first five, the 10th, and the 20th fractions of the wash, as well as the first six fractions of each elution, were selected to measure their absorption at wavelengths 260 nm and 280 nm (Fig. 1). The presence of the complex or individual components in the fractions was determined qualitatively: absorption at 260 nm and 280 nm indicated the presence of 16S rRNA and the protein factor. The presence of ribosomal proteins and RbfA was also confirmed by SDS-PAGE, and the presence of the small subunits (16S rRNA) was confirmed by agarose electrophoresis with ethidium bromide (Fig. 2 and 3). As controls, purified RbfA and free small ribosome subunits were used.
Fig. 1. Optical absorption at 260 nm (1) and 280 nm (2) of NiNTA purification fractions of the 30S–RbfA complex at ratio of 30S/RbfA = 1/1 (a), 1/5 (b), and 1/10 (c) and free 30S ribosomal subunits (d). W1-W20), washing fractions; E1-E6), elution fractions at 100 mM, 150 mM, and 200 mM imidazole.
Figure 1 a-c shows that more than half of the 30S subunits did not bind to the RbfA (W1-W20). It is possible that not all native 30S subunits are able to bind to the factor, or these subunits are damaged and lose their ability to bind. Free 30S subunits also did not bind to the resin in the control experiment (Fig. 1d). Thus, purification of the 30S–RbfA complex by NiNTA allowed us to purify the sample from subunits not bound with RbfA and thereby significantly increase the homogeneity of the sample before analysis by cryo-electron microscopy.
We found that 100 mM imidazole concentration is sufficient for elution of the complex. Using a minimally sufficient concentration of imidazole provides maximum yield of the sample under conditions close to optimal, as well as reducing the risk of additional artifacts and noise on electron micrographs [25]. The greatest amount of the complex is observed in elution fraction E2 with a tenfold excess of RbfA. We used this ratio for subsequent structural studies. For other ratios we tested, the total yield of the complex was ~20% less. Higher 30S/RbfA ratios were not analyzed.
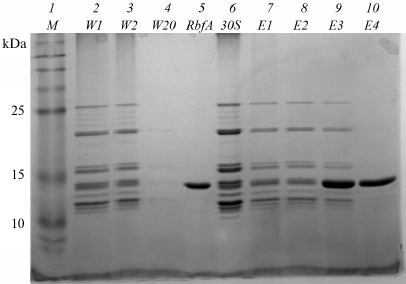
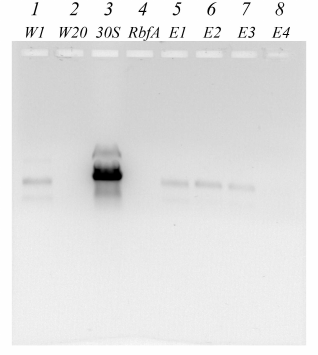
For analysis using electrophoresis methods fractions of the complex reconstituted at molar ratio 1/5 of 30S subunits to RbfA factor were taken. Electropherograms are shown in Figs. 2 and 3, and the data of these figures are correlated with the data presented in Fig. 1. They confirm the reconstitution of the complex. Some fractions of washing (W), elution (E), as well as separately free 30S subunits and purified RbfA as controls were subjected to SDS-PAGE and agarose gels analyses.
Fig. 2. SDS-PAGE of fractions after NiNTA purification of the 30S–RbfA complex (30S/RbfA = 1/5). Lanes: 1) protein ladder; 2-4) washing fractions W1, W2, and W20; 5) RbfA control; 6) 30S control; 7-10) elution fractions E1-E4 with 100 mM imidazole.
Fig. 3. Electrophoretic analysis in agarose gel in the presence of ethidium bromide of fractions after NiNTA purification of the 30S–RbfA complex (30S/RbfA = 1/5). Lanes: 1-2) washing fractions W1, W20; 3) 30S control; 4) RbfA control; 5-8) elution fractions E1-E4 with 100 mM imidazole.
In the first washing fractions, we see the presence of ribosomal proteins on SDS-PAGE and 16S rRNA on agarose gel (the location of the stained protein spots and the fluorescence under ultraviolet light coincide with the 30S and RbfA controls). The last washing fraction does not contain proteins or rRNA. In the first elution fractions we again observe the presence of ribosomal proteins and RbfA on PAGE and 16S rRNA on an agarose gel, this showing that the 30S–RbfA complex was bound to the NiNTA resin. The majority of the complex eluted in the first two fractions. In the E4 fraction on SDS-PAGE (Fig. 2), the presence of only the RbfA protein is observed, which was in excess compared to the ribosome subunits.
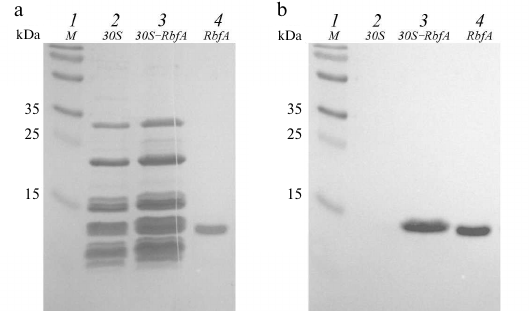
For additional identification of RbfA in the prepared 30S–RbfA complex a Western blot of the elution fraction with the highest concentration of the sample (E2) was performed. We used commercial secondary chemiluminescent antibodies specific for the histidine tag. The only component of our system that has a His-tag is the recombinant RbfA. Figure 4a shows the results of SDS-PAGE of the E2 fraction and control samples – free 30S subunits and RbfA (the gel was stained using Coomassie dye after the protein was transferred to the membrane using the Laemmli protocol [26]).
Fig. 4. Western blot analysis of the 30S–RbfA complex after NiNTA purification: polyacrylamide gel (a); PVDF membrane after binding to antibodies and their activation (b). Lanes: 1) protein ladder; 2) 30S control; 3) 30S–RbfA complex (elution fraction E2); 4) RbfA control.
We see the presence of ribosomal proteins in elution fraction E2, the profile being similar to the control sample of free small subunits. The protein was transferred to a PVDF membrane from this gel. Visualization of the membrane showed emission, which indicates the binding of the antibody to the RbfA with the His-tag. In Figure 4b, we see the presence of protein in the control RbfA sample and in the sample of the 30S–RbfA complex, while in the control 30S sample there is nothing. The molecular weights of RbfA in the control and complex are the same. We confirm the presence of RbfA protein in the complex.
It can be assumed that the binding of RbfA to the 30S subunit is caused by nonspecific interaction of the RbfA protein with rRNA. To confirm the specificity of binding of RbfA to the small subunit, the reconstitution and purification experiment described above was repeated, but instead of 30S subunits with RbfA, 50S subunits were incubated (50S/RbfA = 1/10). Elution from NiNTA was carried out with 100 mM imidazole. As a control for this experiment, free 50S were applied on the NiNTA resin and eluted under similar conditions.
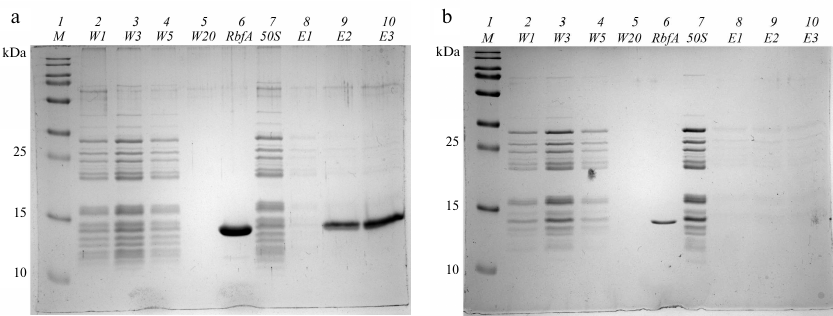
Absorbance values were determined at 260 nm and 280 nm for washing fractions W1-W20 and elution fractions E1-E6. The washing fractions W1, W3, W5, and W20, as well as the elution fractions with the highest concentration of sample (E1-E3), were studied using SDS-PAGE (Fig. 5a). After applying the free 50S subunits to the NiNTA, the same fractions of washing and elution were analyzed by electrophoresis as in the mixture with RbfA (Fig. 5b).
Fig. 5. SDS-PAGE fractions after purification of a mixture of 50S and RbfA (a) and free 50S (b). Lanes: 1) protein ladder; 2-5) washing fractions W1, W3, W5, W20; 6) RbfA control; 7) 50S control; 8-10) elution fractions E1-E3 with 100 mM imidazole.
On the gels shown in Fig. 5 a and b, we see the absence of 50S subunit proteins in the elution fractions E1-E3 both in the experiment with RbfA and in the experiment with free subunits. In the experiment with the RbfA, it is seen that RbfA binds to the NiNTA resin and is washed away from it during elution. 50S subunits are found only in washing fractions (W). We conclude that 50S subunits do not bind to either the NiNTA or the RbfA, i.e., RbfA has an affinity for the 30S subunit. The RbfA specifically binds to the components of the small subunit.
Figure 6 shows an electron micrograph of a sample of the 30S–RbfA complex after purification by NiNTA. Basically, one can notice the individual 30S subunits. There are also a number of subunit aggregates. The homogeneity of the sample allows further data collection. The size of the subunits coincides with data presented in the literature [27] and is about 15 × 20 nm. The presence of the RbfA on the 30S subunit, its structure and localization, as well as the total proportion of subunits bound to the protein, can be determined only after processing and systematization of the data set.
Fig. 6. Microphotograph of a sample of the 30S–RbfA complex obtained using an electron microscope.
The results presented above indicate that a complex of RbfA and the 30S ribosome subunit was obtained and purified for further structural studies using cryo-electron microscopy.
Purified ribosome-binding factor RbfA and the 30S the small subunits of S. aureus ribosomes were prepared. Then, the 30S–RbfA macromolecular complex was reconstituted, the conditions for its additional purification were optimized, the purification itself was carried out, and the sample was characterized and correctly frozen for further studies. A preliminary visual microscopic assessment of the quality of the sample was also carried out. Thus, a sample of the 30S–RbfA complex of S. aureus has obtained for structural studies using cryo-electron microscopy.
Funding. This study was supported by the Russian Foundation for Basic Research (project No. 18-34-00375).
Acknowledgments. We thank the German Academic Exchange Service (DAAD).
Conflict of interest. The authors declare that we have no financial or other conflict of interest in this work.
Compliance with ethical standards. This article does not contain studies involving humans or other animals.
REFERENCES
1.Lowy, F. D. (1998) Medical progress –
Staphylococcus aureus infections, N. Engl. J.
Med., 339, 520-532,
doi: 10.1056/NEJM199808203390806.
2.Kluytmans, J., and Verbrugh, H. (1997) Nasal
carriage of Staphylococcus aureus: epidemiology, underlying
mechanisms, and associated risks, Clin. Microbiol. Rev.,
10, 505-520, doi: 10.1128/CMR.10.3.505.
3.Le Loir, Y., Baron, F., and Gautier, M. (2003)
Staphylococcus aureus and food poisoning, Genet. Mol.
Res., 2, 63-76.
4.Cimolai, N. (2008) MRSA and the environment:
Implications for comprehensive control measures, Eur. J. Clin.
Microbiol. Infect. Dis., 27, 481-493,
doi: 10.1007/s10096-008-0471-0.
5.Wilson, D. N. (2014) Ribosome-targeting antibiotics
and mechanisms of bacterial resistance, Nat. Rev. Microbiol.,
12, 35-48, doi: 10.1038/nrmicro3155.
6.Kaczanowska, M., and Rydén-Aulin, M. (2007)
Ribosome biogenesis and the translation process in Escherichia
coli, Microbiol. Mol. Biol. Rev., 71, 477-494.
7.Shajani, Z., Sykes, M. T., and Williamson, J. R.
(2011) Assembly of bacterial ribosomes, Annu. Rev. Biochem.,
800, 521-526,
doi: 10.1146/annurev-biochem-062608-160432.
8.Starosta, A. L., Lassak, J., Jung, K., and Wilson,
D. N. (2014) The bacterial translation stress response, FEMS,
38, 1172-1201, doi: 10.1111/1574-6976.12083.
9.Datta, P. P. Wilson, D. N., Kawazoe, M., Swami, N.
K., Kaminishi, T., Sharma, M. R., Booth, T. M., Takemoto, C., Fucini,
P., Yokoyama, S., and Agrawal, R. K. (2007) Structural aspects of RbfA
action during small ribosomal subunit assembly, Mol. Cell,
9, 434-445, doi: 10.1016/j.molcel.2007.08.026.
10.Xia, B., Ke, H., Shinde, U., and Inouye M. (2003)
The role of RbfA in 16S rRNA processing and cell growth at low
temperature in Escherichia coli, J. Mol. Biol.,
19, 332, 575-584, doi: 10.1016/S0022-2836(03)00953-7.
11.Huang, Y. J., Swapna, G. V., Rajan, P. K., Ke,
H., Xia, B., Shukla, K., Inouye, M., and Montelione, G. T. (2003)
Solution NMR structure of ribosome-binding factor A (RbfA), a
cold-shock adaptation protein from Escherichia coli,
J. Mol. Biol., 21, 327, 521-536,
doi: 10.1016/S0022-2836(03)00061-5.
12.Rubin, S. M., Pelton, J. G., Yokota, H., Kim, R.,
and Wemmer, D. E. (2003) Solution structure of a putative ribosome
binding protein from Mycoplasma pneumoniae and comparison to a
distant homolog, J. Struct. Funct. Genomics, 4, 235-243,
doi: 10.1023/b:jsfg.0000016127.57320.82.
13.Blokhin, D. S., Bikmullin, A. G., Nurullina, L.
I., Garaeva, N. S., Validov, Sh. Z., Klochkov, V. V., Aganov, A. V.,
Khusainov, I. Sh., Yusupov, M. M. and Usachev, K. S. (2018) Backbone
and side chain NMR assignments for the ribosome binding factor A (RbfA)
from Staphylococcus aureus, Biomolecular NMR Assignments,
13, 27-30, doi: 10.1007/s12104-018-9845-0.
14.Merino, F., and Raunser, S. (2016) Cryo-EM as a
tool for structure-based drug development, Angew. Chem. Int. Ed.
Engl., doi: 10.1002/anie.201608432.
15.Cheng, Y., Grigorieff, N., Penczek, P. A., and
Walz, T. (2015) A primer to single-particle cryo-electron microscopy,
Cell, 161, 438-449,
doi: 10.1016/j.cell.2015.03.050.
16.Binshtein, E., and Ohi, M. D. (2015)
Cryo-electron microscopy and the amazing race to atomic resolution,
Biochemistry, 54, 3133-3141,
doi: 10.1021/acs.biochem.5b00114.
17.Vonk, J., and Mills., D. J. (2017) Advances in
high-resolution cryo-EM of oligomeric enzymes, Curr. Opin. Struct.
Biol., 46, 48-54, doi: 10.1016/j.sbi.2017.05.016.
18.Shimokava-Chiba, N., Muller, C., Fujiwara, K.,
Beckert, B., Koreaki, I., Wilson., D. N. and Chiba, S. (2019) Release
factor-dependent ribosome rescue by BrfA in the Gram-positive bacterium
Bacillus subtilis, Nat. Commun., 10, 5397,
doi: 10.1038/s41467-019-13-408-7.
19.Khusainov, I., Vicens, Q., Ayupov, R., Usachev,
K., Myasnikov, A., Simonetti, A., Validov, Sh., Kieffer, B., Yusupova,
G., Yusupov, M., and Hashem, Y. (2017) Structures and dynamics of
hibernating ribosomes from Staphylococcus aureus mediated by
intermolecular interactions of HPF, EMBO J., 36,
2073-2087, doi: 10.15252/embj.201696105.
20.Li, X., Sun, Q., Jiang, C., Yang, K., Hung, L.,
Zhang, J. and Sacchettini, J. C. (2015) Structure of ribosomal
silencing factor bound to Mycobacterium tuberculosis ribosome,
Structure, 23, 1858-1865,
doi: 10.1016/j.str.2015.07.014.
21.Mishra, S., Ahmed, T., Tyagi, A., Shi, J., and
Bhushan, S. (2018) Structures of Mycobacterium smegmatis 70S
ribosomes in complex with HPF, tmRNA, and P-tRNA, Sci. Rep.,
8, 13587, doi: 10.1038/s41598-018-31850-3.
22.Ayupov, R. Kh., Khusainov, I. Sh., Validov, S.
Z., Yusupova, G. Z., and Yusupov, M. M. (2016) Isolation and
purification of Staphylococcus aureus hibernation-promoting
factor inactivating of the ribosome, Int. J. Pharm. Technol.,
8, 14392-14398.
23.Khusainov, I., Vicens, Q., Bochler, A., Grosse,
F., Myasnikov A., Ménétret, J. F., Chicher, J., Marzi,
S., Romby, P., Yusupova, G., Yusupov, M., and Hashem, Y. (2016)
Structure of the 70S ribosome from human pathogen Staphylococcus
aureus, Nucleic Acids Res., 44, 10491-10504,
doi: 10.1093/nar/gkw933.
24.Weiss, R. L., Kimes, B. W., and Morris, D. R.
(1973) Cations and ribosome structure. III. Effects on the 30S and 50S
subunits of replacing bound Mg2+ by inorganic cations,
Biochemistry, 12, 450-456.
25.Guo, F., and Jiang, W. (2014) Single particle
cryo-electron microscopy and 3-D reconstruction of viruses, Methods
Mol. Biol., 1117, 401-443,
doi: 10.1007/978-1-62703-776-1_19.
26.Laemmli, U. K. (1970) Cleavage of structural
proteins during the assembly of the head of bacteriophage T4,
Nature, 227, 680-685.
27.Vasiliev, V. D. (1974) Morphology of the
ribosomal 30S subparticle according to electron microscopic data,
Acta Biol. Med. Ger., 33, 779-793.