Effect of Benzo(a)pyrene on the Expression of miR-483-3p in Hepatocyte Primary Culture and Rat Liver
S. V. Filippov1,2,a*, A. A. Yarushkin1,3, T. S. Kalinina1, V. Y. Ovchinnikov2, R. A. Knyazev4, and L. F. Gulyaeva1,3
1Institute of Molecular Biology and Biophysics, Federal Research Center of Fundamental and Translational Medicine, 630117 Novosibirsk, Russia2Federal Research Center Institute of Cytology and Genetic, Siberian Branch of the Russian Academy of Sciences, 630090 Novosibirsk, Russia
3Novosibirsk State University, 630090 Novosibirsk, Russia
4Institute of Biochemistry, Federal Research Center of Fundamental and Translational Medicine, 630117 Novosibirsk, Russia
* To whom correspondence should be addressed.
Received May 21, 2019; Revised June 21, 2019; Accepted June 21, 2019
Here, we suggested that the epigenetic mechanism of benzo(a)pyrene (BP) action might be based on the aryl hydrocarbon receptor (AhR)-mediated transcription of the target genes, including miRNAs, that have the dioxin response element (DRE) in their promoters. The effect of BP on the expression of the oncogenic miR-483-3p, its host gene IGF2, and target gene IGF1 in primary hepatocytes and in the liver of Wistar female rats was investigated. The activation of AhR was confirmed using selective AhR inhibitor CH-223191 and by evaluating expression of the target CYP1A1 gene. The lack of coordination between the expression of miR-483-3p and its host gene IGF2 was revealed, which may be due to the presence of the binding site for the estrogen receptor alpha (ERα), which is a negative expression regulator. Our results confirm the existence of the AhR-mediated pathway in the regulation of expression of miR-483-3p, IGF1, and IGF2 under BP exposure, which is of considerable interest for understanding the epigenetic mechanisms of the carcinogenic effect of BP.
KEY WORDS: benzo(a)pyrene, AhR, ERα, DRE, induction, microRNA, target genesDOI: 10.1134/S0006297919100080
Abbreviations: AhR, aryl hydrocarbon receptor; BP, benzo(a)pyrene; DRE, dioxin response element; ERα, estrogen receptor alpha.
Benzo(a)pyrene (BP) is a well-known and actively studied environmental
carcinogen formed by incomplete combustion of hydrocarbons. Besides
acting as a genotoxic carcinogen via formation of adducts with DNA and
subsequent accumulation of mutations, BP activates aryl hydrocarbon
receptor (AhR), a transcription factor from the helix-loop-helix
PER/ARNT/SIM (bHLH-PAS) protein family [1].
Activated AhR interacts with specific binding sites on DNA, dioxin
response elements (DREs) [2], and triggers
transcription of target genes encoding various proteins, such as
cytochrome P450 family 1 (CYP1A/1B) involved in xenobiotic
metabolism and PTPN6 that regulates cell proliferation and
differentiation [3, 4].
Ligand-activated AhR also exhibits the anti-estrogenic activity by
suppressing estradiol-mediated signaling through the estrogen receptor
alpha (ERα), whose target genes control diverse cellular
processes, including proliferation, differentiation, etc. [5].
Other AhR and ERα targets are genes containing in their introns small non-coding RNAs (miRNAs) involved in the post-transcriptional regulation of gene expression in humans and animals [6]. Therefore, we have searched for such genes in silico [7]. The IGF2 gene was of particular interest among the identified genes, because its promoter contains the binding sites for both receptors in rats and humans. Upregulated IGF2 expression is associated with the risk of developing various types of cancer, including colorectal, breast, prostate, and lung cancers [8]. Intron 2 of the IGF2 gene encodes miR-483 precursor that is cut into two copies (miR-483-3p and miR-483-5p) during processing. Both copies exhibit oncogenic properties; however, despite their similarity, these miRNAs are expressed differently and play different roles in various types of cancer [9]. For example, miR-483-3p is overexpressed in prostate cancer [10] and breast cancer [11], while miR-483-5p is overexpressed in oral cancer [12] and esophagus cancer [13].
It remains unclear why the expression level of miR-483 changes during malignant cell transformation. miRNA expression can be regulated by different mechanisms [14], including exposure to xenobiotics. The purpose of this work was to test the hypothesis that BP affects expression of miR-483-3p via AhR activation with subsequent upregulation of the expression of the IGF2 host gene and the IGF1 target gene in the primary culture of hepatocytes and liver of Wistar female rats exposed to BP.
MATERIALS AND METHODS
Cell culture. The study was performed in hepatocytes isolated from a Wistar female rat (body weight, 180 g) obtained from the Animal Facility of the Institute of Clinical Immunology, Siberian Branch of the Russian Academy of Medical Sciences (Novosibirsk, Russia). Hepatocytes were isolated by recirculating enzymatic perfusion using 0.03% collagenase solution (ICN Biomedicals, USA) [15] and separated from non-parenchymal cells by differential centrifugation. Cell viability assessed by the trypan blue (Serva, Germany) exclusion method was at least 90%. The obtained cells were resuspended in RPMI-1640 medium (Biolot, Russia), pH 7.4, containing 20 mM HEPES (ICN Biomedicals), 10% fetal bovine serum (Serva), 2 mM L-glutamine (Vector, Russia), 100 units/ml penicillin, 50 μg/ml gentamicin, 5.6 mM glucose, and 10 nM insulin (Serva). The cells were incubated in 6-well plates (Orange Scientific, USA) coated with collagen in a CO2 incubator (Cole-Parmer, USA) in 5% CO2 at 37°C. The cell density in the primary culture monolayer was 800 cells/mm2.
Primary hepatocyte cultures were incubated with 5 μM BP (Sigma-Aldrich, USA) and 10 μM selective AhR inhibitor CH-223191 (Sigma-Aldrich) for 24 h. BP and CH-223191 were dissolved in dimethyl sulfoxide (DMSO) (Sigma-Aldrich) and then added to the culture medium, so that the final concentration of DMSO was 0.1% (v/v). Cells treated with DMSO only were used as a control.
Animals. Sixteen Wistar female rats weighing 150-180 g were obtained from the Animal Facility of the Institute of Clinical Immunology, Siberian Branch of the Russian Academy of Medical Sciences (Novosibirsk, Russia). The animals were divided into four groups (n = 4 in each group). The experimental animals were injected intraperitoneally with BP (Sigma-Aldrich) dissolved in sunflower oil at 75 mg/kg body weight (0.5 ml per rat) and then sacrificed 24, 48, and 72 h after BP injection; rats of the control group were injected with 0.5 ml of sunflower oil. The animals were sacrificed by decapitation; the liver was removed and used for the isolation of total RNA.
RNA isolation. To isolate total RNA, liver samples were homogenized in 1 ml of Trizol Reagent (Invitrogen, USA); 0.2 ml of chloroform was added, and the aqueous phase was collected. RNA was precipitated with 0.5 ml of isopropyl alcohol, washed twice with 75% ethanol, and dissolved in nuclease-free water. RNA concentration and purity were determined from the absorbance at 260/280 nm with a NanoPhotometer P 360 spectrophotometer (Implen, Germany). Isolated RNA was analyzed by electrophoresis in 1.5% (w/v) agarose gel containing 0.5 μg/ml ethidium bromide at 100 V for 30 min; the images were registered with a V3 Western Workflow system (Bio-Rad Laboratories, USA). Isolated RNA was stored at –70°C prior to the use.
RT-qPCR. cDNA was obtained by reverse transcription on the isolated total RNA using RT M-MuLV-RH kit (Biolabmix, Russia) according to the manufacturer’s recommendations.
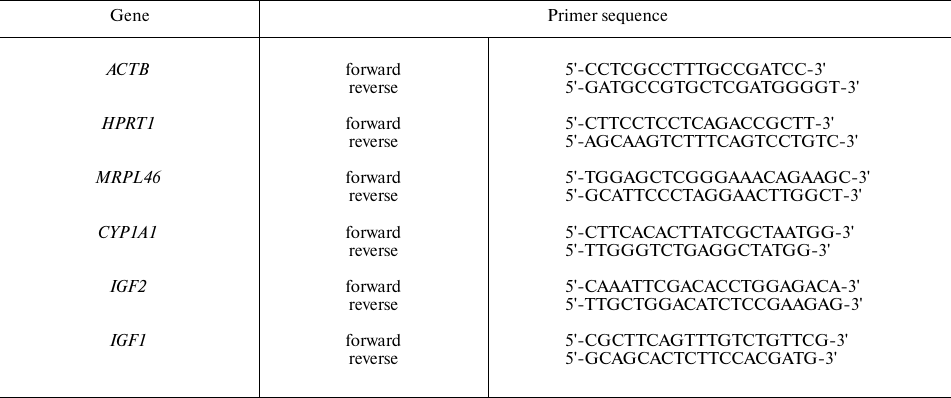
The expression levels of CYP1A1, IGF1, and IGF2 genes were evaluated by qPCR using a BioMaster HS-qPCR SYBR Blue (2×) kit (Biolabmix) as recommended by the manufacturer. ACTB, HPRT1, and MRPL46 were used as the reference genes. The primer sequences are shown in Table 1; the optimal concentration of all primer pairs in the reaction mixture was 300 nM. Amplification was performed with a CFX96™ Touch cycler (Bio-Rad Laboratories) in the following regime: 95°C for 5 min; 40 cycles of 95°C for 15 s, 60°C for 20 s; 72°C for 30 s. All reactions were carried out in triplicate.
Table 1. Primer sequences for qPCR
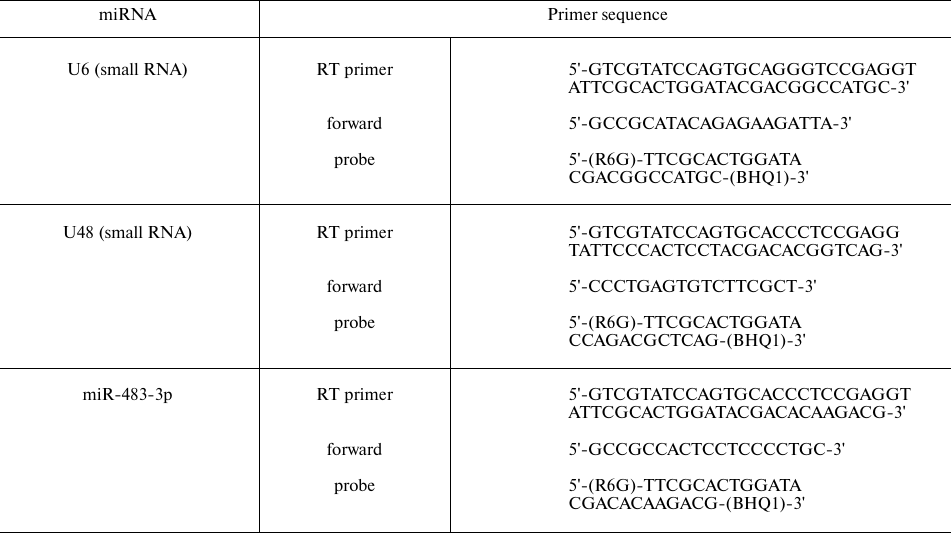
RT-qPCR for miR-483-3p. Reverse transcription on total RNA (40 ng) was performed using M-MuLV-RH reverse transcriptase (Biolabmix) and a special stem-loop primer (Table 2) specific for mature miR-483-3p; the primer sequence was obtained from mirbase.org. PCR was performed in a total volume of 30 μl using the TaqMan technology (R6G fluorophore, BHQ1 quencher) and UDG HS-qPCR Biomaster (2×) kit (Biolabmix) according to the manufacturer’s recommendations. Small nuclear RNAs U6 and U48 were used as the reference genes. The primer sequences are shown in Table 2. Amplification was performed in a CFX96™ Touch cycler in the following regime: 94°C for 5 min; 40 cycles of 94°C for 15 s, 61°C for 20 s; 72°C for 30 s. All reactions were carried out in triplicate.
Table 2. Primer sequences for RT-qPCR of
miRNAs
Bioinformatics analysis of rat genome. To predict the sites for the binding of AhR and ER transcription factors in the putative miR-483-3p promoter, a 10,000-nucleotide (nt) fragment from the start of the sequence coding for the miRNA precursor (according to miRBase v.21) was extracted from the rat genome (Rnor_6.0) and searched for the ER-binding sites (one mutation allowed) and AhR-binding sites (four mutations allowed) using the fqgrep program.
miRNA targets were predicted using the miRNA.org, targetcan.org, and mirDB.org web resources.
Statistical analysis. Statistical processing of the results was carried out with the MS Office software. The results are presented as mean ± standard deviation; the significance of differences between the samples was assessed with the Student’s t-test.
RESULTS
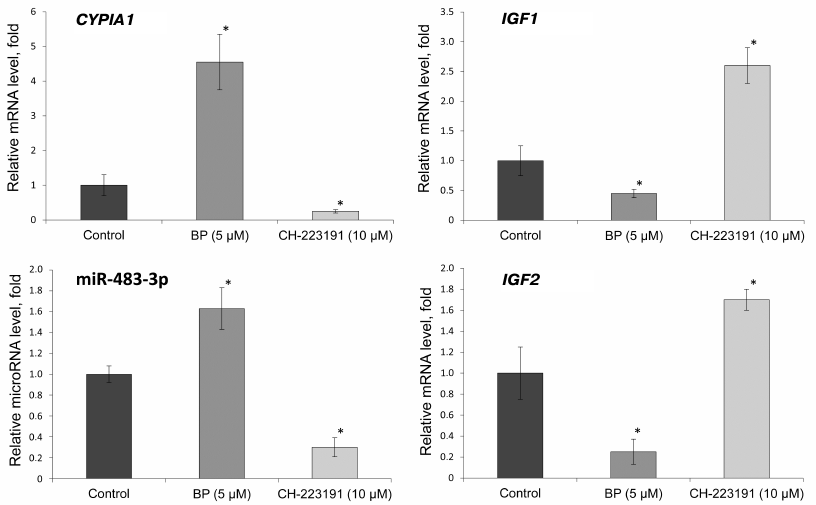
Effect of BP and CH-223191 on the expression of CYP1A1, IGF1, and IGF2 genes and miR-483-3p in the primary culture of rat hepatocytes. To verify whether BP affects the expression of miR-483-3p and IGF1 and IGF2 mRNAs, their relative levels were evaluated by RT-qPCR in the primary culture of rat hepatocytes treated with BP and selective AhR inhibitor CH-223191. To assess the activation of AhR, relative expression of the CYP1A1 gene was evaluated (Fig. 1).
Fig. 1. Relative expression of miR-483-3p and CYP1A1, IGF1, and IGF2 mRNAs in the primary culture of rat hepatocytes incubated with BP (5 μM) and CH-223191 (10 μM) for 24 h; * significant difference with the control group (p < 0.05).
We found that BP increased the expression of the CYP1A1 gene 4.5 times, while the AhR inhibitor decreased the level of the corresponding mRNA 4 times, compared to the control. The relative level of miR-483-3p increased 1.6 times under the action of BP, while the AhR inhibitor decreased it 3 times. The relative expression of the IGF2 gene under the action of BP significantly decreased (5 times); treatment of the cells with the AhR inhibitor increased IGF2 expression 1.7 times. The relative expression level of the miR-483-3p target gene IGF1 decreased 2 times under the action of BP and increased 2.5 times with the inhibition of AhR.
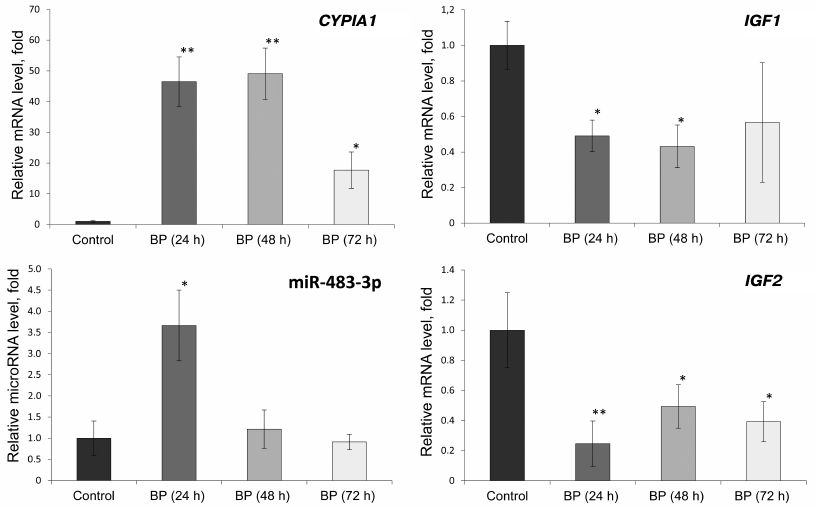
Effect of BP on the expression of CYP1A1, IGF1, and IGF2 genes and miR-483-3p in rat liver. To verify that the observed changes in the gene expression occur in vivo, we studied the levels of CYP1A1, IGF1, and IGF2 mRNAs and miR-483-3p in the liver of rats treated with BP (Fig. 2).
Fig. 2. Relative expression of miR-483-3p and CYP1A1, IGF1, and IGF2 mRNAs in the liver of Wistar rats 24, 48, and 72 h after BP administration (75 mg/kg body weight); *, significant differences with the control group, p < 0.05; **, significant differences with the control group, p < 0.01.
The relative expression of the CYP1A1 gene increased 46 and 49 times under the influence of BP 24 and 48 h, respectively, after its administration. Seventy-two hours after BP injection, the level of the CYP1A1 mRNA was 18 times higher than in the control. The relative content of miR-483-3p increased 3.7 times 24 h after BP administration, but decreased to its initial value after 48 and 72 h. The level of the IGF2 mRNA in the BP-treated rats decreased 2-5 times, while expression of the IGF1 gene decreased 2-2.4 times compared to the control (similar to the results of in vitro experiments).
DISCUSSION
The epigenetic mechanism of chemically induced carcinogenesis is one of the topic problems of modern science. In view of this, miRNAs regulating gene expression at the post-transcriptional level have attracted a considerable interest. Recent studies have shown that many carcinogens cause changes in the expression of miRNAs, which are accompanied by changes in the expression of their target genes [16]. One of such carcinogens is BP, whose mechanism of genotoxic action has been studied quite well. BP is oxidized by cytochromes P450 CYP1A1 and CYP1A2 (mainly in the liver); some of its metabolites, especially electrophilic epoxides, bind to DNA and form adducts with its nitrogenous bases [1]. BP also induces mutational hot spots in DNA [1]. It was found that BP causes alterations in the expression of miRNAs [17], i.e., exhibits the properties of a non-genotoxic carcinogen. However, the exact mechanism of its action in this case has not been established yet. We suggested that BP either changes miRNA expression through the activation of AhR, followed by transcriptional activation of genes containing miRNAs in their sequences, or activates transcription of intergenic miRNAs directly by binding to DREs in their promoters. Bioinformatics analysis of the rat genome revealed 119 miRNAs that can be expressed as a result of AhR activation [7]. To verify our hypothesis, we chose miR-483-3p, which has orthologs in human and rat genomes and is expressed at a sufficiently high level in the liver. The oncogenic properties of this miRNA in hepatocarcinogenesis have been demonstrated in [18, 19].
We found that BP caused a significant increase in the expression of classic AhR target genes both in the primary hepatocyte culture and in the liver of BP-treated rats. The inducing effect of BP was significantly lower in the hepatocyte culture (Figs. 1 and 2), which may be associated with the metabolic features of cell cultures, whose response to monooxygenase inducers depends on multiple factors [20].
Administration of the AhR inhibitor led to a significant decrease in the expression of the CYP1A1 gene compared to the control, which confirmed activation of the AhR and participation of this protein in the regulation of CYP1A1 expression.
Contrary to our expectations, expression of another AhR target gene – IGF2 – was not upregulated by BP administration, but instead decreased almost 5 times in the primary hepatocyte culture (Fig. 1) and 2-5 times in the rat liver (Fig. 2). In contrast, expression of miR-483-3p increased 1.7 times in the hepatocyte culture (Fig. 1) and 3.7 times in the liver (Fig. 2), which was accompanied by a 2-fold decrease in the expression of its target gene IGF1 both in the cell culture and the liver of animals exposed to BP (Figs. 1 and 2).
According to the earlier obtained data, the miR-483-3p host gene IGF2 has the motifs for binding ERα and ERβ in its promoter, along with the AhR-binding site [7]. It is known that ligand-activated AhR negatively regulates the activity of ERα [5]. Moreover, if a promoter of the same gene simultaneously contains an ERE (estrogen response element) and a DRE, the latter can exhibit the properties of the inhibitory domain (iDRE). By binding to the iDRE, the AhR/ARNT (AhR nuclear translocator) complex sterically hinders ERα binding to the ERE [21]. Salisbury et al. have shown that activation of AhR by tetrachlorodibenzo-p-dioxin leads to the downregulation of the IGF2 expression [22], which is in agreement with the data obtained in our work. It can be assumed that this decrease is due to the fact that the DRE in the IGF2 promoter exhibits the properties of iDRE, and that AhR activation results in a reduction of the ERα activity. Ultimately, these two events lead to the downregulation of the IGF2 expression by BP and, conversely, to its upregulation by the AhR inhibitor CH-223191.
However, the expression level of miR-483-3p encoded in the IGF2 intron increased in the liver of female rats 24 h after BP administration and in rat hepatocytes treated with BP for 24 h. At the same time, the AhR inhibitor decreased the expression of this miRNA in hepatocytes. To reveal the reasons for such discrepancy, we searched for the AhR- and ER-binding sites in the proposed promoter of this miRNA and identified the DRE site. Therefore, it can be assumed that the AhR activation by BP led to the miR-483-3p upregulation independently of IGF2. However, 48 and 72 h after BP administration, the levels of miR-483-3p did not differ in the liver of control and treated rats, which might be due to the continuous decrease in the expression of the host gene or to the BP-induced decrease in the ER level, since it was shown that ER performs the function of AhR co-regulator and that ERα activation is necessary to maintain the expression of both AhR itself and its target genes [23, 24].
To assess the impact of changes in the miR-483-3p content on the expression of its target genes, we assessed the levels of the IGF1 mRNA. IGF1 encodes a protein from the family of insulin-like growth factors synthesized mainly in the liver cells. Low serum levels of IGF1 are associated with various liver disorders, including non-alcoholic fatty liver disease [25]. In our experiment, the content of IGF1 mRNA in rat hepatocytes was downregulated by BP and upregulated by the AhR inhibitor, which correlated with the changes in the miR-483-3p expression. Although BP initially decreased the level of IGF1 expression in the rat liver; the content of the IGF1 mRNA in the liver of control and BP-treated rats did not differ 72 h after BP administration, which may be related to the fact that no changes in the levels of miR-483-3p expression were observed at this time point.
Therefore, we have shown that BP administration downregulates expression of the IGF2 gene, whose promoter contains the binding sites for AhR and ERs. At the same time, expression of the oncogenic miR-483-3p encoded in the intron of IGF2 turned out to be independent of this gene; apparently, its transcription can be activated from its own promoter containing the DRE site. We conclude that impairments in the expression of IGF2, miR-483-3p, and its target genes, in particular IGF1, might be one of the mechanisms of the toxic action of BP.
Funding. This study was supported by the Russian Science Foundation (project 15-15-30012).
Conflict of interest. The authors declare no conflict of interest in financial or any other sphere.
Ethical approval. All procedures with the animals were carried out in accordance with the requirements of the “Rules for work with the use of experimental animals” (Order of the USSR Ministry of Higher and Secondary Special Education No. 742; November 13, 1984).
REFERENCES
1.International Agency for Research on Cancer (2012)
Chemical Agents and Related Occupations, IARC Monographs on the
Evaluation of Carcinogenic Risks to Humans, Vol. 100 F.
2.Pieterse, B., Felzel, E., Winter, R., Van Der Burg,
B., and Brouwer, A. (2013) PAH-CALUX, an optimized bioassay for
AhR-mediated hazard identification of polycyclic aromatic hydrocarbons
(PAHs) as individual compounds and in complex mixtures, Environ.
Sci. Technol., 47, 11651-11659; doi: 10.1021/es403810w.
3.Yao, E. F., and Denison, M. S. (1992) DNA sequence
determinants for binding of transformed Ah receptor to a
dioxin-responsive enhancer, Biochemistry, 31, 5060-5067;
doi: 10.1021/bi00136a019.
4.Phadnis-Moghe, A. S., Li, J., Crawford, R. B., and
Kaminski, N. E. (2016) SHP-1 is directly activated by the aryl
hydrocarbon receptor and regulates BCL-6 in the presence of
2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD), Toxicol. Appl.
Pharmacol., 310, 41-50; doi: 10.1016/j.taap.2016.08.014.
5.Gottel, M., Corre, L. L., Dumont, C., Schrenk, D.,
and Chagnon, M. C. (2014) Estrogen receptor α and aryl
hydrocarbon receptor cross-talk in a transfected hepatoma cell line
(HepG2) exposed to 2,3,7,8-tetrachlorodibenzo-p-dioxin,
Toxicol. Rep., 1, 1029-1036; doi:
10.1016/j.toxrep.2014.09.016.
6.Ambros, V. (2004) The functions of animal miRNAs,
Nature, 431, 350-355; doi: 10.1038/nature02871.
7.Ovchinnikov, V. Y., Antonets, D. V., and Gulyaeva,
L. F. (2018) The search of CAR, AhR, ESRs binding sites in promoters of
intronic and intergenic microRNAs, J. Bioinform. Comput. Biol.,
16, 1750029; doi: 10.1142/S0219720017500299.
8.Livingstone, C. (2013) IGF2 and cancer, Endocr.
Relat. Cancer, 20, 321-339; doi: 10.1530/ERC-13-0231.
9.Zhou, W., Yang, W., Ma, J., Zhang, H., Li, Z.,
Zhang, L., Liu, J., Han, Z., Wang, H., and Hong, L. (2018) Role of
miR-483 in digestive tract cancers: from basic research to clinical
value, J. Cancer, 9, 407-414; doi: 10.7150/jca.21394.
10.Korzeniewski, N., Tosev, G., Pahernik, S.,
Hadaschik, B., Hohenfellner, M., and Duensing, S. (2015) Identification
of cell-free microRNAs in the urine of patients with prostate cancer,
Urol. Oncol., 33, 16.e17-16.e22; doi:
10.1016/j.urolonc.2014.09.015.
11.Zhang, M., Liu, D., Li, W., Wu, X., Gao, C., and
Li, X. (2016) Identification of featured biomarkers in breast cancer
with microRNA microarray, Arch. Gynecol. Obstet., 294,
1047-1053; doi: 10.1007/s00404-016-4141-7.
12.Xu, H., Yang, Y., Zhao, H., Yang, X., Luo, Y.,
Ren, Y., Liu, W., and Li, N. (2016) Serum miR-483-5p: a novel
diagnostic and prognostic biomarker for patients with oral squamous
cell carcinoma, Tumor Biol., 37, 447-453; doi:
10.1007/s13277-015-3514-z.
13.Wu, C., Wang, C., Guan, X., Liu, Y., Li, D.,
Zhou, X., Zhang, Y., Chen, X., Wang, J., Zen, K., Zhang, C. Y., and
Zhang, C. (2014) Diagnostic and prognostic implications of a serum
miRNA panel in esophageal squamous cell carcinoma, PLoS One,
9, e92292; doi: 10.1371/journal.pone.0092292.
14.Gulyaeva, L. F., and Kushlinskiy, N. E. (2016)
Regulatory mechanisms of microRNA expression, J. Transl. Med.,
14, 143-153; doi: 10.1186/s12967-016-0893-x.
15.Seglen, P. (1976) Preparation of isolated rat
liver cells, Meth. Cell. Biol., 13, 29-83; doi:
10.1016/S0091-679X(08)61797-5.
16.Pogribny, I. P., Beland, F. A., and Rusyn, I.
(2016) The role of microRNAs in the development and progression of
chemical-associated cancers, Toxicol. Appl. Pharmacol.,
312, 3-10; doi: 10.1016/j.taap.2015.11.013.
17.Gulyaeva, L. F., Chanyshev, M. D., Kolmykov, S.
K., Ushakov, D. S., and Nechkin, S. S. (2016) The effect of xenobiotics
on microRNA expression in the rat liver, Biochemistry (Moscow)
Suppl. Series B Biomed. Chem., 10, 258-263; doi:
10.1134/S1990750816030033.
18.Lupini, L., Pepe, F., Ferracin, M., Braconi, C.,
Callegari, E., Pagotto, S., Spizzo, R., Zagatti, B., Lanuti, P.,
Fornari, F., Ghasemi, R., Mariani-Costantini, R., Bolondi, L.,
Gramantieri, L., Calin, G. A., Sabbioni, S., Visone, R., Veronese, A.,
and Negrini, M. (2016) Over-expression of the miR-483-3p overcomes the
miR-145/TP53 pro-apoptotic loop in hepatocellular carcinoma,
Oncotarget, 7, 31361-31371; doi:
10.18632/oncotarget.8913.
19.Vasuri, F., Fittipaldi, S., De Pace, V.,
Gramantieri, L., Bertuzzo, V., Cescon, M., Pinna, A. D., Fiorentino,
M., D’Errico, A., and Ravaioli, M. (2018) Tissue miRNA-483-3p
expression predicts tumor recurrence after surgical resection in
histologically advanced hepatocellular carcinomas, Oncotarget,
9, 17895-17905; doi: 10.18632/oncotarget.24860.
20.Lnenickova, K., Skalova, L., Stuchlikova, L.,
Szotakova, B., and Matouskova, P. (2018) Induction of
xenobiotic-metabolizing enzymes in hepatocytes by beta-naphthoflavone:
time-dependent changes in activities, protein and mRNA levels, Acta
Pharm., 68, 75-85; doi: 10.2478/acph-2018-0005.
21.Matthews, J., and Gustafsson, J. A. (2006)
Estrogen receptor and aryl hydrocarbon receptor signaling pathways,
Nucl. Recept. Signal., 4, e016; doi:
10.1621/nrs.04016.
22.Salisbury, T. B., Morris, G. Z., Tomblin, J. K.,
Chaudhry, A. R., Cook, C. R., and Santanam, N. (2013) Aryl hydrocarbon
receptor ligands inhibit IGF-II and adipokine stimulated breast cancer
cell proliferation, ISRN Endocrinol., 2013, 104850; doi:
10.1155/2013/104850.
23.Matthews, J., Wihlen, B., Thomsen, J., and
Gustafsson, J. A. (2005) Aryl hydrocarbon receptor-mediated
transcription: ligand-dependent recruitment of estrogen receptor
α to 2,3,7,8-tetrachlorodibenzo-p-dioxin-responsive
promoters, Mol. Cell. Biol., 25, 5317-5328; doi:
10.1128/MCB.25.13.5317-5328.2005.
24.Spink, D. C., Katz, B. H., Hussain, M. M.,
Pentecost, B. T., Cao, Z., and Spink, B. C. (2003) Estrogen regulates
Ah responsiveness in MCF-7 breast cancer cells, Carcinogenesis,
24, 1941-1950; doi: 10.1093/carcin/bgg162.
25.Adamek, A., and Kasprzak, A. (2018) Insulin-like
growth factor (IGF) system in liver diseases, Int. J. Mol. Sci.,
19, 1308; doi: 10.3390/ijms19051308.