Expression of mRNAs for IL-1β, IL-6, IL-10, TNFα, CX3CL1, and TGFβ1 Cytokines in the Brain Tissues: Assessment of Contribution of Blood Cells with and without Perfusion
A. A. Kvichansky1, M. N. Volobueva1, Yu. S. Spivak1, L. V. Tret’yakova1, N. V. Gulyaeva1,a*, and A. P. Bolshakov1
1Institute of Higher Nervous Activity and Neurophysiology, Russian Academy of Sciences, 117485 Moscow, Russia* To whom correspondence should be addressed.
Received March 21, 2019; Revised May 4, 2019; Accepted May 5, 2019
Cytokines are important regulators of brain function under both normal and pathological conditions. Cytokines can be synthesized by resident cells of the central nervous system (CNS) (vascular endothelium, cells of the blood-brain barrier, parenchymal cells of the CNS) or cells in the lumen of blood vessels, as well as introduced with the bloodstream. The ratio between the quantity of cytokines synthesized in the CNS and those entering it from external sources under various conditions remains poorly understood. In this work, we studied the contribution of mRNAs from non-resident cells to the common pool of cytokine (TNFα, IL-1β, IL-6, IL-10, CX3CL1, and TGFβ1) mRNAs in the rat neocortex, hippocampus, dura matter, pia matter, and choroid plexus. We also evaluated the representation of various populations of resident and non-resident immune cells based on the expression of marker genes (Ncf1, Tbx21, Foxp3, RORγc). The removal of blood by transcardial perfusion led to a decrease in the quantity of the TNFα mRNA in the neocortex and hippocampus and of the IL-1β, IL-6, and IL-10 mRNAs in the dura mater. The mRNA levels of other cytokines in studied structures were not affected by perfusion. Our findings suggest that mRNAs present in the blood can make a significant contribution to the mRNA levels of some cytokines in the CNS; therefore, preliminary perfusion of brain tissue is a necessary stage of experimental design for correct estimation of mRNA content in the brain.
KEY WORDS: cytokines, IL-1β, IL-6, IL-10, TNFα, CX3CL1, TGFβ1, perfusion, hippocampus, neocortexDOI: 10.1134/S0006297919080066
Abbreviations: CX3CL1, C-X3-C chemokine ligand 1 (fractalkine); IL, interleukin; TNFα, tumor necrosis factor alpha; TGFβ1, transforming growth factor beta 1.
Neuroinflammation is a specific form of inflammatory process typical for
the central nervous system (CNS). It was first described as a response
to various types of brain damage, including trauma, exposure to
toxicants, hypoxia, and infection. However, it was shown later that the
processes characteristic of neuroinflammation occur in the norm, e.g.,
in synaptic plasticity and neurogenesis, which indicates their adaptive
role [1].
An increase in the mRNA levels of proinflammatory cytokines (IL-1β, IL-6, TNFα), which are expressed in the brain mostly by the microglia, is used as a standard marker of active neuroinflammation [2]. A balance between the concentrations of pro- and anti-inflammatory cytokines plays an important role in the neuroinflammation regulation [3]. mRNAs for the anti-inflammatory cytokines IL-10 and TGFβ (TGFβ1, TGFβ2, TGFβ3) have been found in various cells of the CNS [2, 4, 5]. A special role belongs to fractalkine (CX3CL1). This cytokine is synthesized mostly by neurons in the CNS [2] and regulates migration of macrophages into the CNS and their transformation into the microglia, especially during the brain formation. In mature brain, CX3CL1 acts as an anti-inflammatory mediator providing negative feedback from neurons to the microglia [6]. Assessment of mRNA expression levels for various pro- and anti-inflammatory cytokines is commonly used in the studies on the neuroinflammatory response development.
Brain meninges can be an important source of cytokines to the local bloodstream, since they contain a large number of immune cells. Besides, blood vessels that supply the brain run through the meninges. It was shown that dura mater is an important part of the so-called glymphatic system that regulates interactions between immune and nervous systems [7]. Major veins and arteries responsible for the blood supply to the brain are located in the subarachnoid space. The subarachnoid space also contains venous sinuses associated with the immune system cells regulating multiple brain functions [8]. The choroid plexus is another anatomical structure of the brain populated by immune system cells regulating numerous processes in the CNS [9].
Cytokines in the CNS can be of endogenous (synthesized by the brain resident cells) or exogenous origin (synthesized by nonresident cells in the lumen of blood vessels or outside the CNS). Despite the fact that cytokine mRNAs in the brain are synthesized by both parenchymal and blood cells, the vast majority of studies on the cytokine expression in the brain assume a priori that the main source of cytokines in the brain tissue is the parenchyma and not the blood cells. However, the contribution of parenchymal and blood cells to the pool of cytokine mRNAs in the brain has not been evaluated. Such evaluation might be important for the interpretation of experimental data, since the levels of mRNAs for proinflammatory cytokines in the brain are relatively low (especially, in the norm) and contamination with blood can significantly increase experimentally measured levels of cytokine mRNAs and skew the ratio between these values, thereby introducing an unpredictable error to the study results. The above considerations prompted us to conduct this methodological study.
Here, we investigated the effect of blood removal from the rat brain vessels and meninges by perfusion on the relative quantities of mRNAs for the main neuroinflammation-associated cytokines. We also analyzed the effect of perfusion on the relative quantity of mRNAs for the markers of immune cell subpopulations involved in the neuroinflammation regulation in the brain and its meninges.
MATERIALS AND METHODS
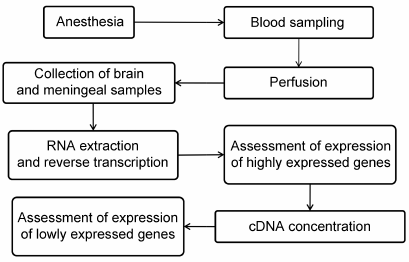
Experimental procedures. Male Wistar rats weighing 250-300 g from the Stolbovaya nursery, Moscow Region, were used. The rats from the experimental group were anesthetized by injection of 10% (w/v) chloral hydrate (Sigma, USA) at a dose of 9 ml/kg body weight and then subjected to transcardial perfusion with ice-cold 0.01 M phosphate buffered saline (PBS, pH 7.4) (Pan-Eco, Russia) for 20 min. Blood samples (100 μl) were collected immediately after the right atrium opening. After perfusion, the rats were decapitated; their brains were removed, washed with ice-cold PBS, and dried on filter paper. Tissue samples of the neocortex, hippocampus, dura mater, pia and arachnoid meninges adjacent to the cortex, pia and arachnoid meninges adjacent to the hippocampus, and choroid plexus were collected (see Fig. 1 for the experimental scheme). The rats of the control group were subjected to the same manipulations except for transcardial perfusion.
Fig. 1. Experimental scheme.
RNA isolation and reverse transcription. Collected tissue and blood samples were placed in tubes containing 500 μl of ExtractRNA reagent (Evrogen, Russia). RNA isolation was performed in accordance with the manufacturer’s recommendations. To remove traces of genomic DNA, RNA samples were treated with DNase I (Thermo Scientific, USA). Reverse transcription was performed using the MMLV RT kit reagent kit (Evrogen) using murine RNase Inhibitor (New England Biolabs, USA) as recommended by the manufacturer. An equimolar mixture of random decaprimer (Evrogen) and oligo(dT)15 primer (Evrogen) was used; the concentration of each primer in the reaction was 1 μM. After reverse transcription, the reaction mixture was diluted 8-fold with deionized water.
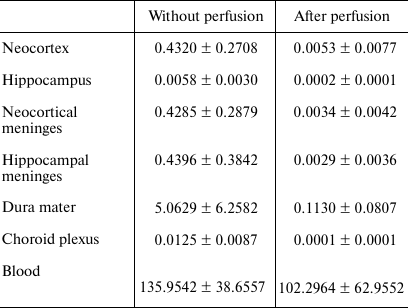
qPCR. Relative quantities of mRNAs for the genes of interest were evaluated with a Bio-Rad CFX-384 real-time PCR station (Bio-Rad, USA) using a qPCRmix-HS SYBR + LowROX mix for PCR (Evrogen) according to the manufacturer’s recommendations. Relative quantities of mRNAs for highly represented genes of pro-inflammatory cytokines (Il1b, Il6, Tnf), anti-inflammatory cytokines (Tgfb1 and Cx3cl1), and the marker of monocytes (Ncf1) [10] were normalized to the geometric mean of the mRNA expression levels for the Ywhaz and Hprt1 genes. Expression levels of the lowly expressed genes for the anti-inflammatory cytokine IL-10 (Il10) and a set of lymphocyte markers, such as Tbx21 (Th1 lymphocytes [11]), Foxp3 (Treg lymphocytes [12]), and RORγc (Th17 lymphocytes [13]), were normalized to the Osbp1 mRNA after 5-fold concentration of cDNA by precipitation with an isopropanol/glycogen mixture (Thermo Scientific) that allowed to increase the concentration of the target cDNA and reduce the random error of the method. To assess the quality of the DNase treatment for all the samples and genes, we ran a negative control with the product of DNase I treatment. To assess the quality of perfusion, the relative quantity of the Alas2 mRNA (erythrocyte marker) was determined by normalization to the mRNA expression of the Ywhaz and Hprt1 genes. Samples from the perfused rats, in which the relative quantity of Alas2 mRNA was less than 10 times lower than the average value in the control group, were excluded from the study (the average values in the groups are shown in Table 1).
Table 1. Effect of perfusion on the relative
quantity of Alas2 mRNA
Primers for qPCR were designed for the mRNA sequences (Table 2) from the NCBI database using the PrimerSelect software package (DNASTAR Lasergene). The genes used for normalization were selected based on the results of transcriptome analysis [14].
Table 2. Primers used for qPCR

Data analysis. Gene expression was analyzed by the EΔΔCt method.
The data in Table 1 are shown as mean ± SD. The data in the graphs are shown as grouped point diagrams. The significance of differences between groups was evaluated using the Mann–Whitney method with the Statistica 10 software (n ≥ 6 in all groups).
RESULTS AND DISCUSSION
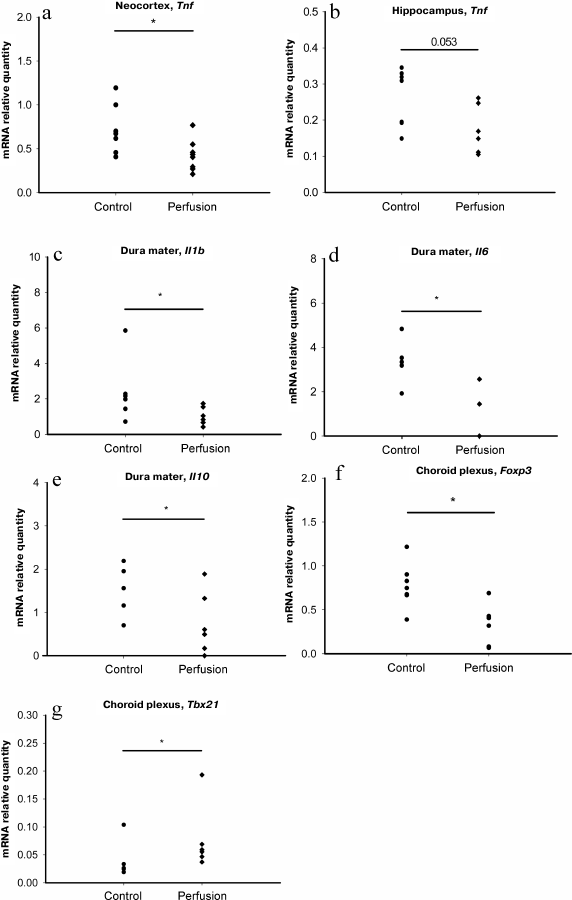
We found that transcardial perfusion led to a decrease in the relative quantity of Tnf mRNA in the neocortex and hippocampus, (Fig. 2, a and b), while the relative quantities of mRNAs for other potentially exogenous cytokines, as well as markers of monocytes and T helper cells in these brain regions were unaffected (Table 3). In the dura mater, perfusion resulted in the decrease in the relative quantities of mRNAs for the Il1b, Il6 and Il10 genes, but not for the marker genes (Fig. 2, c-e, and Table 3).
Fig. 2. Effect of perfusion on the content of mRNAs for major neuroinflammation-associated cytokines and marker genes of immune cell subpopulations: a) Tnf mRNA in the neocortex; b) Tnf mRNA in the hippocampus; c) Il1b mRNA in the dura mater; d) Il6 mRNA in the dura mater; e) Il10 mRNA in the dura mater; f) Foxp3 mRNA in the choroid plexus; g) Tbx21 mRNA in the choroid plexus; * p ≤ 0.05, Mann–Whitney test.
Table 3. Effect of perfusion on the relative
quantities of mRNAs for cytokines and markers of cell
subpopulations
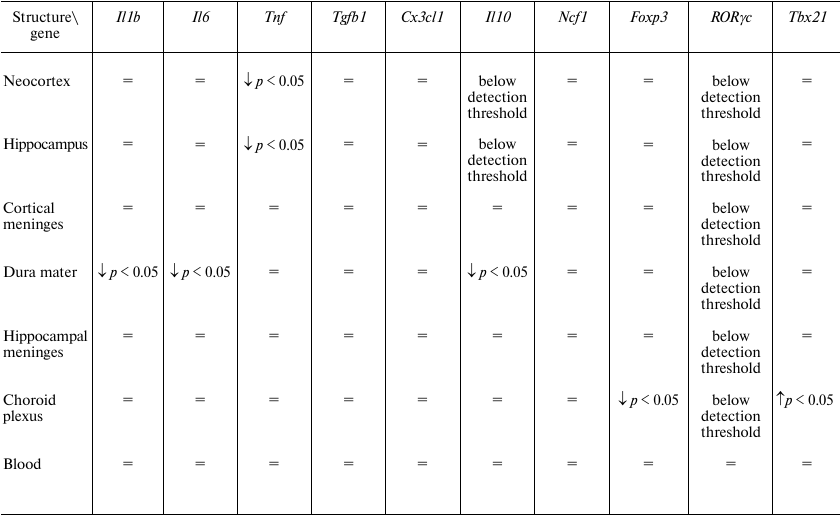
Note: =, no significant differences.
These data indicate that, in the forebrain and dura mater, mRNAs for most immune mediators are synthesized by the resident cells. However, a decrease in the mRNA levels for the Tnf gene in the neocortex and hippocampus and for the Il1b, Il6, and Il10 genes in the dura mater suggests that non-resident cells make a considerable contribution to the mRNA pool of these genes in the corresponding structures. At the same time, perfusion did not affect relative quantity of mRNAs for the markers of monocytes (Ncf1) and some subtypes of T cells (Tbx21, Foxp3, RORγc). It is possible that the sources of mRNAs removed by perfusion were blood cells that did not belong to studied cell populations, e.g., neutrophils or Th2 lymphocytes.
A decrease in the relative quantity of Foxp3 mRNA was detected in the choroid plexus (Fig. 2f); however, the relative quantity of mRNA for IL-10, the main cytokine of Treg lymphocytes, was not affected by perfusion [15], likely because choroid plexus contains a large number of resident cells expressing IL-10, and removal of Treg lymphocytes from the choroid plexus vessels did not affect the level of this cytokine. Paradoxically, the relative quantity of mRNA for the Tbx21 gene in the choroid plexus increased after perfusion (Fig. 2g).
Therefore, we have shown that the presence of blood cells in brain tissues, meninges, and choroid plexus considerably contributes to the mRNA levels of various immune mediators and markers of cell subpopulations in these brain regions. This contribution varies for different genes and anatomical structures. In general, it is impossible to predict the contribution of exogenous cells to the total mRNA pool, especially since this contribution may not be the same under normal and pathological conditions. In this regard, we recommend perfusion of laboratory animals as an important step before collection of biological material in the qPCR studies of neuroinflammatory response in the brain and its meninges.
Funding. This work was supported by the Russian Foundation for Basic Research (project 18-015-00314a).
Conflict of interest. The authors declare no conflict of interest.
Ethical approval. All applicable international, national, and/or institutional guidelines for the care and use of animals were followed. The experimental protocol was approved by the Ethics Committee of the Institute of Higher Nervous Activity and Neurophysiology of the Russian Academy of Sciences.
REFERENCES
1.Estes, M. L., and McAllister, A. K. (2014)
Alterations in immune cells and mediators in the brain: it’s not
always neuroinflammation! Brain Pathol., 24, 623-630,
doi: 10.1111/bpa.12198.
2.Zeisel, A., Munoz-Manchado, A. B., Codeluppi, S.,
Lonnerberg, P., La Manno, G., Jureus, A., Marques, S., Munguba, H., He,
L., Betsholtz, C., Rolny, C., Castelo-Branco, G., Hjerling-Leffler, J.,
and Linnarsson, S. (2015) Brain structure. Cell types in the mouse
cortex and hippocampus revealed by single-cell RNA-seq, Science,
347, 1138-1142, doi: 10.1126/science.aaa1934.
3.Meyer, U., Murray, P. J., Urwyler, A., Yee, B. K.,
Schedlowski, M., and Feldon, J. (2008) Adult behavioral and
pharmacological dysfunctions following disruption of the fetal brain
balance between pro-inflammatory and IL-10-mediated anti-inflammatory
signaling, Mol. Psychiatry, 13, 208-221, doi:
10.1038/sj.mp.4002042.
4.Zhang, Y., Chen, K., Sloan, S. A., Bennett, M. L.,
Scholze, A. R., O’Keeffe, S., Phatnani, H. P., Guarnieri, P.,
Caneda, C., Ruderisch, N., Deng, S., Liddelow, S. A., Zhang, C.,
Daneman, R., Maniatis, T., Barres, B. A., and Wu, J. Q. (2014) An
RNA-sequencing transcriptome and splicing database of glia, neurons,
and vascular cells of the cerebral cortex, J. Neurosci.,
34, 11929-11947, doi: 10.1523/JNEUROSCI.1860-14.2014.
5.Pisanu, A., Lecca, D., Mulas, G., Wardas, J.,
Simbula, G., Spiga, S., and Carta, A. R. (2014) Dynamic changes in pro-
and anti-inflammatory cytokines in microglia after PPAR-γ agonist
neuroprotective treatment in the MPTPp mouse model of progressive
Parkinson’s disease, Neurobiol. Dis., 71, 280-291,
doi: 10.1016/j.nbd.2014.08.011.
6.Rogers, J. T., Morganti, J. M., Bachstetter, A. D.,
Hudson, C. E., Peters, M. M., Grimmig, B. A., Weeber, E. J., Bickford,
P. C., and Gemma, C. (2011) CX3CR1 deficiency leads to impairment of
hippocampal cognitive function and synaptic plasticity, J.
Neurosci., 31, 16241-16250, doi:
10.1523/JNEUROSCI.3667-11.2011.
7.Louveau, A., Herz, J., Alme, M. N., Salvador, A.
F., Dong, M. Q., Viar, K. E., Herod, S. G., Knopp, J., Setliff, J. C.,
Lupi, A. L., Da Mesquita, S., Frost, E. L., Gaultier, A., Harris, T.
H., Cao, R., Hu, S., Lukens, J. R., Smirnov, I., Overall, C. C.,
Oliver, G., and Kipnis, J. (2018) CNS lymphatic drainage and
neuroinflammation are regulated by meningeal lymphatic vasculature,
Nat. Neurosci., 21, 1380-1391, doi:
10.1038/s41593-018-0227-9.
8.Filiano, A. J., Gadani, S. P., and Kipnis, J.
(2017) How and why do T cells and their derived cytokines affect the
injured and healthy brain? Nat. Rev. Neurosci., 18,
375-384, doi: 10.1038/nrn.2017.39.
9.Marques, F., and Sousa, J. C. (2015) The choroid
plexus is modulated by various peripheral stimuli: implications to
diseases of the central nervous system, Front. Cell. Neurosci.,
9, 136, doi: 10.3389/fncel.2015.00136.
10.Ding, X., Zhang, M., Gu, R., Xu, G., and Wu, H.
(2017) Activated microglia induce the production of reactive oxygen
species and promote apoptosis of co-cultured retinal microvascular
pericytes, Graefe’s Arch. Clin. Exp. Ophthalmol.,
255, 777-788, doi: 10.3389/fncel.2015.00136.
11.Ferreira, L., Hamey, F., Teichmann, S. A.,
Cvejic, A., Macaulay, I. C., Svensson, V., Labalette, C., and Voet, T.
(2016) Single-cell RNA-sequencing reveals a continuous spectrum of
differentiation in hematopoietic cells, Cell Rep., 14,
966-977, doi: 10.1016/j.celrep.2015.12.082.
12.Prieto Martin, P., Bending, D., Ono, M., Ducker,
C. B., Crompton, T., and Paduraru, A. (2018) A temporally dynamic Foxp3
autoregulatory transcriptional circuit controls the effector Treg
programme, EMBO J., 37, e99013, doi:
10.15252/embj.201899013.
13.Zhao, Y., Balato, A., Fishelevich, R., Chapoval,
A., Mann, D. L., and Gaspari, A. A. (2009) Th17/Tc17 infiltration and
associated cytokine gene expression in elicitation phase of allergic
contact dermatitis, Br. J. Dermatol., 161, 1301-1306,
doi: 10.1111/j.1365-2133.2009.09400.x.
14.Dobryakova, Y. V., Kasianov, A., Zaichenko, M.
I., Stepanichev, M. Y., Chesnokova, E. A., Kolosov, P. M., Markevich,
V. A., and Bolshakov, A. P. (2018) Intracerebroventricular
administration of 192IgG-saporin alters expression of
microglia-associated genes in the dorsal but not ventral hippocampus,
Front. Mol. Neurosci., 10, doi:
10.3389/fnmol.2017.00429.
15.Williams, A., Zandee, S. E. J., Mair, I.,
Anderton, S. M., O’Connor, R. A., and Leech, M. D. (2017)
IL-10-producing, ST2-expressing Foxp3+ T-cells in multiple sclerosis
brain lesions, Immunol. Cell Biol., 95, 484-490, doi:
10.1038/icb.2017.3.