Properties of Malic Enzyme from the Aerobic Methanotroph Methylosinus trichosporium
O. N. Rozova1, V. N. Khmelenina1,a*, I. I. Mustakhimov1, S. Y. But1, and Yu. A. Trotsenko1
1G. K. Skryabin Institute of Biochemistry and Physiology of Microorganisms, Russian Academy of Sciences, Federal Research Center “Pushchino Scientific Center for Biological Research of the Russian Academy of Sciences”, 142290 Pushchino, Moscow Region, Russia* To whom correspondence should be addressed.
Received August 14, 2018; Revised December 12, 2018; Accepted December 12, 2018
Recombinant malic enzyme from the aerobic methanotroph Methylosinus trichosporium was obtained by heterologous expression in Escherichia coli and purified by affinity metal-chelating chromatography. The homohexameric enzyme of 6×80 kDa catalyzed the reversible reaction of oxidative decarboxylation of malate to pyruvate in the presence of mono- and divalent cations and NADP+ as a cofactor. The kcat/Km ratio indicated much higher catalytic efficiency of the malate decarboxylation reaction as compared with the pyruvate carboxylation reaction. Analysis of the protein sequence revealed that the C-region of the enzyme contains a large domain homologous to phosphoacetyltransferase, but no phosphoacetyltransferase activity was detected either for a full chimeric malic enzyme or for the C-end fragment obtained as a separate protein. This C-end domain promoted activity of the malic enzyme.
KEY WORDS: malic enzyme, methanotrophs, Methylosinus trichosporiumDOI: 10.1134/S0006297919040060
Abbreviations: DTNB, 5,5′-dithiobis(2-nitrobenzoic acid); ME, malic enzyme; PEP, phosphoenolpyruvate; THF, tetrahydrofolate.
Malic enzyme (ME) is widely distributed in macro- and microorganisms. ME
catalyzes the oxidative decarboxylation of malate to pyruvate and
CO2 with the reduction of NAD+ or
NADP+ [1, 2]:
Malate + NAD(P)+ ⇆ Pyruvate + NAD(P)H2 + HCO3+.
Malic enzymes are divided into three different categories based on their substrate specificity and cofactor dependency: NAD+-dependent decarboxylating oxaloacetate ME (EC 1.1.1.38), NAD(P)+-dependent ME non-decarboxylating oxaloacetate (EC 1.1.1.39), and NADP+-dependent ME (EC 1.1.1.40) [1-3]. Some malic enzymes use both pyridine nucleotides, but they have different specificities for cofactors. Although it is believed that malic enzymes catalyze a reversible reaction, they differ in their ability to carboxylate pyruvate – from the complete absence of the reaction to its prevalence over the decarboxylation reaction [4, 5].
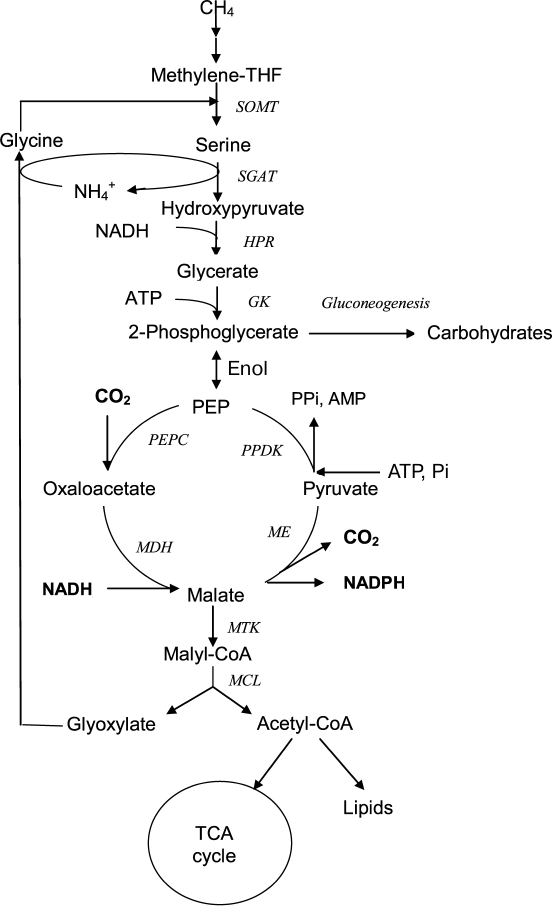
Aerobic bacteria using methane as a growth substrate (methanotrophs) have an increasing potential for biotechnology; therefore, methods for genetic modification and correction of the metabolism of promising strains are widely used for its successful use. In turn, this requires knowledge of the main biochemical pathways and enzyme properties. The obligate methanotroph Methylosinus trichosporium OB3b is a member of the Alphaproteobacteria class and one of the model organisms for studying methylotrophy as the mode for microbial life and for valuation of metabolic potential of these bacteria. Methylosinus trichosporium uses the serine pathway for C1 assimilation in which the C3 compound (serine) is the primary product resulting from the condensation of methylene tetrahydrofolate (methylene-THF) and glycine (Fig. 1). Malate, one of the central metabolites of this pathway, is synthesized via transformation of serine through a series of reactions including transamination with glyoxylate, reduction of hydroxypyruvate into glycerate, glycerate phosphorylation, and carboxylation of phosphoenolpyruvate (PEP) into oxaloacetate [6, 7]. The last of these reactions is performed by highly active PEP carboxylase represented by two isoforms [8], and malate is synthesized from oxaloacetate by NADH-dependent malate dehydrogenase [7]. Further transformation of malate in the serine pathway is associated with the formation of malyl-CoA and the decomposition of this compound into acetyl-CoA and glyoxylate, which is a precursor of glycine, a one-carbon compound acceptor. Thus, the decarboxylating activity of malic enzyme leads to a loss of the C–C bond formed during C1 assimilation. This work aimed at studying the catalytic properties of the recombinant malic enzyme in order to understand the regulation of this metabolic region in the aerobic methanotroph Ms. trichosporium.
Fig. 1. Putative participation of malic enzyme in central metabolism of Ms. trichosporium. SOMT, serine oxymethyltransferase; SGAT, serine glyoxylate aminotransferase; HPR, hydroxypyruvate reductase; GK, glycerate kinase; PEPC, PEP carboxylase; MDH, malate dehydrogenase; ME, malic enzyme; PPDK, pyruvate, phosphate dikinase; MTK, ATP-dependent malate thiokinase; MCL, malyl-CoA-lyase; methylene-THF, methylenetetrahydrofolate.
MATERIALS AND METHODS
Bacteria and growth conditions. Methylosinus trichosporium OB3b (VKM B-2117, ATCC 35070) was grown at 28°C under a methane–air atmosphere (1 : 1 v/v) in nitrate mineral medium “P” [9] containing (g/liter): KNO3 (1.0), MgSO4·7H2O (0.2), CaCl2 (0.02), Na2HPO4·5H2O (1.5), KH2PO4 (0.7), Trilon B (Na2EDTA) (0.005), FeSO4·7H2O (0.002), ZnSO4·7H2O (0.0001), MnCl2·4H2O (0.00003), CoCl2·6H2O (0.0002), CuSO4·5H2O (0.0001), NiCl2·6H2O (0.00002), Na2MoO4·2H2O (0.00003) in a thermostated shaker (200 rpm; New Brunswick Scientific, USA). Escherichia coli BL21(DE3) (Novagen, Germany) was grown at 37°C in selective Luria–Bertani agar or broth [10]. For growth of plasmid-bearing cells of E. coli, ampicillin was added at concentration of 100 μg/ml.
DNA manipulations. Plasmid isolation, digestion by restriction enzymes, agarose gel electrophoresis, ligation, and transformation of E. coli cells were performed according to described methods [10]. Restriction enzymes, T4 DNA-ligase, Pfu and Taq DNA-polymerases, dNTP mixture, and Page Ruler Prestained Protein Ladder for SDS-PAGE were purchased from Thermo Scientific (USA).
Preparing and purification of malic enzyme (ME). Chromosomal DNA from Ms. trichosporium cells was prepared as previously described [11]. The dme gene (ID 2507408727) encoding putative ME from Ms. trichosporium (IMG https://img.jgi.doe.gov) was amplified by PCR using primers N-dme-Nde (5′-TCCATATGGCGGAGAAGCCGCGCATGGACC) and C-dme-Hind (5′-ATAAGCTTCCCCCCGACGCCGAAGGCCGCCAGC) containing recognition sites for endonucleases NdeI and HindIII, respectively. For expression of dme, vector pET22b:dme was constructed and transferred to E. coli BL21(DE3) cells. The synthesis of the enzyme was induced by the addition of 0.5 mM isopropyl-1-thio-β-D-galactopyranoside (IPTG) at A600 = 0.6-0.7. After growth at 18°C for 15 h, the cells were harvested by centrifugation (30 min at 8°C and 5000g) and stored at –20°C. The His6-tagged protein was purified by affinity chromatography on a Ni2+-nitrilotriacetic acid (Ni-NTA) agarose column as described earlier [12]. The purified enzyme was stored in 40% glycerol at –20°C.
For cloning of N-terminal sequence of the enzyme-encoding gene (mae fragment), the forward primer N-dme-Nde (see above) and the reverse primer 5′-TAAGCTTATCCAGCGCGGCGACCGGATCGCGCC were used. For cloning of the C-terminal region of the gene (patr fragment) the primers PaTR74-Nde-F (5′-TACATATGCATACGATCTACGATCGCGTGCGGC) and C-dme-Hind were used. The cloning and expression of mae and patr fragments and purification of the protein were carried out by methods described above.
Determination of molecular masses. The quaternary form of the enzymes were analyzed by non-denaturing gel electrophoresis by using pore-limiting gradient of polyacrylamide (4-30%) [13] with protein marker set (Sigma-Aldrich, Germany): thyroglobulin (660 kDa, dimer), ferritin (440 kDa, 24 subunits), catalase (232 kDa, tetramer), lactate dehydrogenase (140 kDa, tetramer), bovine serum albumin (67 kDa, monomer).
Assay of malic enzyme activity. Activity of the malic enzyme in the direction of malate decarboxylation was determined by measuring NADP+ reduction velocity at 30°C in 1 ml of the reaction mixture containing 50 mM potassium phosphate buffer, pH 7.5, 2.5 mM MgCl2, 0.3 mM NADP+, and ~1 μg of the enzyme. The reaction was initiated by addition of 10 mM malic acid disodium salt. In the direction of pyruvate carboxylation, the enzyme activity was tested by measuring NADPH oxidation in 1 ml reaction mixture containing 50 mM Tris-HCl, pH 7.5, 2.5 mM MgCl2, 0.25 mM HADPH, 50 mM KHCO3, 25 mM sodium pyruvate, and ~9 μg of the protein.
The ability of ME to catalyze oxaloacetate decarboxylation was tested by two methods. (i) Spectrophotometrically by following the decrease of oxaloacetate absorption at 280 nm [14] in reaction mixture containing 50 mM potassium phosphate buffer (pH 7.5) or MES-NaOH (pH 5.0), 2.5 mM MgCl2, 1-10 mM oxaloacetate, and 50 μg ME in the presence or absence of 0.1 mM NADP+ or NADPH. (ii) By testing the formation of pyruvate in this reaction by HPLC on a Reprosil-Pur c18-AQ column (5 μm, 250 × 10 mm) (Dr. Maisch, Germany) using 1 mM H2SO4 and 8 mM Na2SO4 as the mobile phase at 25°C and flow rate of 1 ml/min.
The phosphotransacetylase activities of ME and its C-terminal fragment were tested by registering the formation of 5-thio(2-nitrobenzoic acid) as a result of the interaction between 5,5′-dithiobis(2-nitrobenzoic acid) (DTNB) with the sulfhydryl groups of CoA-SH formed during the reaction [15]. The reaction mixture (1 ml) contained 50 mM potassium phosphate buffer, pH 7.5, 0.1 mM DTNB, 2.5 mM MgCl2, and 0.2 mM acetyl-CoA. The reaction was started by adding 10-50 μg of the enzyme. The measurements were carried out at 412 nm.
To study pH dependence of the enzyme activity, the following buffers were used (50 mM): glycine-NaOH (pH 9.0-10.5), CHES-NaOH (pH 8.5-10.0), Tris-HCl (pH 7.6-8.9), potassium phosphate (pH 6.0-8.0), and MES-NaOH (pH 5.0-7.0). The dependence of ME activity on mono- and divalent cations was tested by using aqueous solutions of KCl, NH4Cl, NaCl (at a final concentration of 50 mM) and MgCl2, MnCl2, CoCl2 (1 mM). Fructose, glucose, glucose-6-phosphate, fructose-6-phosphate, fructose-1,6-bisphosphate (at concentration 5 mM), pyruvate, phosphoenolpyruvate, hydroxypyruvate, oxaloacetate, glycerate, α-ketoglutarate, isocitrate, citrate, serine, aspartate (1 mM), ATP, ADP, AMP, PPi (2 mM) were tested as potential inhibitors or activators of the decarboxylation reaction. To test effect of divalent metals on the enzyme activity, the aqueous solutions of CuCl2, RbCl2, CdCl2, NiCl2, SnCl2, CoCl2, BaCl2, ZnCl2, or CaCl2 at a final concentration of 1 mM were added in the standard reaction mixture containing 1 mM MnCl2 (instead of 2.5 mM MgCl2). To determine the thermal stability, aliquots of the concentrated enzyme in Eppendorf (Germany) tubes were incubated at 30, 40, 50, 60, and 70°C from 5 min to 3 h. After heating, an aliquot was diluted 50-fold by the cooled buffer, and residual activity was determined at 30°C. The percentage of residual activity was calculated by comparison with the non-incubated enzyme. To search for the optimal temperature for activity, the reaction velocity was measured at 10-70°C. For calculation of Km, the enzyme activity was measured by varying substrates in the concentration range of 0.156-10 mM (malate), 0.0047-0.25 mM (NADP+), 1.56-25 mM (pyruvate), and 0.0156-0.375 mM (NADPH). Km and Vmax values were calculated using SigmaPlot (v.10). Protein concentrations were assayed by a modification of the Lowry method [16]. NADPH oxidation/formation rates were followed at 340 nm on a Shimadzu UV-1700 spectrophotometer (Japan).
RESULTS
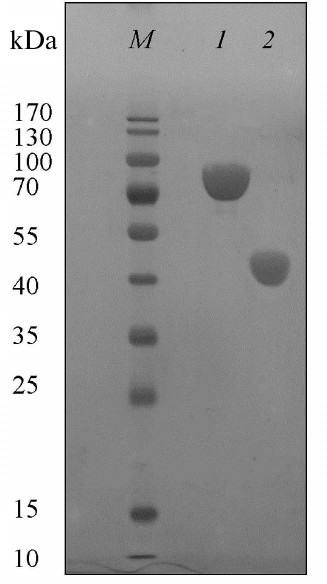
Preparation and purification of the malic enzyme. The dme gene encoding putative malic enzyme from the Ms. trichosporium genome was expressed in E. coli BL21(DE3). The recombinant protein was purified from the E. coli cell extract by single-stage metal chelating chromatography. SDS-PAGE of the protein revealed a single band corresponding to a molecular mass of ~80 kDa, which corresponded with the theoretically calculated (81.5 kDa) (Fig. 2). According to electrophoresis under non-denaturing conditions, the protein mass is 480 kDa, which indicates its hexameric organization. Earlier, dimeric, tetrameric, and octameric forms have been detected in microbial malic enzymes. NADP+-ME from E. coli, as well as NADP+-ME and NAD(P)+-ME from Sinorhizobium meliloti, were found to be homooctamers [17, 18], while NADP+-ME from Bradyrhizobium japonicum was a dimer or tetramer depending on pH values (pH 8.0 and 7.2, respectively) [19].
Fig. 2. SDS-PAGE of Ms. trichosporium malic enzyme (1) and ME1 (N-end fragment of Ms. trichosporium ME) (2). M, markers of molecular masses.
Catalytic properties. The malic enzyme catalyzed the decarboxylation of malate to pyruvate using NADP+ as a cofactor. It did not use NAD+ and did not show activity of oxaloacetate decarboxylation. The enzyme activity was strongly dependent on both mono- and divalent cations: K+ (or NH4+) and Mn2+ (or Mg2+) (Table 1). At the same time, in the presence of 50 mM KCl, 50 mM NaCl inhibited its activity by 40%, and KHCO3, but not NaHCO3, served as a CO2 donor for carboxylation of pyruvate. The enzyme was active in wide ranges of pH (pH 6.0-9.0) and temperature (20-70°C), demonstrating maximum of activity at pH 7.0 and 65°C. ME showed moderate thermal stability: the enzyme activity was not decreased after incubating the protein for 3 h at 30-40°C, but decreased by half after heating for 1 h at 50°C, and 80% activity was lost after 5 min heating at 60°C.
Table 1. Influence on K+,
NH4+, Na+ (50 mM), and divalent metals
(1 mM) on the activity of the recombinant malic enzyme from Ms.
trichosporium
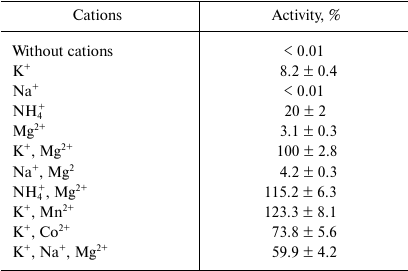
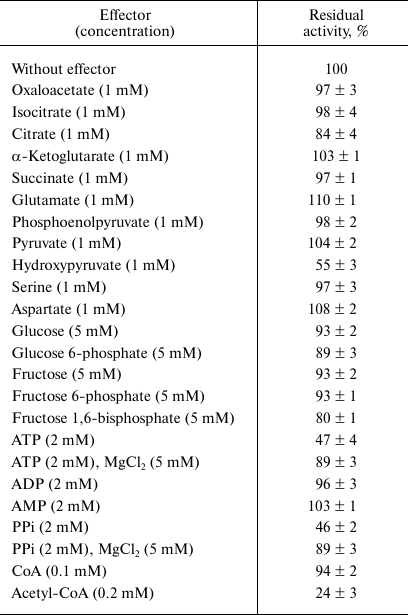
The dependence of the activity on the concentration of substrates obeyed Michaelis–Menten kinetics. At 30°C and optimal pH, apparent Km values were 2.7 ± 0.3 mM for malate, 64 ± 9 μM for NADP+, 6.0 ± 0.8 mM for pyruvate, and 47 ± 4 μM for NADPH. The enzyme demonstrated 4.7-fold higher activity in the direction of malate decarboxylation compared with pyruvate carboxylation (Table 2). As followed from the kcat/Km values, the efficiency of the enzyme in the decarboxylation reaction was an order of magnitude higher than that in the carboxylation reaction. Hydroxypyruvate at concentration of 1 mM inhibited activity of ME by 45% (Table 3). Acetyl-CoA (0.1 mM) exerted the greatest inhibitory effect, in the presence of which the residual activity was 24%. ATP and PPi at a concentration of 2 mM inhibited the enzyme by 50%, but the activity was completely recovered if Mg2+ concentration increased to 5 mM. In the presence of Mn2+, Cd2+ ions almost completely inhibited the activity (5.9% residual activity), and Sn2+ ions reduced activity by 31%. The other metals tested (see “Materials and Methods”) did not have a significant effect on the ME activity.
Table 2. Kinetic parameters of the Ms.
trichosporium malic enzyme and its N-terminal fragment
(ME1)
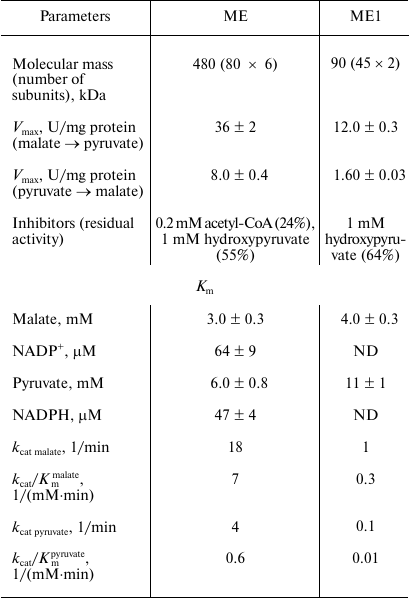
Note: ND, not determined.
Table 3. Activity of the Ms.
trichosporium malic enzyme in the presence of some metabolites
Effect of the C-terminal domain on activity of the malic enzyme. In the amino acid sequence of Ms. trichosporium ME, the N-terminal fragment consisting of 437 a.a. homologous to malic enzymes (ME1) and an extended C-terminal sequence of 322 a.a. corresponding to phosphoacetyltransferases (EC 2.3.1.8) were found. A similar domain structure has been previously described for NADP+-dependent malic enzymes from bacteria, such as E. coli and Sinorhizobium meliloti, as well as from plants [17, 18, 20].
By cloning of the DNA sequences, two separate proteins, ME1 and the putative phosphoacetyltransferase, were obtained and purified. The specific activities of ME1 in decarboxylation and carboxylation directions were 12 and 1.6 U/mg protein, respectively. At the same time, the apparent Km values for carbon substrates were higher for ME1 as compared to the initial two-domain protein (Table 2). Unlike the full-length protein, acetyl-CoA did not affect the ME1 activity, while the inhibiting effect of hydroxypyruvate was maintained. Neither the complete chimeric protein nor the phosphoacetyltransferase domain transfer acetyl groups and formation of CoA-SH from acetyl-CoA.
Protein ME1 had a molecular weight of 90 kDa, which corresponded to a dimeric structure, whereas the phosphoacetyltransferase fragment was a hexamer with a molecular mass of 210 kDa. Obviously, the C-terminal 322 a.a. fragment was responsible for optimal oligomerization of the malic enzyme. A similar assumption has been made for two malic enzymes from Sinorhizobium meliloti [17]. Phosphoacetyltransferase activity was not detected for any of the studied chimeric malic enzymes.
DISCUSSION
In this work, we first described the malic enzyme from the obligate methanotroph Ms. trichosporium. The enzyme belongs to the group of NADP+-dependent malic enzymes. Both divalent metal ions (Mn2+ or Mg2+) and monovalent cations (K+ or NH4+) were necessary for the enzyme activity, whereas Na+ ions had an inhibitory effect. Activation by NH4+ cations has been found to be a characteristic feature of NADP+-dependent malic enzymes (for example, ME from Clostridium thermocellum) [21] and of the NAD+-dependent ones (from Streptococcus bovis) [22]. However, K+ ions had only a small stimulating effect on the NAD+-ME from S. bovis. Evidences on the Na+ inhibition on the malic enzyme has not been reported in the literature.
Like other NADP+ enzymes, ME from Ms. trichosporium catalyzed a reversible reaction, while the activity of pyruvate carboxylation was significantly lower than malate decarboxylation. In addition, the enzyme from Ms. trichosporium had a high Km for pyruvate (~6 mM) relative to its physiological intracellular concentrations. Although the pyruvate content in cells of this methanotroph can reach high values (>1 mM [23]), such concentration still could not support effective CO2 assimilation. High values of apparent Km for pyruvate have been found for NADP+-ME from E. coli and Corynebacterium glutamicum (6.21 and 13.8 mM, respectively) [18, 24]. Interestingly, only the enzyme from the hyperthermophilic archaea Thermococcus kodakaraensis had lower Km to pyruvate than to malate (7.3 and 16.9 mM, respectively) [5], although the catalytic efficiencies of the decarboxylation and carboxylation reactions were almost the same. In addition, similarly to NADP+-ME from Clostridium thermocellum [21] and Thermococcus kodakaraensis [5], ME from Ms. trichosporium could not decarboxylate oxaloacetate.
ME activity from Ms. trichosporium was inhibited by intermediates of the carbon assimilation pathway – hydroxypyruvate and acetyl-CoA. Typically, the activities of bacterial malic enzymes were affected by the TCA intermediates. Malate, succinate, and fumarate activated, but acetyl-CoA inhibited the NAD(P)+-dependent enzyme from Sinorhizobium meliloti, while NADP+-ME did not undergo any regulation [25]. NADP+-ME activity from E. coli was inhibited by fumarate, oxaloacetate, and acetyl-CoA, whereas glutamate, aspartate, glucose-6-phosphate, and acetyl phosphate activated this enzyme [18].
Malic enzyme from Ms. trichosporium consisted of two fragments. The N-terminal polypeptide obtained as independent protein had low activity and was not inhibited in the presence of acetyl-CoA. Since the diminished version of the malic enzyme (in the absence of the C-terminal fragment) was a dimer, but not a hexamer, unlike the full-length protein, the role of the C-terminal sequence in the optimal enzyme configuration was suggested. Considering the absence of inhibition, the “chimeric” NADP+-dependent malic enzyme from Sinorhizobium meliloti by acetyl-CoA, the impact of the additional fragment in interaction of effectors with the enzyme requires further study.
According to the kinetic properties, the malic enzyme from Ms. trichosporium catalyzed malate decarboxylation more efficiently than pyruvate carboxylation; therefore, it can be considered as a “lipogenic” enzyme that produces NADPH, which is necessary for the synthesis of fatty acids and steroids. Such a function was proposed for a number of bacterial NADP+-ME [1, 2, 26].
In Ms. trichosporium, malate is formed by carboxylation of phosphoenolpyruvate (PEP) by highly active PEP carboxylase [8] followed by reduction of the oxaloacetate by NADH-dependent malate dehydrogenase [7]. The subsequent decarboxylation of malate by the malic enzyme leads to the synthesis of NADPH (Fig. 1). Consequently, of the activity of the three listed enzymes, PEP is transformed into pyruvate, which is accompanied by the consumption of NADH and the formation of NADPH:
PEP + NADH + NADP+ → Pyruvate + NAD+ + NADPH.
In the case of the functioning of pyruvate, phosphate dikinase (ID 2507410009), PEP can be regenerated with the consumption of an ATP molecule and formation of PPi and AMP in a total reaction of the futile cycle:
NADH + NADP+ + ATP + Pi → NADPH + NAD+ + AMP + PPi.
The substitution of ATP for PPi in this cycle correlates with an important metabolic role of PPi in methanotrophs [27]. Methylosinus trichosporium can synthesize NADPH by the NADP+-dependent isocitrate dehydrogenase [8]. However, the function of the malic enzyme as a source of NADPH does not seem to be redundant, since this methanotroph has high requirements for NADPH, which is necessary for the synthesis of fatty acids and the formation of a system of intracytoplasmic membranes, where methane is oxidized by the particulate (membranous) methane monooxygenase.
Funding
This work was carried out with financial support of the Russian Science Foundation (project no. 18-14-00326).
Acknowledgements
The authors are thankful to Evrogen (Russia) for synthesis of the oligonucleotides.
Conflict of Interest
The authors declare no conflict of interest.
Ethical Approval
This article does not include research carried out by the authors with the participation of people or animals.
REFERENCES
1.Chang, G. G., and Tong, L. (2003) Structure and
function of malic enzymes, a new class of oxidative decarboxylases,
Biochemistry, 42, 12721-12733.
2.Sauer, U., and Eikmanns, B. J. (2005) The
PEP–pyruvate–oxaloacetate node as the switch point for
carbon flux distribution in bacteria, FEMS Microbiol. Rev.,
29, 765-794.
3.Viljoen, M., Subden, R. E., Krizus, A., and Van
Vuuren, H. J. (1994) Molecular analysis of the malic enzyme gene
(mae2) of Schizosaccharomyces pombe, Yeast,
10, 613-624.
4.Liguori, M., Tessarolo, D., Abbruzzese, C., and
Giacanelli, M. (1995) NAD+/NADP+-dependent malic
enzyme: evidence of a NADP+ preferring activity in human
skeletal muscle, Biochem. Mol. Med., 56, 14-18.
5.Fukuda, W., Ismail, Y. S., Fukui, T., Atomi, H.,
and Imanaka, T. (2005) Characterization of an archaeal malic enzyme
from the hyperthermophilic archaeon Thermococcus kodakaraensis
KOD1, Archaea, 1, 293-301.
6.Trotsenko, Y. A., and Murrell, J. C. (2008)
Metabolic aspects of aerobic obligate methanotrophy, Adv. Appl.
Microbiol., 63, 183-229.
7.Rozova, O. N., Khmelenina, V. N., Bocharova, K. A.,
Mustakhimov, I. I., and Trotsenko, Y. A. (2015) Role of
NAD+-dependent malate dehydrogenase in the metabolism of
Methylomicrobium alcaliphilum 20Z and Methylosinus
trichosporium OB3b, Microorganisms, 3, 47-59.
8.Matsen, J. B., Yang, S., Stein, L. Y., Beck, D.,
and Kalyuzhnaya, M. G. (2013) Global molecular analyses of methane
metabolism in methanotrophic Alphaproteobacterium, Methylosinus
trichosporium OB3b. Part I. Transcriptomic study, Front.
Microbiol., 7, 1-13.
9.Galchenko, V. F. (2001) Methanotrophic
Bacteria [in Russian], GEOS, Moscow.
10.Sambrook, J., and Russell, D. W. (2001)
Molecular Cloning: A Laboratory Manual, 3rd Edn., Cold Spring
Harbor Laboratory, New York.
11.Kalyuzhnaya, M., Khmelenina, V. N., Kotelnikova,
S., Holmquist, L., Pedersen, K., and Trotsenko, Y. A. (1999)
Methylomonas scandinavica sp. nov., a new methanotrophic
psychrotrophic bacterium isolated from deep igneous rock ground water
of Sweden, Syst. Appl. Microbiol., 22, 565-572.
12.Reshetnikov, A. S., Rozova, O. N., Khmelenina, V.
N., Mustakhimov, I. I., Beschastny, A. P., Murrell, J. C., and
Trotsenko, Y. A. (2008) Characterization of the pyrophosphate-dependent
6-phosphofructokinase from Methylococcus capsulatus Bath,
FEMS Microbiol. Lett., 288, 202-210.
13.Slater, G. G. (1969) Stable pattern formation and
determination of molecular size by pore-limit electrophoresis, Anal.
Chem., 41, 1039-1041.
14.Sender, P. D., Martin, M. G., Peiru, S., and
Magni, C. (2004) Characterization of an oxaloacetate decarboxylase that
belongs to the malic enzyme family, FEBS Lett., 570,
217-222.
15.Bock, A. K., Glasemacher, J., Schmidt, R., and
Schonheit, P. (1999) Purification and characterization of two extremely
thermostable enzymes, phosphate acetyltransferase and acetate kinase,
from the hyperthermophilic eubacterium Thermotoga maritima,
J. Bacteriol., 181, 1861-1867.
16.Shacterle, G. R., and Pollack, R. L. (1973) A
simplified method for quantitative assay of small amounts of protein in
biological material, Anal. Biochem., 51, 654-657.
17.Mitsch, M. J., Voegele, R. T., Cowie, A.,
Osteras, M., and Finan, T. M. (1998) Chimeric structure of the
NAD(P)+- and NADP+-dependent malic enzymes of
Rhizobium (Sinorhizobium) meliloti, J. Biol. Chem.,
273, 9330-9336.
18.Bologna, F. P., Andreo, C. S., and Drincovich, M.
F. (2007) Escherichia coli malic enzymes: two isoforms with
substantial differences in kinetic properties, metabolic regulation,
and structure, J. Bact., 189, 5937-5946.
19.Chen, F., Okabe, Y., Osano, K., and Tajima, S.
(1997) Purification and characterization of the NADP-malic enzyme from
Bradyrhizobium japonicum A1017, Biosc. Biotech. Biochem.,
61, 384-386.
20.Troconi, M. A., Andreo, C. S., and Drincovich, M.
F. (2018) Chimeric structure of plant malic enzyme family: different
evolutionary scenarios for NAD- and NADP-dependent isoforms, Front.
Plant Sci., 9, 1-15.
21.Taillefer, M., Rydzak, T., Levin, D. B., Oresnik,
I. J., and Sparling, R. (2015) Reassessment of the
transhydrogenase/malate shunt pathway in Clostridium
thermocellum ATCC 27405 through kinetic characterization of malic
enzyme and malate dehydrogenase, Appl. Environ. Microbiol.,
81, 2423-2432.
22.Kawai, S., Suzuki, H., Yamamoto, K., Inui, M.,
Yukawa, H., and Kumagai, H. (1996) Purification and characterization of
a malic enzyme from the ruminal bacterium Streptococcus bovis
ATCC 15352 and cloning and sequencing of its gene, Appl. Environ.
Microbiol., 62, 2692-2700.
23.Yang, S., Matsen, J. B., Konopka, M.,
Green-Saxena, A., Clubb, J., Sadilek, M., Orphan, V. J., Beck, D., and
Kalyuzhnaya, M. G. (2013) Global molecular analyses of methane
metabolism in methanotrophic Alphaproteobacterium, Methylosinus
trichosporium OB3b. Part II. Metabolomics and
13C-labeling study, Front. Microbiol., 4,
70.
24.Gourdon, P., Baucher, M.-F., Lindley, N. D., and
Guyonvarch, A. (2000) Cloning of the malic enzyme gene from
Corynebacterium glutamicum and role of the enzyme in lactate
metabolism, Appl. Environ. Microbiol., 66, 2981-2987.
25.Driscollt, B. T., and Finan, T. M. (1997)
Properties of NAD+- and NADP+-dependent malic
enzymes of Rhizobium (Sinorhizobium) meliloti and differential
expression of their genes in nitrogen-fixing bacteroids,
Microbiology, 143, 489-498.
26.Ratledge, C. (2014) The role of malic enzyme as
the provider of NADPH in oleaginous microorganisms: a reappraisal and
unsolved problems, Biotechnol. Lett., 36, 1557-1568.
27.Khmelenina, V. N., Rozova, O. N., Akberdin, I.
R., Kalyuzhnaya, M. G., and Trotsenko, Y. A. (2018).
Pyrophosphate-dependent enzymes in methanotrophs: new findings and
views, in Methane Biocatalysis: Paving the Way to Sustainability
(Kalyuzhnaya, M. G., and Xing, X. H., eds.) Springer, International
Publishing AG, Switzerland, pp. 83-98.