Antirestriction Protein ArdB (R64) Interacts with DNA
A. A. Kudryavtseva1,a*, I. S. Okhrimenko1, V. S. Didina1, G. B. Zavilgelsky2, and I. V. Manukhov1,2
1Moscow Institute of Physics and Technology, 141707 Dolgoprudny, Moscow Region, Russia2State Research Institute of Genetics and Selection of Industrial Microorganisms, Kurchatov Institute National Research Center, 117545 Moscow, Russia
* To whom correspondence should be addressed.
Received November 15, 2019; Revised January 14, 2020; Accepted January 17, 2020
The antirestriction ArdB protein inhibits the endonuclease activity of type I restriction/modification (RM) systems in vivo; however, the mechanism of inhibition remains unknown. In this study, we showed that recombinant ArdB from Escherichia coli cells co-purified with DNA. When overexpressed in E. coli cells, a portion of ArdB protein formed insoluble DNA-free aggregates. Only native ArdB, but not the ArdBΔD141 mutant lacking the antirestriction activity, co-purified with DNA upon anion-exchange and affinity chromatography or total DNA isolation from formaldehyde-treated cells. These observations confirm the hypothesis that ArdB blocks DNA translocation via the R subunits of the R2M2S complex of type I RM enzymes.
KEY WORDS: antirestriction, transmissible plasmid, R64, ArdBDOI: 10.1134/S0006297920030074
Abbreviations: GST, glutathione S-transferase; RM, restriction/modification (system).
The antirestriction proteins of the ArdB family selectively inhibit the
endonuclease activity of type I restriction/modification (RM) enzymes,
thereby allowing conjugative plasmids to overcome the restriction
barrier. The ardB gene encoding the antirestriction ArdB protein
was found for the first time in the transmissible pKM101 plasmid (IncN
incompatibility group) [1]. ArdB specifically
inhibits the endonuclease activity of type I RM enzymes in vivo
[1, 2] but does not form
complexes with type I RM enzymes in vitro [2]. The C-terminal aspartate residue (D141)
plays the key role in the antirestriction activity of ArdB [3]. Balabanov et al. [4] showed
that ArdB loses its ability to protect unmodified λ.0 phage DNA
in UV-irradiated cells of Escherichia coli K12 from hydrolysis
by EcoKI restriction endonuclease, provided that the cells contain a
considerable amount of unmodified chromosomal. Considering the effect
of unmodified DNA on the antirestriction activity of ArdB, we put
forward a hypothesis that ArdB blocks the translocation of unmodified
DNA via R subunits of the R2M2S complex of type I
RM enzymes [4].
Here, we investigated the ability of ArdB encoded by the ardB gene located in the transmissible plasmid R64 (IncI1 incompatibility group) to bind DNA in vivo and, as a consequence, the possibility of ArdB copurification with DNA under different conditions.
MATERIALS AND METHODS
Strains and cultivation conditions. The following E. coli K-12 strains were obtained from the All-Russian Collection of Industrial Microorganisms of the State Research Institute of Genetics and Selection of Industrial Microorganisms, Kurchatov Institute National Research Center (GosNIIgenetika): E. coli AB1157, E. coli TG1 (thi relA supE44 hsdR17 hsdM Δ(lac-proAB)[F′ traD36 proAB lacIqZ ΔM15]), and E. coli BL21(DE3).
The bacteria were grown in flasks (150 ml) with LB medium on a shaker (New Brunswick Scientific, USA) at 37°C and 200 rpm. Agar medium was prepared with 1.5% agar. The transformed cells were grown in the medium containing 100 μg/ml ampicillin. The optical density (OD) of bacterial suspension was measured at 590 nm with a KFK-2MP photocolorimeter (ProfMT, Russia). The biosynthesis of ArdB in E. coli BL21(DE3) cells was carried out in the lactose-containing induction medium as described by Pokrovsky et al. [5] and Studier [6].
The gene coding for chimeric GST-ArdB protein (glutathione S-transferase fused with ArdB) was expressed from the pGEX-ArdB plasmid in E. coli AB1157 cells in LB medium; protein expression was induced with 1 mM IPTG when the cells reached OD590 0.8.
Genes for the ArdB and ArdBΔD141 proteins fused with the Flag-tag were expressed from the pFlag-ArdB and pFlag-ArdBΔD141 plasmids, respectively, in E. coli AB1157 cells in LB medium without the inducer.
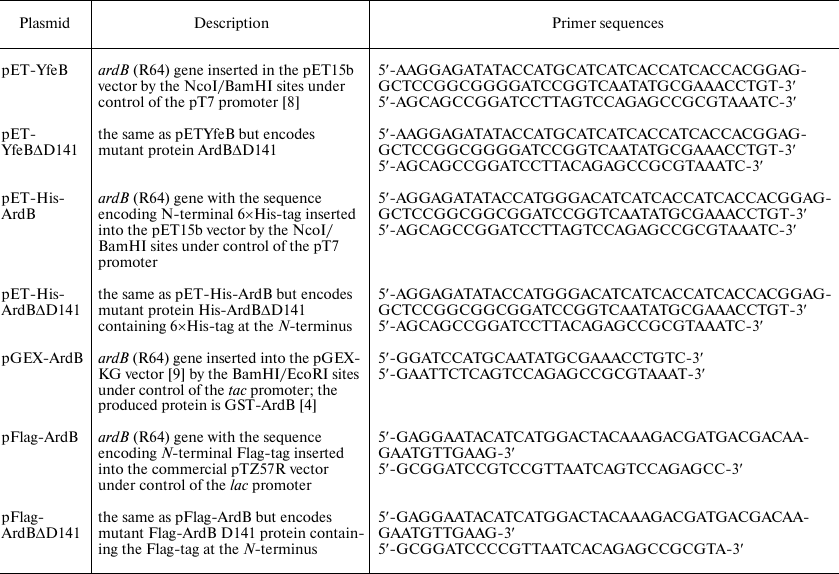
Plasmids. Transmissible R64 (IncI1) plasmid was used as a source of the ardB gene. DNA cleavage with restriction endonucleases, ligation of DNA fragments, electrophoresis in agarose gel, and isolation of DNA fragments from agarose gel were performed according to standard techniques [7]. The resulting constructs are presented in the table.
Plasmids carrying the ardB gene and its variants
Escherichia coli BL21(DE3) cells transformed with the pET15b plasmid (Novagen, USA) were used as a negative control for the ardB expression from the T7 promoter.
Antirestriction activity of ArdB proteins. The antirestriction activity of ArdB proteins encoded by the pGST-ArdB, pFlag-ArdB, and pFlag-ArdBΔD141 constructs was assayed by the phage technique. For this, λ.0 phage (containing unmodified DNA) was titrated using E. coli K12 AB1157 cells with the active type I RM system (EcoKI) as a host strain. The efficiency of plating (EOP) of the λ.0 phage was defined as a ratio of phage titer in E. coli AB1157 cells carrying a plasmid with the ardB gene and the same cells without the plasmid or containing a vector lacking the ardB gene.
Ion-exchange chromatography. Escherichia coli BL21(DE3) pET-YfeB cells were lyzed with a M-100P microfluidizer (Microfluidics International Corporation, USA) in 50 mM Tris-HCl buffer (pH 8.0) at 1500 bar. The lysates were centrifuged for 10 min at 15,000g, and the supernatant was applied on chromatographic ion-exchange sorbents Q-Sepharose® FastFlow (GE Healthcare, USA) or SPS-bio-Q (Technosorbent, Russia) at 4°C by the batch technique. The pellet was resuspended in 20 mM Tris-OH (pH ~ 10.0), and the resulting solution was centrifuged for 10 min at 15,000g. The transition of aggregated ArdB from the inclusion bodies into the soluble phase was controlled by electrophoresis in 15% polyacrylamide gel under denaturing conditions (SDS-PAGE) with Coomassie G-250 staining. The pH of the supernatant containing solubilized ArdB was adjusted to 8.0 with HCl. Ion-exchange column chromatography of solubilized ArdB was also performed on Q-Sepharose® FastFlow or SPS-bio-Q sorbents. ArdB was eluted with a linear gradient of NaCl (0 to 1 M in 20 volumes of the sorbent) in 50 mM Tris-HCl (pH 8.0) at a rate of 6 ml/min at 4°C. Eluted fractions were analyzed by SDS-PAGE.
Affinity chromatography. Flag-ArdB protein was isolated using Anti-FLAG® M2 Magnetic Beads (M8823; Sigma-Aldrich, USA) according to the manufacturer’s instructions; protein samples in 50 mM Tris-HCl buffer (pH 8.0), 150 mM NaCl were incubated with the sorbent for 1 h at room temperature under continuous slow stirring on an orbital shaker. The beads with the adsorbed protein were precipitated with a magnet; the supernatant was removed; the beads were then washed with the loading buffer (20 volumes of the sorbent). Proteins were eluted with 100 mM glycine (pH 3.0), and an equal volume of 100 mM Tris-HCl buffer (pH 8.0), 300 mM NaCl, was added to the eluted fraction to adjust pH to 8.0.
The pET-His-ArdB construct encoded ArdB fused with the polyhistidine (6×His) tag at the N-end through the Gly-Gly-Ser-Gly-Gly-Gly-Ser-Gly linker. Cell lysates (20 mM phosphate buffer, pH 8.0, 300 mM NaCl) containing soluble ArdB fraction were applied on a column with TALON® Metal Affinity Resin (Clontech, USA), and the column was washed with 20 mM phosphate buffer (pH 8.0) containing 300 mM NaCl and 10 mM imidazole. The target protein was eluted with 20 mM phosphate buffer (pH 8.0) containing 300 mM NaCl and 500 mM imidazole.
The GST–ArdB protein containing GST linked to the N-terminus via the thrombin-sensitive linker Leu-Val-Pro-Arg-Gly-Ser-Pro-Gly [9], was isolated with a Pierce™ GST Protein Interaction Pull-Down Kit (ThermoScientific, USA). The cells were lyzed in PBS (pH 7.5), 150 mM NaCl. After centrifugation, clarified lysates containing soluble ArdB were added to the glutathione-modified sorbent and incubated with slow stirring for 1 h at 4°C. Then, the sorbent was washed three times with PBS, and the target protein was eluted with 50 mM Tris-HCl containing 50 mM glutathione.
Fractions obtained by affinity chromatography were analyzed by SDS-PAGE and mass-spectrometric fingerprinting. The presence of DNA in the samples was assayed by electrophoresis in 1% agarose gel with ethidium bromide staining using electrode buffer containing RNase A.
The content of target proteins in the fractions after chromatographic separation was determined by densitometric analysis of the gels using the TotlLAb program. The intensity of ArdB bands in the gel in each fraction was normalized to the fraction volume. The sum of staining intensities of ArdB bands was taken as 100%.
DNA–protein conjugation with formaldehyde. Escherichia coli BL21(DE3) cells without the plasmid or with plasmids carrying the ardB (wild-type ArdB) or ardB-ΔD141 (mutant ArdBΔD141) genes under control of the pT7 promoter were incubated for 40 min at room temperature in 30% formaldehyde [10, 11]. Next, total DNA was isolated with a Wizard® Genomic DNA Purification Kit (Promega, USA), and the samples were analyzed in 1% agarose and 15% polyacrylamide gels.
Mass-spectrometry analysis was used for identification of ArdB proteins. Proteins were isolated from the gel after SDS-PAGE, treated with trypsin [12-16], and identified by the MALDI-TOF (matrix-assisted laser description ionization-time of flight) mass spectrometry with a MALDI-TOF Reflex III system (Bruker, Germany) at the Center of Proteomic Research, Orekhovich Institute of Biomedical Chemistry, Russian Academy of Sciences. The resulting mass fingerprints were analyzed using the SwissProt database [17].
RESULTS
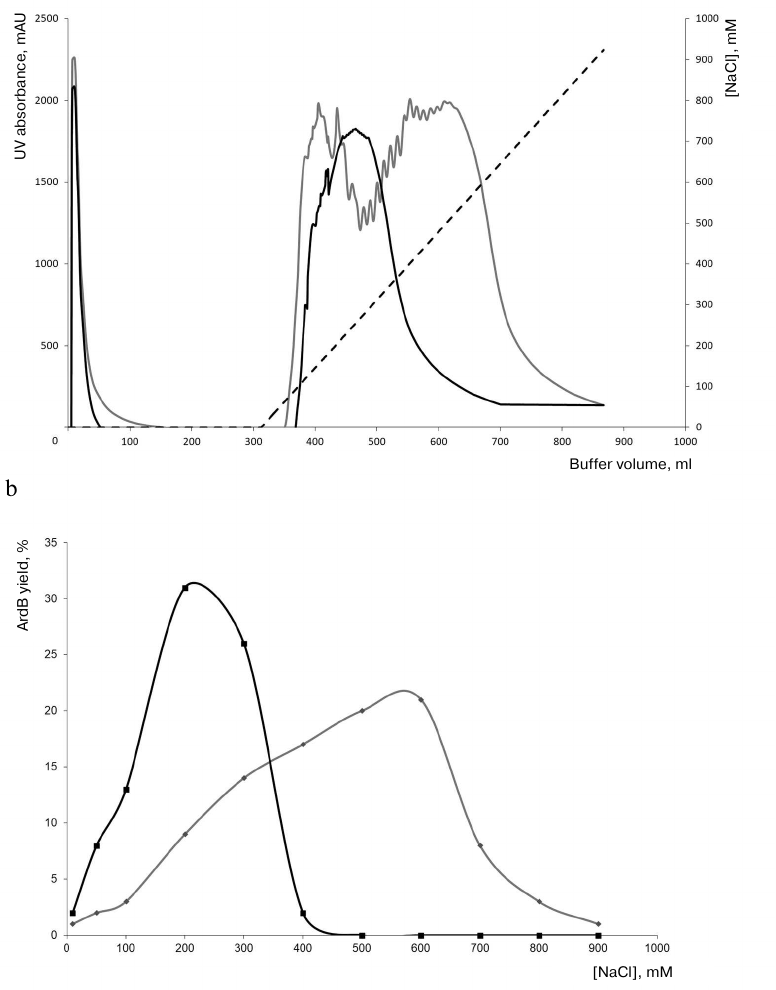
The results of ArdB purification by ion-exchange chromatography are shown in Fig. 1. Escherichia coli BL21(DE3) pET-YfeB cells were cultivated at 37°C in the induction medium with lactose for 20 h. When the ardB gene was expressed from the phage T7 promoter, a significant portion of the produced protein formed aggregates (nonclassical inclusion bodies). Ion-exchange chromatography was performed both for the soluble ArdB fraction and the aggregates dissolved in untitrated 20 mM Tris base solution.
Fig. 1. a) Elution profiles (absorbance at 280 nm) of soluble and insoluble ArdB forms fractionated on ion-exchange sorbent (with quaternary amines as charged group). b) The content of ArdB protein in the fractions eluted from the ion-exchange sorbent at different NaCl concentrations. Gray curve, soluble ArdB fraction; solid curve, dissolved ArdB inclusion bodies; dotted line, linear gradient of NaCl concentration in the eluent.
Figure 1 shows that ArdB from dissolved aggregates was eluted from the sorbent at a comparatively low salt concentration (maximum elution at 200 mM NaCl), which was expected for the protein with the estimated isoelectric point of 4.97. One should note a comparative compactness of the elution peak, probably, indicating protein homogeneity. However, ArdB isolated from the soluble fraction of the cell lysate was eluted as a broader peak at the NaCl concentration of 200-700 mM. The resulting elution profiles suggest that soluble ArdB can be bound to DNA, resulting in the increase of distributed electrostatic negative charge and enhanced adsorption of the ArdB protein complex with DNA on the ion-exchange sorbent, which required higher NaCl concentrations for its elution. DNA heterogeneity in the ArdB–DNA complex might be a reason for a considerable broadening of the chromatographic peak during the elution.
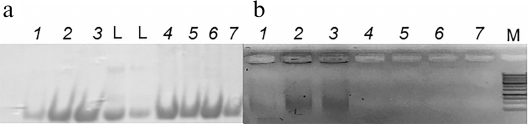
Figure 2 shows the results of analysis of ArdB fractions after ion-exchange chromatography by electrophoresis in polyacrylamide and agarose gels. After chromatographic purification, ArdB-containing fractions demonstrated comparatively low quantities of admixed proteins of the producer strain. Analysis by agarose gel electrophoresis (Fig. 2b) revealed the presence of DNA in ArdB fractions obtained by ion-exchange chromatography of soluble ArdB (lanes 1-3), but not of ArdB dissolved from the aggregates (lanes 4-7).
Fig. 2. Analysis of ArdB fractions obtained by ion-exchange chromatography by electrophoresis in SDS-polyacrylamide gel (a) and 1% agarose gel (b). Lanes: 1-3) fractions obtained by chromatography of soluble ArdB and eluted from the sorbent with 200, 500 and 600 mM NaCl, respectively; 4-7) fractions obtained by chromatography of ArdB from dissolved aggregates and eluted from the sorbent with 150, 200, 300 and 350 mM NaCl, respectively; L, 14.5-kDa protein marker (RNase A; Evrogen, Russia); M, 1-kb DNA ladder (Evrogen).
DNA binding by ArdB was verified in the experiments on the isolation of ArdB and its mutant form ArdBΔD141 lacking the antirestriction activity by different types of affinity chromatography.
Since plasmids actively expressing ardB gene are often lost by the cells, the antirestriction activity of chimeric proteins was preliminarily confirmed in vivo by inoculation with unmodified λ phage.
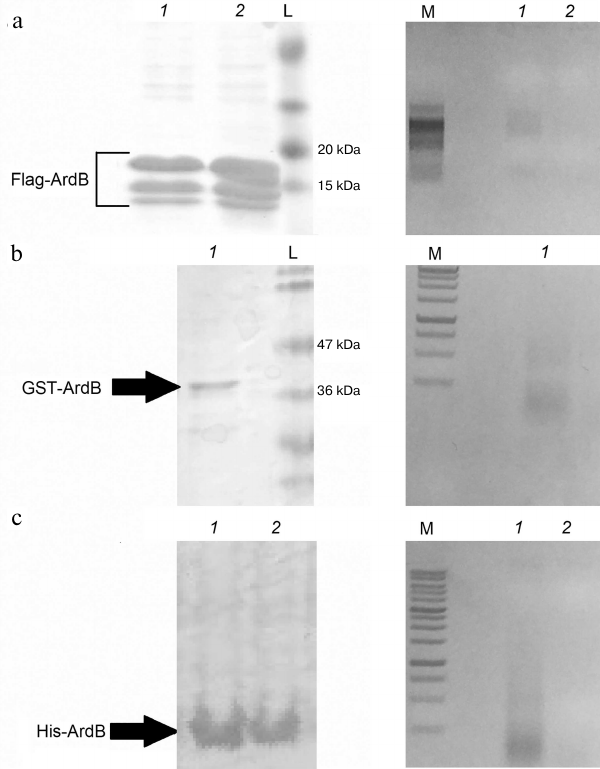
Figure 3 shows the results of affinity chromatography of chimeric Flag-ArdB, Flag-ArdBΔD141, GST-ArdB, His-ArdB, and His-ArdBΔD141 proteins. The presence of ArdB and ArdBΔD141 in the corresponding bands in polyacrylamide gel (Fig. 3, left panel) after affinity chromatography was confirmed by mass-spectrometric fingerprint analysis. The presence of DNA in the eluates was demonstrated by electrophoresis in 1% agarose gel with ethidium bromide staining.
Fig. 3. Analysis of fractions after affinity chromatography of chimeric ArdB proteins by electrophoresis in SDS-polyacrylamide gel (right) and 1% agarose gel (left): a) Flag-ArdB (lanes 1) and Flag-ArdBΔD141 (lanes 2); b) GST-ArdB (lanes 1); c) His-ArdB (lanes 1) and His-ArdBΔD141 (lanes 2). L, pre-stained Protein MW markers (a) and PageRuler Plus (b) (Thermo Scientific); M, 100+ bp (a) and 1 kb (b, c) DNA ladders (Evrogen).
As one can see from Fig. 3, DNA copurified with ArdB when different affinity sorbents were used, such as anti-Flag magnetic beads, glutathione-Sepharose, and cobalt resin. DNA copurified only with the tagged Flag-ArdB and His-ArdB forms of the wild-type protein (lanes 1), but not with the Flag-ArdBΔD141 and His-ArdBΔD141 (lanes 2) mutants. Note that DNA in the samples was represented by different fragments within a range of 1000 bp.
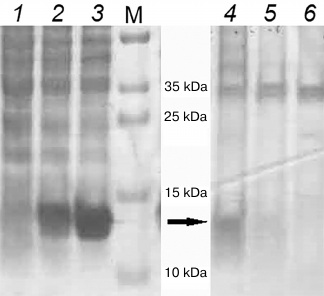
The interaction between ArdB and DNA was verified by obtaining covalent ArdB–DNA conjugates using formaldehyde, i.e., by formation of covalent bonds between the primary amino groups of the protein and nucleic acid [10, 11]. Escherichia coli strains BL21(DE3) (without the plasmid), BL21(DE3) (pET-YfeB) with the native ardB gene, and BL21(DE3) (pET-YfeBΔD141) with the mutant ardB-ΔD141 gene, were cultivated to OD590 0.8 and treated with formaldehyde, followed by isolation of total DNA. Isolation of chromosomal DNA was controlled by electrophoresis in agarose gel. Proteins copurified with DNA were detected by SDS-PAGE (Fig. 4, lanes 4-6) after ultrasonic pretreatment of the samples in order to degrade large DNA fragments. The 14-kDa bands were cut out of the gel and treated with trypsin; the resultant peptides were eluted and identified as ArdB peptides by peptide fingerprinting and comparison with the Swiss-Prot database.
Fig. 4. PAGE of E. coli BL21(DE3) cell lysates and isolated chromosomal DNA. Lanes 1-3, lysates of cells not treated with formaldehyde: 1) BL21(DE3) pET15; 2) BL21(DE3) pET-YfeB; 3) BL21(DE3) pET-YfeBΔD141. Lanes 4-6, chromosomal DNA isolated from formaldehyde-treated cells: 4) BL21(DE3) pET-YfeB; 5) BL21(DE3) pET-YfeBΔD141; 6) BL21(DE3) pET15; M, PageRuler Plus markers (Thermo Scientific).
A large number of proteins copurified with DNA after cell treatment with formaldehyde. However, only the wild-type ArdB copurified with total DNA (Fig. 4, lane 4), but not the mutant protein lacking the antirestriction activity (Fig. 4, lane 5). The ArdBΔD141 protein lacking the C-terminal aspartate was incapable of interacting with DNA, which might be the reason for the absence of antirestriction activity in this protein.
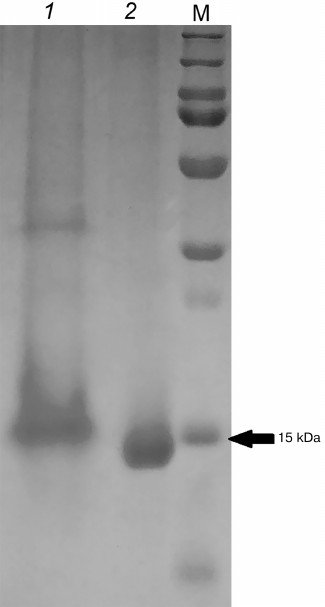
Direct interaction between ArdB and DNA was confirmed by the gel retardation assay (Fig. 5). For this purpose, the samples obtained by total DNA isolation from formaldehyde-treated cells (Fig. 4, lane 4) and ArdB purification from the aggregates (DNA-free; Fig. 2, lane 5) were analyzed by electrophoresis in the same polyacrylamide gel.
Fig. 5. PAGE of proteins copurified with DNA after isolation from formaldehyde-treated cells (1) and obtained by ArdB purification from the aggregates (2). M, PageRuler Plus markers (Thermo Scientific).
Figure 5 shows that the electrophoretic mobility of the ArdB protein in the sample obtained by total DNA isolation was less than the mobility of protein samples isolated from ArdB aggregates by ion-exchange chromatography. The presence of ArdB in the 15-kDa band was confirmed by mass-spectrometric fingerprinting. This result can be considered as direct indication of the interaction between DNA and ArdB in vivo.
DISCUSSION
DNA was identified in the ArdB fractions obtained by protein purification by ion-exchange chromatography. Similar results were obtained for the Flag-, 6×His-, and GST-tagged ArdB proteins purified by different affinity chromatography techniques. Since different sorbents were used, DNA co-elution of together with the target proteins cannot be attributed simply to DNA binding to the sorbent. Total DNA isolation from formaldehyde-treated cells were performed to compare DNA binding by the wild-type ArdB and the C-terminal aspartate mutant ArdBΔD141 lacking the antirestriction activity. ArdBΔD141 did not bind DNA, which might explain the absence of the antirestriction activity in this protein. Since the electrophoretic mobility in polyacrylamide gel of ArdB in the samples obtained by total DNA isolation was lower than the mobility of ArdB purified from protein aggregates by ion-exchange chromatography, the interaction between DNA and ArdB in vivo can be considered proven.
Zavilgelsky et al. [18] determined the Kd (~10–8 M) of the DNA-mimicking protein ArdA by studying the inhibition of the endonuclease activity of type I RM enzymes in vivo. Balabanov et al. [19] carried out comparative measurements of in vivo Kd values for ArdA and ArdB proteins encoded in the R64 plasmid. It was shown that Kd characterizing the activity of ArdB (~5·10–7 M) was ~50-fold higher than that of ArdA. According to Serfiotis-Mitsa et al. [2], no ArdB activity was detected in vitro and, consequently, no protein binding to DNA took place. However, it should be noted that in this work, ArdA and ArdB were compared in equimolar concentrations; probably, further in vitro experiments should be performed with the higher ArdB concentrations.
The data on the existence of ArdB complexes with chromosomal DNA obtained in this work confirm the hypothesis proposed by Balabanov et al. [4] that ArdB blocks the translocation of unmodified DNA via the R subunits of the R2M2S complex of type I RM enzyme. As a result, only the restricting (endonuclease) but not the modifying (methylase) activity of the enzyme is inhibited.
Funding. The work was supported by the Russian Foundation for Basic Research (projects nos. 18-34-00753 and 19-04-00495) and State Order no. 6.9899.2017/BCh.
Conflict of interest. The authors declare no conflict of interest in financial or any other area.
Compliance with ethical standards. This article does not contain any studies involving animals or human participants performed by any of the authors.
REFERENCES
1.Belogurov, A. A., Delver, E. P., and Rodzevich, O.
V. (1993) Plasmid pKM101 encodes two nonhomologous antirestriction
proteins (ArdA and ArdB) whose expression is controlled by homologous
regulatory sequences, J. Bacteriol., 175, 4843-4850, doi:
10.1128/jb.175.15.4843-4850.1993.
2.Serfiotis-Mitsa, D., Herbert, A. P., Roberts, G.
A., Soares, D. C., White, J. H., Blakely, G. W., Uhrin, D., and Dryden,
D. T. (2010) The structure of the KlcA and ArdB proteins reveals a
novel fold and antirestriction activity against type I DNA restriction
systems in vivo but not in vitro, Nucleic Acids
Res., 38, 1723-1737, doi: 10.1093/nar/gkp1144.
3.Kudryavtseva, A. A., Osetrova, M. S., Livinyuk, V.
Ya., Manukhov, I. V., and Zavilgelsky, G. B. (2017) The
C-terminal residue of aspartic acid (D141) is necessary of the
antirestriction activity of protein ArdB (R64), Mol. Biol.
(Moscow), 51, 831-835.
4.Balabanov, V. P., Kudryavtseva, A. A., Melkina, O.
E., Pustovoit, K. S., Khrulnova, S. A., and Zavilgelsky, G. B. (2019)
ArdB protective activity for unmodified λ phage against EcoKI
restriction decreases in UV-treated Escherichia coli, Curr.
Microbiol., 76, 1374-1378, doi:
10.1007/s00284-019-01755-z.
5.Pokrovsky, V. S., Anisimova, Yu. N., Davydov, Z.
D., Bazhenov, S. V., Bulushova, N. V., Zavilgelsky, G. B., Kotova, V.
Y., and Manukhov, I. V. (2019) Methionine gamma lyase from
Clostridium sporogenes increases the anticancer effect of
doxorubicin in A549 cells and human cancer xenografts, Invest. New
Drugs, 37, 201-209, doi: 10.1007/s10637-018-0619-4.
6.Studier, F. W. (2005) Protein production by
auto-induction in high density shaking cultures, Protein Expr.
Purif., 41, 207-234, doi: 10.1016/j.pep.2005.01.016.
7.Sambrook, J., and Russell, D. W. (2001)
Molecular Cloning: A Laboratory Manual, Cold Spring Harbor
Laboratory Press, N. Y.
8.Goryanin, I. I., Kudryavtseva, A. A., Balabanov, V.
P., Biryukova, V. S., Manukhov, I. V., and Zavilgelsky, G. B. (2018)
Antirestriction activities of KlcA (RP4) and ArdB (R64) proteins,
FEMS Microbiol. Lett., 365, doi:
10.1093/femsle/fny227.
9.Guan, K. L., and Dixon, J. E. (1991) Eukaryotic
proteins expressed in Escherichia coli: an improved thrombin
cleavage and purification procedure of fusion protein with glutathione
S-transferase, Anal. Biochem., 192, 262-267, doi:
10.1016/0003-2697(91)90534-z.
10.Solomon, M. J., and Varshavsky, A. (1985)
Formaldehyde-mediated DNA–protein crosslinking: a probe for in
vivo chromatin structures, Proc. Natl. Acad. Sci. USA,
82, 6470-6474, doi: 10.1073/pnas.82.19.6470.
11.Hoffman, E. A., Frey, B. L., Smith, L. M., and
Auble, D. T. (2015) Formaldehyde crosslinking: a tool for the study of
chromatin complexes, J. Biol. Chem., 290, 26404-26411,
doi: 10.1074/jbc.R115.651679.
12.Han, M. J., Yoon, S. S., and Lee, S. Y. (2001)
Proteomic analysis of metabolically engineered Escherichia coli
producing poly(3-hydroxybutyrate), J. Bacteriol., 183,
301-308, doi: 10.1128/JB.183.1.301-308.2001.
13.Rosenfeld, J., Capdevielle, J., Guillemoi, J.,
and Ferrara, P. (1992) In-gel digestion of proteins for internal
sequence analysis after one- or two-dimensional gel electrophoresis,
Anal. Biochem., 203, 173-179, doi:
10.1016/0003-2697(92)90061-b.
14.Patterson, S. D., and Aebersold, R. (1995)
Mass-spectrometric approaches for the identification of gel-separated
proteins, Electrophoresis, 16, 1791-1814, doi:
10.1002/elps.11501601299.
15.Bazhenov, S. V., Khrulnova, S. A., Konopleva, M.
N., and Manukhov, I. V. (2019) Seasonal changes in luminescent
intestinal microflora of the fish inhabiting the Bering and Okhotsk
seas, FEMS Microbiol. Lett., 366, pii: fnz040, doi:
10.1093/femsle/fnz040.
16.Manukhov, I. V., Melkina, O. E., Goryanin, I. I.,
Baranova, A. V., and Zavilgelsky, G. B. (2010) The N-terminal
domain of the Aliivibrio fischeri LuxR is a target of the GroEL
chaperonin, J. Bacteriol., 192, 549-551, doi:
10.1128/JB.00754-10.
17.Bairoch, A., and Apweiler, R. (2000) The
SWISS-PROT protein sequence database and its supplement TrEMBL in 2000,
Nucleic Acids Res., 28, 45-48, doi:
10.1093/nar/28.1.45.
18.Zavilgelsky, G. B., Kotova, V. Yu., and
Rastorguev, S. M. (2008) Comparative analysis of antirestriction
activity of ArdA (ColIbP9) and Ocr (T7) proteins, Biochemistry
(Moscow), 73, 1124-1130.
19.Balabanov, V. P., Pustovoit, K. S., and
Zavilgelsky, G. B. (2012) Comparative analysis of antirestriction
activity of R64 ArdA and ArdB proteins, Mol. Biol. (Moscow),
46, 269-275.