Analysis of Glycosyl-Enzyme Intermediate Formation in the Catalytic Mechanism of Influenza Virus Neuraminidase Using Molecular Modeling
E. M. Kirilin1,a and V. K. Švedas1,2,b*
1Belozersky Institute of Physico-Chemical Biology, Lomonosov Moscow State University, 119991 Moscow, Russia2Lomonosov Moscow State University, Faculty of Bioengineering and Bioinformatics, 119991 Moscow, Russia
* To whom correspondence should be addressed.
Received February 3, 2020; Revised March 2, 2020; Accepted March 2, 2020
Using classical molecular dynamics, constant-pH molecular dynamics simulation, metadynamics, and combined quantum mechanical and molecular mechanical approach, we identified an alternative pathway of glycosyl-enzyme intermediate formation during oligosaccharide substrate conversion by the influenza H5N1 neuraminidase. The Asp151 residue located in the enzyme mobile loop plays a key role in catalysis within a wide pH range due to the formation of a network of interactions with water molecules. Considering that propagation of influenza virus takes place in the digestive tract of birds at low pH values and in the human respiratory tract at pH values close to neutral, the existence of alternative reaction pathways functioning at different medium pH can explain the dual tropism of the virus and circulation of H5N1 viral strains capable of transmission from birds to humans.
KEY WORDS: influenza, neuraminidase, glycosyl-enzymeDOI: 10.1134/S0006297920040094
Abbreviations: QM/MM, combined quantum mechanics and molecular mechanics.
Highly pathogenic strains of the influenza H5N1 virus (bird flu) are not
only a serious problem for the populations of wild and domestic birds.
They can also cause disease outbreaks in humans with a mortality of
~60%. According to the World Health Organization, the peak of the H5N1
viral infection had been observed from 1998 to 2015 [1]; however, the mechanism of virus adaptation to
humans is still under investigation [2], since the
development of dual (bird/human) tropism is a very dangerous stage of
the virus evolution. As a rule, the virus infects host cells via
interaction with glycan receptors – oligosaccharide fragments
with terminal sialic acid – on the cell membrane. Neuraminidase
is a vital enzyme of the virus lifecycle that catalyzes hydrolysis of
sialylated glycans with the following release of mature influenza
virions. Neuraminidase acts in concert with hemagglutinin (HA), which
is responsible for the primary recognition of the host cell glycans and
is involved in the virulence mechanism of the influenza virus [3]. Based on the antigenic properties, there are nine
types of neuraminidases that are classified into group 1
(neuraminidases N1, N4, N5, and N8) and group 2 (neuraminidases N2, N3,
N6, N7, and N9) [4]. Neuraminidases from the H5N1
virus strain isolated from humans and birds display low activity at
strongly acidic pH [5], while neuraminidases from
other avian influenza strains maintain high activity in strongly acidic
environment, as the virus propagation in birds occurs in their
digestive tract. The majority of seasonal human influenza strains
display low activity at acidic pH [6, 7]; the virus propagates in the human respiratory
tract, where pH is close to neutral (pH 6.5-7.0) or slightly acidic (pH
5.7-6.0) [8]. Hence, adaptation of neuraminidases
of the H5N1 strain in birds has likely made these enzymes similar to
neuraminidases from human seasonal influenza viruses, which, in turn,
could result in the high probability of their direct transfer from
birds to humans. It was established that glycosidases from the GH34
family, which also includes influenza virus neuraminidase [9, 10], act via the double
replacement mechanism, in which one of the catalytic residues (Tyr406
activated by Glu277) plays a role of a nucleophile during formation of
the covalent glycosyl-enzyme intermediate, while the second residue
(Asp151) serves as a proton donor for stabilization of the leaving
group. This mechanism imposes specific requirements on the functioning
of Asp151 at neutral pH values. The microenvironment of the enzyme
active site, the nature of the adjacent residues, and isolation from
bulk water molecules could modulate the acid-base properties of the
carboxyl group. Similar mechanism has been demonstrated for some
glycosidases, in which the binding of the oligosaccharide substrate
results in the active site closing and displacement of solvent
molecules, thus creating the opportunity for the protonated Asp151 to
interact directly with the leaving group [11].
However, according to the X-ray diffraction data, Asp151 in the H5N1
neuraminidase is located in the so-called mobile loop-150 (a.a.
147-152) [4] in the substrate-binding region. It is
directly accessible to the solvent and, therefore, cannot perform the
required functional role at pH 5.7-6.5.
So, how does the Asp151 of the catalytic site of H5N1 neuraminidase function at neutral pH? How does the binding of oligosaccharide substrate in the enzyme active site affect the pKa value of the Asp151 residue? What is the conformational state of the substrate in the active site that ensures the most efficient enzyme catalysis and formation of the glycosyl-enzyme intermediate? In this study, we attempted to answer these questions using a complex approach including classic molecular dynamics, metadynamics, combined quantum mechanics and molecular mechanics (QM/MM), and simulation of titration of ionizable residues.
MATERIALS AND METHODS
Substrate structure. α-Neu5Ac-2,3-β-Gal-1,4-β-Glc-O-Me trisaccharide (typical terminal fragment of glycan receptors in birds) was used as a substrate. The structure of this compound was obtained with the help of Glycam Biomolecule Builderserver [12] using the Amber force field.
Dihedral angles of glycosidic bonds. Dihedral angles of glycosidic bonds were determined using the following procedure. Starting with the terminal sialic acid residue, φ1 and ψ1, φ2 and ψ2, φ1 and ψ1 were determined from the O–C2–O3–C3 and C2–C3–O3–C2 atoms; φi and ψi for the rest of glycosidic bonds were determined as O–C1–O4′–C4′ and C1–O4′–C4′–C3 for the 1→4 bonds and O–C1–O3′–C3′ and C1–O3′–C3′–C2′ for the 1→3 bonds, where the non-reducing end of the glycosidic bond is denoted by an apostrophe.
Molecular dynamics. Substrate structure was prepared using the leap program from the AmberTools package [13]. A system was created for the enzyme and enzyme–substrate complex that included the TIP3P solvent [14], in which the distance from the edge of periodic box to any atom of solute was no less than 15 Å. Cl– and Na+ ions (up to 0.1 M concentration) were added to the system to create conditions close to physiological ones. The total charge of the system was adjusted to zero by adding additional ions. Energy minimization was carried out for each system (2500 steps) followed by heating to 300 K for 60 ps, lifting of limitations from heavy atoms for 290 ps, and relaxation for 5 ns in the NPT ensemble with a gradual adjustment of the density to the constant value of 1.04 g/cm3. Standard simulation in solution was performed with the Amber14 program at 300 K with the integration step of 2 fs under conditions of the NVT ensemble. The system temperature was controlled using a Langevin thermostat [15]. The duration of each trajectory was 1 µs; the dynamic frames were saved every 10 ps.
Accelerated molecular dynamics calculations were carried out with the in-built functions in the Amber14 program using parameters identical to the above mentioned and by setting the following specific values for thresholds and parameters: EthreshD = 13.42, alphaD = 3.2, EthreshP = –20203.56, alphaP = 602.24.
Metadynamics. Metadynamics calculations were carried out using the Amber14 package modified with PLUMED (v. 2.5) [16]. The height of the Gaussian hills in metadynamics was set to the initial value of 0.6 kcal/mol; the width was 0.01 or 0.0075 for proton transfer. New potentials were added every 100 steps at the integration step of 0.25 fs. The temperature of simulation was 300 K. Collective variables were limited from both sides by the quadratic wall potential. Water molecules in the quantum region were also limited by the quadratic potential for the coordination number around the oxygen atom of the leaving group at the distance of 7 Å. Ten parallel calculations with shared metadynamics potential (multiple-walkers technique) were used to accelerate calculations [17].
QM/MM. QM/MM calculations were carried out using the AmberTools package. The SCC-DFTB method was used for describing the quantum region, which has been well established for QM/MM modeling of reactions catalyzed by other enzymes of the sialidase family [18, 19]. The residues Tyr406, Glu277, Arg292, and Asp151 were included in the quantum region, as well as the monomers of sialic acid and the following galactose residue together with 14 water molecules at the distance of 7 Å from the oxygen atom of the leaving group during formation of the covalent glycosyl-enzyme intermediate.
Simulation of titration of ionizable residues in explicit solvent. The pH-REMD method incorporated into Amber14 was used to calculate pKa values of Asp151; simulation of 12 replicas was conducted in the pH range from 2 to 7.5. Replica exchange attempts were performed every 5000 steps; an attempt of re-protonation was conducted every 200 steps, and relaxation was conducted every 300 steps. The pKa values were calculated from the adapted Henderson–Hasselbalch equation taking into account the calculated probability of formation of the protonated and deprotonated forms [20].
Clustering methods. Substrate structure clustering was carried out based on the values of dihedral angles between carbohydrate monomers using the following modification:
description of the phase space of ui angles in the general case of a L-mer box Pc:
Pc = {u ∈ RL; 0 ≤ u1 < 2π}, 1 ≤ i < L,
through transformation:
The product of description is a torus Te (L-mer surface on a unit sphere S2L–1). The description is a local isometry with preserved distance measure [21].
The transformed variables were used for clustering using the non-parametric Bayesian method incorporated into the dpMMlowVar program [22]. Optimization of angular parameter for this method was performed in the range [–0.6; –0.3] with a step of 0.01 based on the silhouette estimation function. The principal component method for the transformed angle values with identification of the first three principal components was used for the visualization of clustering results.
RESULTS AND DISCUSSION
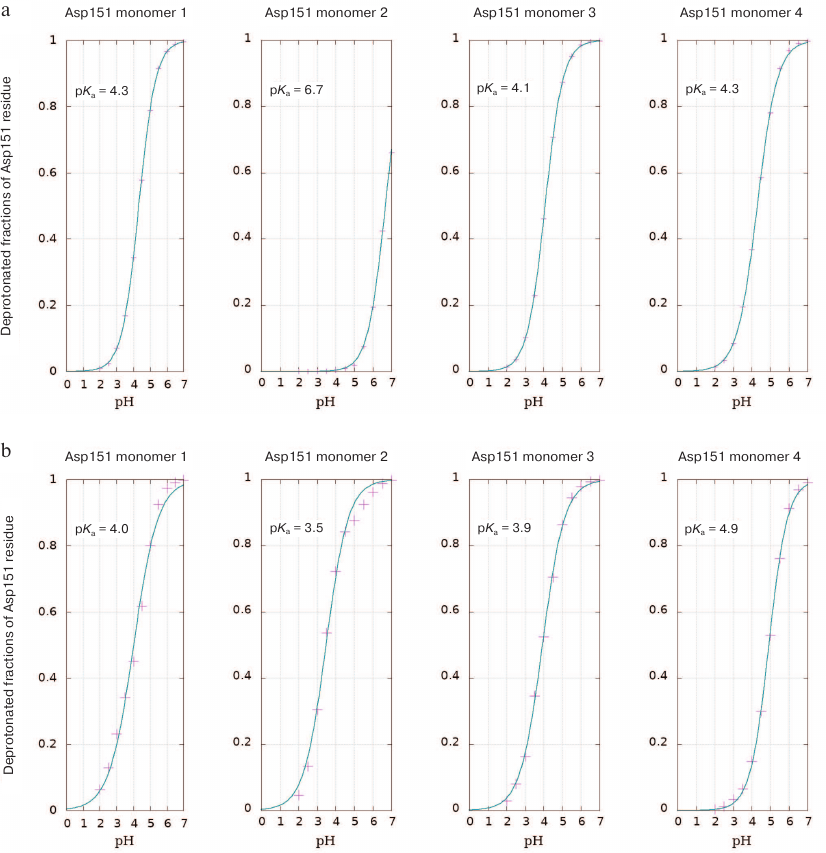
Calculation of Asp151 pKa. The shift in the Asp151 pKa value in the active site of neuraminidase N1 upon substrate binding was determined using QM/MM in an explicit solvent and by calculating the Poisson–Boltzmann electrostatic field according to the discreet protonation model, which considers the ionization state of an investigated residue depending on its current orientation, accessibility to the solvent, and medium pH. The method allows conducting a number of parallel simulations in a wide pH range. In the course of experiment, we performed sampling of the system taking into account dynamic transitions between the conformations with protonated and non-protonated Asp151 carboxyl group. Considering that neuraminidase functions as a tetramer, pKa values were calculated independently for four aspartic acid residues (Fig. 1). The three Asp151 residues in the free enzyme were found to be deprotonated at neutral conditions (pKa values are in the range 4.1-4.3), while one residue (denoted as Asp2 in Fig. 1) displayed higher pKa value of 6.7. Analysis of the loop-150 location in the subunit with abnormally high pKa value of 6.7 (but not in other subunits) demonstrated formation of carboxyl-carboxylate pair between Asp151 and Glu119 during modeling at highly acidic pH values (pH 2.0). No such pair has been observed in the enzyme crystal structure. It is possible that in this case, transition to the denatured catalytically inactive state took place.
Fig. 1. Titration curves of Asp151 residues in the four subunits of H5N1 neuraminidase homotetramer with the calculated pKa values (see Materials and Methods): a) free enzyme; b) enzyme complex with oligosaccharide substrate bound in the active site.
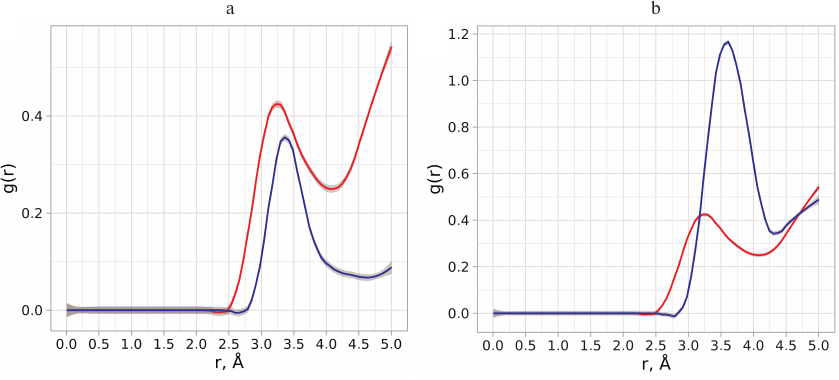
Neuraminidases from various sources differ in the structure of their active site and environment of ionizable residues, as well as in the dynamic behavior of the loop-150 containing Asp151 [23]. This can be the reason for different influence of substrate binding on the pKa values of residues in enzymes from neuraminidase family. Calculations of aspartate residue protonation/deprotonation in the enzyme–substrate complex formed by neuraminidase N1 showed that the pKa value of Asp151 was in the range of 3.5-4.9, which indicated the existence of various stabilizing structural topologies of the oligosaccharide substrate in the active site (see Conformational state of oligosaccharide substrate upon the enzyme binding). The most abundant substrate conformation has pKa 4.9. At the same time, neuraminidase N1 displayed no significant shift in the pKa value upon substrate binding (unlike pKa shift to 8.0-9.0 observed for neuraminidase N9) [11]. At the first glance, the calculated pKa values for Asp151 in neuraminidase N1 produced by modeling contradict the possibility of effective catalysis by this residue in neutral media via direct proton transfer to the leaving group of the substrate. However, this conclusion seems not as accurate, if we consider other possibilities for the leaving group stabilization. Thus, if the carboxyl group of Asp151 is constantly accessible to the solvent molecules, the Grotthuss mechanism of proton transfer is possible, which has been already observed for other enzyme systems [24, 25]. Moreover, the deprotonated form of aspartate can stabilize water molecule in the neutral from or in the form of hydronium ion in the vicinity of the leaving group and, hence, facilitate the proton transfer. In order to evaluate the possibility of such mechanism, we analyzed the distribution of water molecules around oxygen atoms of the leaving group and carboxyl carbon of Asp151 in the free enzyme and in the enzyme–substrate complex determined based on the molecular dynamics trajectories at pH values 5.5, 6.0, 6.5, and 7.0 (Fig. 2, a and b).
Fig. 2. a) Radial distribution function of water molecules (red curve) around the oxygen atom of the leaving group and distribution function of distances from the oxygen atom of the leaving group to the oxygen atoms of Asp151 carboxyl group (blue curve). b) Radial distribution function of water molecules around the carboxyl carbon of Asp151 in the free enzyme (red curve) and enzyme with the bound oligosaccharide substrate (blue curve).
Analysis of the distance distribution between the oxygen atoms of the carboxyl group and oxygen atom of the leaving group revealed that water molecules have direct access to the cleaved bond and could be not only bridge between the Asp151 residue and the leaving group, but also act as proton donors. Introduction of hydronium ion in such proton transfer chain could be facilitated by the surrounding residues in the enzyme, as well as by the possibility of optimal orientation and stabilization of the entire system [26]. In order to evaluate the energy barriers for the formation of the glycosyl-enzyme intermediate via the alternative mechanism, it was necessary to perform conformational analysis of positions of carbohydrate substrate in the active site of neuraminidase to produce the starting structure of the enzyme–substrate complex.
Conformational state of the oligosaccharide substrate upon enzyme binding. The conformational space of the oligosaccharide substrate bound in the active site of neuraminidase N1 was determined by analysis of molecular dynamics trajectories produced by simulation of Asp151 titration at different pH values (pH 5-7). The values of dihedral angles of glycosidic bonds in the course of simulation were extracted, transformed (see Materials and Methods), and used for searching the clusters of stabilized structures (topologies) (Fig. 3).
Fig. 3. Sampling of conformational space of the oligosaccharide substrate in the neuraminidase N1 active sited followed by clustering. For clarification, the data are visualized for the first three principal components during analysis of eight variable dihedral angles that characterize oligosaccharide conformation; the clusters are color-coded.
The identified oligosaccharide topologies and the corresponding values of dihedral angles are presented in the table. Similarly conducted search for additional structural clusters using accelerated molecular dynamics methods revealed no new topologies. The substrate bound in the enzyme active site can assume three possible topologies (table). The predominant topology 1 is characterized by the following values of dihedral angles: φ1 = 84.29, ψ1 = 98.90, φ2 = 95.70, and ψ2 = 105.71. Populations of topologies 2 and 3 in the active site comprise 8 and 14%, respectively. Furthermore, topology 2 is oriented along the active site identically to topology 1, while topology 3 indicates the existence of an alternative binding cavity. It must be mentioned that the structure of the trisaccharide similar to topology 1 was observed upon substrate binding with neuraminidase N2 from the H3N2 strain (crystal structure 4GZW [27]).
Characteristic values of dihedral angles of the oligosaccharide
substrate in neuraminidase N1 active site
Formation of glycosyl-enzyme intermediate. Formation of the glycosyl-enzyme intermediate was modeled using metadynamics. The structure of the enzyme–substrate complex with the dihedral angles and conformation of the neuraminic acid ring 5S1 characteristic for topology 1 (predominant topology for oligosaccharide substrate binding in the enzyme active site, see previous section) was used as a starting structure [28]. The following collective variables, in the phase space of which substrate hydrolysis and formation of the intermediate glycosyl-enzyme product occur, were used: (i) CV1 characterizing the cleavage of glycosidic bond formed by the sialic acid residue in the substrate and formation of new bond with Tyr406 and representing the difference in the distance between C1SIA–OTyr406 and C1SIA–O3LB; (ii) CV2 reflecting activation of nucleophilic residues via proton transfer to Glu277; (iii) CV3 characterizing proton transfer to the leaving group from one of the water molecules or Asp151 (Fig. 4a). CV3 was determined as the minimal distance from glycosidic oxygen of the leaving group to one of the protons of water molecules in the quantum region (eight molecules) or proton in Asp151 (by using this approach we considered the probability of the fastest and energetically favorable proton transfer pathway either from water molecules or Asp151).
Fig. 4. Structures of basic stationary states during formation of the covalent glycosyl-enzyme intermediate in the reaction catalyzed by H5N1 neuraminidase. Gal, leaving group; Neu, sialic acid residue. a) Structure of the enzyme–substrate complex; colored dashed lines, main components of collective variables including the pathway of proton transfer to Glu277 and activation of nucleophilic Tyr406 residue (pink), cleaved and formed bonds (yellow), and any process of proton transfer from adjacent water molecules or Asp151 (blue); b) transition state structure; c) formation of glycosyl-enzyme.
It must be noted that we included the possibility of Asp151 protonation into the starting structure, which allowed us to take into consideration not only the mechanism of proton transfer from the hydronium ion coordinated by the Asp151 carboxylate anion, but also theoretical direct proton transfer to the leaving group. Hence, the selected starting quantum region represented an identical starting point for the two possible pathways of proton transfer to the leaving group of the hydrolyzed substrate.
Using QM/MM with three selected collective variables, we modeled hydrolysis of oligosaccharide substrate by the enzyme and determined the energy surface of this process (Fig. 5). Based on the Pauling criterion of the bond order, the process corresponded to the dissociative ANDN mechanism with the barrier of ~14 kcal/mol (Fig. 5, a and b). Sialic acid residue underwent structural transition from the 5S1 ring conformation in the enzyme–substrate complex (MC; Fig. 4a) to the E4-3H4 conformation in the transition state (TS; Fig. 4b) and 1C4 conformation (IC; Fig. 4c) in the intermediate glycosyl-enzyme, the position of which in the energy diagram was higher than the initial state by 7 kcal/mol (Fig. 5, a and b). In the course of enzymatic reaction, the proton for neutralization of the leaving group was transferred from the nearest water molecule that formed a bridge between the glycosidic bond of the substrate and Asp151. It must be mentioned that Asp151 can play a dual role, similar to the water molecule. When the carboxyl group in Asp151 is protonated, water molecule could act as a shuttle and simultaneously accept proton from Asp151 during proton transfer to the leaving group. In the case when the Asp151 carboxyl group is deprotonated, it can coordinate hydronium ion, which serves as a proton donor for the leaving group. The probability of one or another pathway depends on a number of conditions: medium acidity, loop-150 mobility, as well as the features of the active site organization in the enzyme mutants. The participation in the catalytic mechanism of Asp151 with the protonated carboxyl group seems less favorable, as the loop-150 limits the mobility of the residue and hinders formation of direct proton transfer chain: these states were not identified during independent initiation of molecular dynamics trajectories. Nevertheless, such mechanism cannot be ruled out for the enzyme functioning in an acidic medium. At the same time, proton transfer from the hydronium ion coordinated by the negatively charged carboxyl group of Asp151 established in this work can be universal and occur in a wide pH range, which could explain the dual tropism of the virus and circulation of H5N1 stains capable of direct transfer from birds to humans, as it has been observed in the recent disease outbreaks from 1998 to 2015.
Fig. 5. a) Free energy surface of the covalent glycosyl-enzyme intermediate formation in the reaction of oligosaccharide substrate hydrolysis catalyzed by neuraminidase from the influenza virus H5N1 constructed based on collective variables of the glycosidic bond cleavage at the sialic acid residue and formation of a new bond with Tyr406 (CV1), activation of nucleophilic residue Tyr406 (CV2), and proton transfer from one of the water molecules or Asp151 to the leaving group of the substrate (CV3). MC, enzyme complex with the substrate; TS, transition state prior to the glycosyl-enzyme formation; IC, glycosyl-enzyme intermediate. b) Simplified energy diagram of the covalent glycosyl-enzyme intermediate formation.
The catalytic mechanism of neuraminidases from various influenza viruses, in particular, the ability of these enzymes to function in different media, is interesting from both fundamental and practical viewpoints. The conformational state of the oligosaccharide substrate, as well as the formation and following hydrolysis of the glycosyl-enzyme intermediate, play an important role in the glycan transformation. Molecular modeling of the mechanism of action of neuraminidase N1 allowed us to reveal the affinity of the enzyme active site to a certain substrate conformer and to establish conformational changes taking place in the course of enzymatic catalysis. We also analyzed possible pathways for the formation of glycosyl-enzyme intermediate in media with different pH values, which are directly associated with the virus adaptation and propagation in the human respiratory tract and with the transfer of the virus strains from birds to humans. Understanding the functioning of neuraminidases from different influenza virus strains under different conditions as a key virulence factor is an important step in both prophylactics and fight against influenza and other infections with similar mechanisms.
Funding. This work was supported by the Russian Foundation for Basic Research (project No. 18-34-00953).
Acknowledgements. The study was performed using equipment from the Center of Collective Use of super high-performance computational resources of the Lomonosov Moscow State University [29].
Conflict of interest. The authors declare that they have no conflict of interest.
Ethical approval. This article does not contain studies with human participants or animals performed by any of the authors.
REFERENCES
1.URL: https://www.who.int/influenza/human_animal_interface/H5N1_cumulative_table_archives/en/
2.Welkers, M. R. A., Pawestri, H. A., Fonville, J.
M., Sampurno, O. D., Pater, M., Holwerda, M., Han, A. X., Russell, C.
A., Jeeninga, R. E., Setiawaty, V., de Jong, M. D., and Eggink, D.
(2019) Genetic diversity and host adaptation of avian H5N1 influenza
viruses during human infection, Emerg. Microbes Infect.,
8, 262-271, doi: 10.1080/22221751.2019.1575700.
3.Mitnaul, L. J., Matrosovich, M. N., Castrucci, M.
R., Tuzikov, A. B., Bovin, N. V., Kobasa, D., and Kawaoka, Y. (2000)
Balanced hemagglutinin and neuraminidase activities are critical for
efficient replication of influenza A virus, J. Virol.,
74, 6015-6020, doi: 10.1128/JVI.74.13.6015-6020.2000.
4.Russell, R. J., Haire, L. F., Stevens, D. J.,
Collins, P. J., Lin, Y. P., Blackburn, G. M., Hay, A. J., Gamblin, S.
J., and Skehel, J. J. (2006) The structure of H5N1 avian influenza
neuraminidase suggests new opportunities for drug design,
Nature, 443, 45-49, doi: 10.1038/nature05114.
5.Takahashi, T., Nidom, C. A., Quynh Le, M. T.,
Suzuki, T., and Kawaoka, Y. (2012) Amino acid determinants conferring
stable sialidase activity at low pH for H5N1 influenza A virus
neuraminidase, FEBS Open Bio, 2, 261-266,
doi: 10.1016/j.fob.2012.08.007.
6.Suzuki, T., Takahashi, T., Saito, T., Guo, C.-T.,
Hidari, K. I.-P. J., Miyamoto, D., and Suzuki, Y. (2004) Evolutional
analysis of human influenza A virus N2 neuraminidase genes based on the
transition of the low-pH stability of sialidase activity, FEBS
Lett., 557, 228-232,
doi: 10.1016/S0014-5793(03)01503-5.
7.Takahashi, T., Kurebayashi, Y., Ikeya, K., Mizuno,
T., Fukushima, K., Kawamoto, H., Kawaoka, Y., Suzuki, Y., and Suzuki,
T. (2010) The low-pH stability discovered in neuraminidase of 1918
pandemic influenza A virus enhances virus replication, PLoS One,
5, doi: 10.1371/journal.pone.0015556.
8.Fischer, H., and Widdicombe, J. H. (2006)
Mechanisms of acid and base secretion by the airway epithelium, J.
Membr. Biol., 211, 139-150,
doi: 10.1007/s00232-006-0861-0.
9.Koshland, D. E. (1953) Stereochemistry and the
mechanism of enzymatic reactions, Biol. Rev., 28,
416-436.
10.Guce, A. I., Clark, N. E., Salgado, E. N.,
Ivanen, D. R., Kulminskaya, A. A., Brumer, H., and Garman, S. C. (2010)
Catalytic mechanism of human α-galactosidase, J. Biol.
Chem., 285, 3625-3632,
doi: 10.1074/jbc.M109.060145.
11.Yu, H., and Griffiths, T. M. (2014)
pKa cycling of the general acid/base in glycoside
hydrolase families 33 and 34, Phys. Chem. Chem. Phys.,
16, 5785-5792, doi: 10.1039/C4CP00351A.
12.Woods, R. J. (2008) Biomolecule Builder,
GLYCAM Web, Complex Carbohydrate Research Center, University of
Georgia.
13.Case, D. A., Babin, V., Berryman, J., Betz, R.
M., Cai, Q., et al. (2014) Amber 14, University of California,
San Francisco.
14.Mahoney, M. W., and Jorgensen, W. L. (2000) A
five-site model for liquid water and the reproduction of the density
anomaly by rigid, nonpolarizable potential functions, J. Chem.
Phys., 112, 8910-8922, doi: 10.1063/1.481505.
15.Paterlini, M. G., and Ferguson, D. M. (1998)
Constant temperature simulations using the Langevin equation with
velocity Verlet integration, Chem. Phys., 236, 243-252,
doi: 10.1016/S0301-0104(98)00214-6.
16.Tribello, G. A., Bonomi, M., Branduardi, D.,
Camilloni, C., and Bussi, G. (2014) PLUMED 2: new feathers for an old
bird, Comp. Phys. Commun., 185, 604-613,
doi: 10.1016/j.cpc.2013.09.018.
17.Raiteri, P., Laio, A., Gervasio, F. L.,
Micheletti, C., and Parrinello, M. (2006) Efficient reconstruction of
complex free energy landscapes by multiple walkers metadynamics, J.
Phys. Chem. B, 110, 3533-3539,
doi: 10.1021/jp054359r.
18.Pierdominici-Sottile, G., Horenstein, N. A., and
Roitberg, A. E. (2011) Free energy study of the catalytic mechanism of
Trypanosoma cruzi trans-sialidase. From the Michaelis complex to
the covalent intermediate, Biochemistry, 50, 10150-10158,
doi: 10.1021/bi2009618.
19.Bueren-Calabuig, J. A., Pierdominici-Sottile, G.,
and Roitberg, A. E. (2014) Unraveling the differences of the hydrolytic
activity of Trypanosoma cruzi trans-sialidase and Trypanosoma
rangeli sialidase: a quantum mechanics-molecular mechanics modeling
study, J. Phys. Chem. B, 118, 5807-5816,
doi: 10.1021/jp412294r.
20.Harris, R. C., and Shen, J. (2019)
GPU-accelerated implementation of continuous constant pH molecular
dynamics in Amber: pKa predictions with single-pH
simulations, J. Chem. Inf. Model., 59, 4821-4832,
doi: 10.1021/acs.jcim.9b00754.
21.Costa, S. I. R., Torezzan, C., Campello, A., and
Vaishampayan, V. A. (2013) Flat tori, lattices and spherical codes, 1-8
in 2013 Information Theory and Applications Workshop (ITA),
doi: 10.1109/ITA.2013.6503002.
22.Straub, J., Campbell, T., How, J. P., and Fisher,
J. W. (2015) Small-variance Nonparametric Clustering on the
Hypersphere, The IEEE Conf. on Computer Vision and Pattern
Recognition (CVPR), pp. 334-342.
23.Amaro, R. E., Minh, D. D. L., Cheng, L. S.,
Lindstrom, W. M. Jr., Olson, A. J., Lin, J.-H., Li, W. W., and
McCammon, J. A. (2007) Remarkable loop flexibility in avian influenza
N1 and its implications for antiviral drug design, J. Am. Chem.
Soc., 129, 7764-7765, doi: 10.1021/ja0723535.
24.Riccardi, D., König, P., Prat-Resina, X.,
Yu, H., Elstner, M., Frauenheim, T., and Cui, Q. (2006) “Proton
holes” in long-range proton transfer reactions in solution and
enzymes: a theoretical analysis, J. Am. Chem. Soc.,
128, 16302-16311, doi: 10.1021/ja065451j.
25.Parke, C. L., Wojcik, E. J., Kim, S., and
Worthylake, D. K. (2010) ATP hydrolysis in Eg5 kinesin involves a
catalytic two-water mechanism, J. Biol. Chem., 285,
5859-5867, doi: 10.1074/jbc.M109.071233.
26.Mohammed, O. F., Pines, D., Dreyer, J., Pines,
E., and Nibbering, E. T. (2005) Sequential proton transfer through
water bridges in acid-base reactions, Science, 310,
83-86.
27.Zhu, X., McBride, R., Nycholat, C. M., Yu, W.,
Paulson, J. C., and Wilson, I. A. (2012) Influenza virus neuraminidases
with reduced enzymatic activity that avidly bind sialic acid receptors,
J. Virol., 86, 13371-13383,
doi: 10.1128/JVI.01426-12.
28.Kirilin, E. M., and Švedas, V. K. (2018)
Study of the conformational variety of the oligosaccharide substrates
of neuraminidases from pathogens using molecular modeling, Moscow
Univ. Chem. Bull., 73, 39-45,
doi: 10.3103/S0027131418020050.
29.Sadovnichy, V., Tikhonravov, A., Voevodin, V.,
and Opanasenko, V. (2017) “Lomonosov”: supercomputing at
Moscow State University, Contemporary High Performance Computing,
doi: 10.1201/9781351104005-11.