REVIEW: Archaeal Flagella as Biotemplates for Nanomaterials with New Properties
S. N. Beznosov, M. G. Pyatibratov, and O. V. Fedorov*
Institute of Protein Research, Russian Academy of Sciences, 142290 Pushchino, Moscow Region; E-mail: fedorov@vega.protres.ru* To whom correspondence should be addressed.
Received May 30, 2017; Revision received July 6, 2017
At the end of 1980s, regions of the polypeptide chain of bacterial flagella subunits (flagellins) responsible for different properties of these protein polymers were identified by structural studies. It was found that the N- and C-terminal regions are responsible for the polymerization properties of subunits, and the central region is responsible for antigenic properties of the flagellum. Soon after that, it was proposed to use variability of the central flagellin domain for directed modification to impart new properties to the flagellum surface. Such studies of flagella and other polymeric structures of bacterial origin thrived. However bacterial polymers have some shortcomings, mainly their instability to dissociating effects. This shortcoming is absent in archaeal flagella. A limiting factor was the lack of the three-dimensional structure of archaeal flagellins. A method was developed that allowed modifying flagella of the halophilic archaeon Halobacterium salinarum in a peptide that connects positively charged ions. Later, corresponding procedures were used that allowed preparing the anode material for a lithium-ion battery whose characteristics 4-5-fold exceeded those of batteries commonly used in industrial production. We describe other advantages of archaeal flagella over bacterial analogs when used in nanotechnology.
KEY WORDS: Archaea, flagella, archaella, protein modification, biotemplated nanomaterialsDOI: 10.1134/S0006297918140067
In recent decades, the number of publications describing the use of various supramolecular structures of biological origin (such as viruses, flagella, pili) for nanotechnological purposes has been growing exponentially. This research field is recognized as very promising and is rapidly developing [1-7]. The use of biotemplated nanomaterials for medical applications is being actively discussed [8].
In such studies, biological structures are used as scaffolds for binding a given ligand. In the used supramolecular structures, their constituent protein molecules are modified with an additional peptide insert having a high affinity for the ligand. Thus, an ordered arrangement of this ligand on the surface of the biopolymer matrix can be achieved, and nanostructured materials having the required properties (conductive, magnetic, catalytic, sorption, etc.) can be obtained.
One of the most suitable structures for such modifications are prokaryotic flagella, and this was first discussed previously [9]. Flagella of prokaryotes are long helical filaments extending from the cell surface; they are composed of protein subunits called flagellins. Although archaeal flagella (archaella) look like the bacterial analogs, they are principally different in structure, assembly mechanisms, and evolutionary origin [10-12]. The main role of the flagellum is to provide motility of a cell in a liquid environment, but several cases of flagella playing a role as a sensor organelle sensitive to environmental changes and modulating intracellular processes have been published. Moreover, flagella have been shown to be involvement in adhesion [13-16].
Archaeal flagella might have several advantages over their bacterial analogs when used for nanotechnological purposes. On one hand, most archaea live in harsh environments; on the other hand, archaeal flagella have multicomponent structure, which might make it possible to obtain polyfunctional materials. The development of techniques for directed modification of archaeal flagellins [17, 18] and the recently published spatial structure of the methanogenic archaea Methanospirillum hungatei archaellum [19] open broad possibilities for directed modification of the archaellum surface and, therefore, generation of a new class of nanomaterials with various properties.
MODIFICATION OF BACTERIAL BIOPOLYMERS
In early studies it was shown that in flagella of various bacteria, the N- and C-terminal sequences of flagellins responsible for polymerization are rather conserved (even copolymerization of flagellins from different strains is possible), while the central parts of the flagellin polypeptide chains differ significantly in different bacteria and form the flagellar surface, being responsible for their antigenic properties [9, 20].
It was claimed that “it is possible to construct artificial supramolecular structures using one flagellin domain as a universal polymerization block and by varying the other domain by insertions and deletions” [9]. There is a possibility to modify the central part of the flagellin polypeptide chain through insertion of additional peptide loops, which will be exposed to the flagellar surface and will provide new properties to it, while the conservative N- and C-terminal sequences responsible for flagella polymerization will not be changed.
However, gene engineering methods were not sufficiently developed at that time to modify the surface of polymeric structures. Works aimed at generation of bacterial structures with given properties started to progress only 15 years later. The main idea of these works was to deliver to the surface of the biopolymer-composing subunits additional peptide loops that would be able to bind a specific ligand. Upon ligand binding by such matrices and, in some cases, after a set of additional chemical reactions, fibrous nanomaterials with ligand-specific properties (conducting, semiconducting, magnetic, catalytic, adsorbing, etc.) could be obtained.
Nanofibers based on bacterial flagella were obtained using this approach [4, 21]. A system based on the structural data for the bacterial flagellin from Escherichia coli was developed. Flagellin consists of five domains: D1-D5, where variable D2- and D3-domains are exposed to the surface and can be modified by point mutations, deletions, and insertions of peptide loops without affecting the function of the flagellin. So-called “FliTrx flagellin peptide display” consists of partial exchange of the D2- and D3-domains of E. coli flagellin with thioredoxin protein from the same organism. This protein is used to select the amino acid sequences generating loops exposed to the solvent. All manipulations are carried out in a plasmid bearing the sequence of the thioredoxin gene followed by the sequence encoding a peptide loop. This expression plasmid was used to transform E. coli strain with knockout of the endogenous flagellin gene. The resulting cells expressed the modified flagellin. Using this approach, different peptide sequences were inserted, e.g. Arg-X-X-Arg tetrapeptides that bind ions and metal oxides [22], histidine loops “His-loop” (-Gly-His-His-His-His-His-His-), and similar “Glu-Asp loop” (-Asp-Glu-Asp-Glu-Asp-Glu-Gly-) and “Arg-Lys loop” (-Arg-Lys-Arg-Lys-Arg-Lys-Arg-) that bind metal ions and provide anionic or cationic properties depending on composition [23]. In similar studies, peptide loops of different length (up to 19 a.a.) and structure were inserted directly into the central domain of the E. coli flagellin between residues Asp242 and Tyr245. Flagellin was modified with polyhistidine and polycysteine loops (or with the avidin tag (AviTag), which binds biotin (BioFliC) during synthesis and allows binding to many other ligands through other avidin fragments) and the peptide fragment bearing the TEV protease cleavage site. It was demonstrated (by the digestion of the modified flagellin with the corresponding protease as well) that these peptide loops are exposed on the flagellar surface and are suitable for decoration with different ligands [24, 25].
Moreover, long amino acid sequences binding large biological molecules were successfully inserted into bacterial flagellin. Thus, fibronectin-binding D-repeats from fibronectin-binding protein A (FnBPA) were inserted into the E. coli flagellin [26]. The sizes of peptide insertions were 39, 77, and 115 a.a., and in all cases the inserted peptides gave to flagella the ability to bind fibronectin. Similarly, the sequence of collagen-binding adhesin YadA from Yersinia enterocolitica was inserted. Collagen-binding peptides (from 42 to 302 a.a. long) were also inserted [26]. At the same time, the bacterial flagellum is a thoroughly studied structure, and researchers knew exactly where large peptides can be inserted without any deleterious effect on the structure of the flagella.
The most significant results showing the possibility of technological use of biopolymer-based nanomaterials were obtained by Angela Belcher’s group (Massachusetts Institute of Technology, USA). To produce functional nanofibers, they used genetically controlled filamentous bacteriophage M13 [1]. In the experiment, four glutamic acid residues were inserted into the N-terminal region of the major viral protein p8. The virus modified in this way was incubated with an aqueous solution of cobalt chloride, and after interaction with NaBH4 and subsequent spontaneous oxidation, they obtained nanowires of cobalt oxide (Co3O4). These nanowires were used to create test samples of anodes for lithium-ion batteries, which demonstrated higher performance [1]. The capacity of such anodes was 2-3 times higher than that of generally used carbon anodes. Using the same technique of mineralization of modified M13 virus, they obtained material for cathodes of lithium-ion batteries [7], as well as the catalytic material for fuel [27] and solar [28] cells. Amino acid sequences that would bind the required substance and form the nanomaterial naturally had such properties (tetraglutamate sequence binding positively charged metal ions) or were detected using the phage display technique (gold-binding motif LKAHLPPSRLPS [1], zinc sulfate-binding peptide CNNPMHQNC [29]).
Though most publications on the development of nanomaterials based on modified bacterial polymers deal with bacterial flagella and viruses, it is worth mentioning studies that used other bacterial polymers. Some of these polymers can be purposefully modified as, for example, pilins, or their composition can be changed as, for example, in DNA or RNA sequences. By polymerization of bacterial pilins in the presence of various chemical compounds, nanomaterials based on Pseudomonas aeruginosa pili were obtained [2, 30]. Nucleic acids are widely used to generate nanomaterials [31, 32]. By changing the nucleotide sequence, it is possible to create DNAs having required binding properties. Such DNA molecules will either bind certain molecules or will be able to organize themselves in ordered structures [33, 34]. Not only filamentous biopolymers can be used to generate nanomaterials, but any structures with nanosize organization (cell walls, organelles) [35]. Besides protein and nucleic acid polymers, sugar polymers can also be used, but in this case there is no possibility to modify specific binding properties of the polymer. Thus, the material for anodes for lithium-ion batteries with high capacity was obtained based on alginate from brown algae [36]. Biological lipids being amphiphilic, chemically diverse, and able to be organized in complex structures can also be tools for nanotechnology [37].
Although nanomaterials were generally obtained using bacteria-derived polymers, it should be mentioned that bacterial viruses and flagella are subject to dissociation at temperatures exceeding physiological ones and they poorly resist non-physiological salt concentrations and pH, which may impose certain restrictions in their use, especially for nanotechnology.
MODIFICATION OF ARCHAEAL FLAGELLINS
Earlier [38], we described in detail features of archaella that give them certain advantages over flagella and viruses of bacterial origin currently used in nanotechnology. The main advantages are as follows:
1) The flagella of many archaea have high resistance to dissociating effects [39].
2) Many archaella consist of several (usually, 2-5) types of subunits. The multicomponent composition makes it possible to specifically modify various flagellins to produce polyfunctional materials.
3) The flagella of some archaea (Halobacterium) are capable of ordered side-to-side association with each other, forming so-called “superflagella” [40]. In addition, flagella of some archaea possess adhesive properties to certain types of surfaces [14, 41], which is important for nanotechnological applications.
One of the reasons why archaella have not been used for nanotechnological purposes has been the lack of detailed data on their spatial structure. The first experiments with insertion of a peptide loop into archaeal flagellin were done in 2002 in Jarrell’s laboratory. To localize the minor flagellin FlaA in the flagellum of the methanogenic archaeon Methanococcus voltae, the hemagglutinin peptide (HA tag, YPYDVPDYA) was inserted into this flagellin. Localization of this flagellin was analyzed using commercial antibodies specific to the HA-peptide. The insertion was done in the central variable part of the flagellin between residues 132 and 133. Addition of hemagglutinin peptide to FlaA has no effect on cell motility or on assembly of the flagella. However, modified FlaA-flagellin of M. voltae was detected in the intact flagella only by immunoblotting, while immunoelectron microscopy was unsuccessful [42]. Apparently, the conformational state of the insert prevented its interaction with the antibodies. It was necessary to find more suitable places for the insertion, which would make it available for the corresponding ligands without interfering with the polymerization properties of the flagellin. Thus, another approach was required to identify the regions of the polypeptide chain exposed to the flagellar surface and suitable for the required insertions. This approach was found, experimentally proven, and shown to be suitable for the directed insertion of specific binding groups onto the surface of flagella of the halophilic archaea [38]. For the development of nanomaterials with new properties based on archaeal flagella, the halophilic archaeon Halobacterium salinarum was chosen. This archaeon was one of the first species for which methods of genetic engineering allowing manipulation of the archaeal genome were developed. The flagellum of H. salinarum consists of five flagellins (A1, A2, B1, B2, B3) encoded by genes of two operons, which gives great opportunities to modify the flagellar surface. The method of directed modification of H. salinarum flagellins with insertions of amino acid residues supposes that the inserts should not affect formation and functions of the flagellum but should be available for detection. Moreover, it becomes possible to obtain flagella-based nanomaterials by insertion of peptide loops that can bind a specific ligand. As there was no crystal structure of H. salinarum flagellins, other data were used to define the regions of the polypeptide chain of flagellar subunits exposed to the surface and suitable for insertion of modifying loops.
It is known that H. salinarum flagella undergo glycosylation and that glycosylation takes place on the cell’s exterior after the supramolecular structure of the flagellum is already assembled, i.e. on its surface [43]. Having analyzed the probable sites of N-glycosylation (NXT(S)), we found two closely located sites and inserted a FLAG peptide between them (Fig. 1).
Fig. 1. Amino acid sequence of A1-flagellin of H. salinarum. The glycosylation sites are highlighted in gray; amino acid residues 90 and 91 between which the FLAG peptide was inserted are underlined.
Modified flagella kept the polymeric structure, and the presence of the FLAG peptide on the surface of the flagella was detected using commercial anti-FLAG antibodies (Fig. 2).
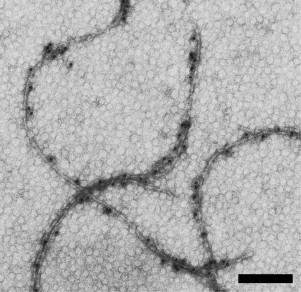
Fig. 2. Electron microscopic images of H. salinarum flagella: modified flagella with FLAG peptide inserted in A2-flagellin. Samples were labeled with specific antibodies and protein A conjugated to colloidal gold particles (10 nm) and negatively stained with uranyl acetate. The scale bar is 200 nm.
It should be mentioned that the region that we have chosen for the insertion modifying the flagellar surface is conserved in all five H. salinarum flagellins, i.e. it can be used to modify all of them. This can be used to improve the characteristics of a polymer by increasing the number of FLAG peptide insertions and to answer the fundamental question about the distribution of flagellins in the H. salinarum flagellum by insertion of the FLAG peptide in different subunits [17].
To demonstrate the acquired properties, modified H. salinarum flagella were used to obtain a new generation of electrode materials. For this, peptide loops binding nanosize metal oxides used as the anode material for lithium-ion batteries (LIB) were inserted on the surface of the flagella.
In the electrochemical literature, it is claimed that the use of nanosize materials with selective binding properties will make it possible to stabilize the bound nanoparticles and prevent their aggregation. It is assumed that the use of nanomaterials will provide the dynamic development of LIB, including improvement of their specific capacity, reliability, stability during cycling, and allow higher charging rate [44].
The same FLAG peptide was chosen as an insert for binding of anode material. This peptide contains five negatively charged aspartate residues, which allows it to bind positively charged metal ions. Three types of flagella bearing the FLAG-modified A1-, A2-, or B2-flagellins were obtained. The FLAG-modified flagella were then tested for binding of positively charged ions. When such flagella are incubated in the presence of metal ions, subsequent treatment with sodium borohydride results in flagella with metal oxide particles bound to their surface. Using this approach, H. salinarum flagella covered with cobalt oxide nanoparticles bound by FLAG peptide were obtained [45]. Thus, a method of mineralization of the surface of modified H. salinarum flagella with electrode materials was developed.
This sample, when tested as anode material, showed considerable improvement of electrochemical characteristics compared to traditional electrode materials. While the standard capacity of graphite generally used as the anode for LIB is within 200-250 mAh/g (at the theoretical limit of 372 mAh/g), the new anode material shows capacity more than 450 mAh/g [45].
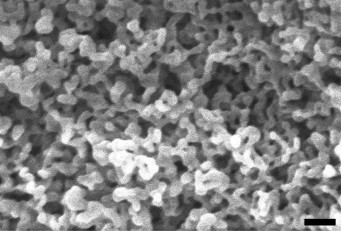
Later, H. salinarum flagella bearing FLAG-modified A1- and/or A2-flagellins and mineralized by iron oxide were obtained. These nanostructured materials showed increased electrochemical capacity (more than 1000 mAh/g), which is more than four times higher than the capacity of graphite used for industrial production of LIB [18]. A similar approach was used to obtain H. salinarum flagella mineralized with iron phosphate. The electron microscopic analysis of this material (Fig. 3) shows its large specific surface area and its percolating network, which makes it rather promising as an LIB cathode.
Fig. 3. Scanning electron micrographs of H. salinarum flagella modified with FLAG peptide mineralized with iron phosphate (done using scanning electron microscope JEOL). The scale bar is 100 nm.
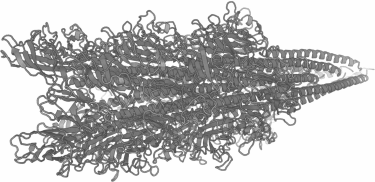
It is possible to increase the number of FLAG peptides exposed on the surface of flagella, and, consequently, to increase the number of binding sites for the electrode materials, since not all five flagellins forming the flagellum were modified. Moreover, other sites in flagellins suitable for insertions can be chosen. The recently solved spatial structure of the flagellum of the methanogenic archaeon Methanospirillum hungatei with 3.4 Å resolution [19] can also be used to predict probable structures of flagella of other archaea using methods of molecular modeling. The predicted structure of the flagellum from H. salinarum is shown on Fig. 4; this structure can be used to search for the surface-exposed regions of the flagellum suitable for additional inserts.
Fig. 4. A model of the structure of the H. salinarum flagella fragment generated for A2-flagellin using molecular modeling [46] with the structure of the Methanospirillum hungatei flagellin [19] as a template.
Apart from the FLAG peptide, loops with other amino acid sequences can be inserted into flagellins, providing other properties to the modified flagellum. For example, experiments with bacterial polymers showed that introduction of the LKAHLPPSRLPS sequence binding colloidal gold [1] or DMPRTTMSPPPR sequence binding carbon nanotubules [7] improves the electrochemical characteristics of electrode materials. Halobacterium salinarum flagella modified with the gold-binding peptide have also been obtained [17]. It is possible that better electrochemical characteristics will require a different ratio between the number of electrode material binding sites and the number and the location of inserts improving conductivity of the material.
Experiments aimed at the development of nanomaterials with new properties through modification of the surface of natural biopolymers have been further developed. For the first time, polymeric structures of an organism from another domain of life – Archaea – were successfully used as the biopolymer to be modified. As an example, the surface of flagella from the archaeon H. salinarum was modified by the insertion of a peptide able to reversibly bind metal ions. Flagella-modifying insertions for other nanotechnological purposes can be made. In the case of ligand binding, the required amino acid sequences can be determined using the well-known method of phage display [47].
With constant progress in determining of spatial structures of archaeal flagellins, the tasks of finding regions suitable for a modifying insert will become easier. However, it is already possible to recommend such regions in the primary structure of some halophilic archaea basing on homology to the region of the insertion found for H. salinarum.
Acknowledgments
This work was supported by the Russian Foundation for Basic Research (projects Nos. 12-04-92691-IND_a and 14-04-01604-a).
REFERENCES
1.Nam, K. T., Kim, D., Yoo, P. J., Chiang, C.,
Meethong, N., Hammond, P. T., Chiang, Y., and Belcher, A. M. (2006)
Virus-enabled synthesis and assembly of nanowires for lithium ion
battery electrodes, Science, 312, 885-888.
2.Audette, G. F., and Hazes, B. (2007) Development of
protein nanotubes from a multi-purpose biological structure, J.
Nanosci. Nanotechnol., 7, 2222-2229.
3.Gazit, E. (2007) Use of biomolecular templates for
the fabrication of metal nanowires, FEBS J., 274,
317-322.
4.Kumara, M. T., Tripp, B. C., and Muralidharan, S.
(2007) Self-assembly of metal nanoparticles and nanotubes on
bioengineered flagella scaffolds, Chem. Mater., 19,
2056-2064.
5.Yu, B., Giltner, C. L., Schaik, E. J., Bautista, D.
L., Hodges, R. S., Audette, G. F., Li, D. Y., and Irvin, R. T. (2007) A
novel biometallic interface: high affinity tip-associated binding by
pilin-derived protein nanotubes, J. Bionanosci., 1,
73-83.
6.Atabekov, J. G. (2008) Using viral structures as
nanotechnology instruments, Nanotechnologies in Russia,
3, 128-137.
7.Lee, Y. J., Yi, H., Kim, W., Kang, K., Yun, D. S.,
Strano, M. S., Ceder, G., and Belcher, A. M. (2009) Fabricating
genetically engineered high-power lithium ion batteries using multiple
virus genes, Science, 324, 1051-1055.
8.Ghosh, D., Lee, Y., Thomas, S., Kohli, A. G., Yun,
D. S., Belcher, A. M., and Kelly, K. A. (2012) M13-templated magnetic
nanoparticles for targeted in vivo imaging of prostate cancer,
Nat. Nanotechnol., 10, 677-682.
9.Fedorov, O. V., and Efimov, A. V. (1990) Flagellin
as an object for supramolecular engineering, Prot. Eng.,
3, 411-413.
10.Jarrell, K. F., Bayley, D. P., and Faguy, D. M.
(1993) Structure, molecular sequence analysis and genetics of the
flagella of the domain Archaea: comparison with bacterial flagella,
Curr. Top. Mol. Genet., 1, 15-31.
11.Fedorov, O. V., Pyatibratov, M. G., Kostyukova,
A. S., Osina, N. K., and Tarasov, V. Y. (1994) Protofilament as a
structural element of flagella of haloalkaliphilic archaebacteria,
Can. J. Microbiol., 40, 45-53.
12.Jarrell, K. F., and Albers, S. V. (2012) The
archaellum: an old motility structure with a new name, Trends
Microbiol., 20, 307-312.
13.Wang, Q., Suzuki, A., Mariconda, S., Porwollik,
S., and Harshey, R. M. (2005) Sensing wetness: a new role for the
bacterial flagellum, EMBO J., 24, 2034-2042.
14.Nather, D. J., Rachel, R., Wanner, G., and Wirth,
R. (2006) Flagella of Pyrococcus furiosus: multifunctional
organelles, made for swimming, adhesion to various surfaces, and
cell–cell contacts, J. Bacteriol., 188,
6915-6923.
15.Haiko, J., and Westerlund-Wikstrom, B. (2013) The
role of the bacterial flagellum in adhesion and virulence,
Biology, 2, 1242-1267.
16.Friedlander, R. S., Vogel, N., and Aizenberg, J.
(2015) Role of flagella in adhesion of Escherichia coli to
abiotic surfaces, Langmuir, 31, 6137-6144.
17.Beznosov, S. N., Pyatibratov, M. G., Veluri, P.
S., Mitra, S., and Fedorov, O. V. (2013) A way to identify archaellins
in Halobacterium salinarum archaella by FLAG tagging, Cent.
Eur. J. Biol., 8, 828-834.
18.Beznosov, S. N., Veluri, P. S., Pyatibratov, M.
G., Chatterjee, A., MacFarlane, D. R., Fedorov, O. V., and Mitra, S.
(2015) Flagellar filament bio-templated inorganic oxide
materials – towards an efficient lithium battery anode,
Sci. Rep., 5, 7736.
19.Poweleit, N., Ge, P., Nguyen, H. H., Loo, R. R.
O., Gunsalus, R. P., and Zhou, Z. H. (2016) CryoEM structure of the
Methanospirillum hungatei archaellum reveals structural features
distinct from the bacterial flagellum and type IV pilus, Nat.
Microbiol., 2, 16222.
20.Fedorov, O. V., Kostyukova, A. S., and
Pyatibratov, M. G. (1988) Architectonics of a bacterial flagellin
filament subunit, FEBS Lett., 241, 145-148.
21.Kumara, M. T., Tripp, B. C., and Muralidharan, S.
(2007) Layer-by-layer assembly of bioengineered flagella protein
nanotubes, Biomacromolecules, 8, 3718-3722.
22.Thai, C. K., Dai, H., Sastry, M. S., Sarikaya,
M., Schwartz, D. T., and Baneyx, F. (2004) Identification and
characterization of Cu2O- and ZnO-binding polypeptides by
Escherichia coli cell surface display: toward an understanding
of metal oxide binding, Biotechnol. Bioeng., 87,
129-137.
23.Kumara, M. T., Srividya, N., Muralidharan, S.,
and Tripp, B. C. (2006) Bioengineered flagella protein nanotubes with
cysteine loops: self-assembly and manipulation in an optical
trap, Nano Lett., 6, 2121-2129.
24.Woods, R. D., Takahashi, N., Aslam, A., Pleass,
R. J., Aizawa, S., and Sockett, R. E. (2007) Bifunctional nanotube
scaffolds for diverse ligands are purified simply from Escherichia
coli strains coexpressing two functionalized flagellar genes,
Nano Lett., 7, 1809-1816.
25.Deplanche, K., Woods, R. D., Mikheenko, I. P.,
Sockett, R. E., and Macaskie, L. E. (2008) Manufacture of stable
palladium and gold nanoparticles on native and genetically engineered
flagella scaffolds, Biotechnol. Bioeng., 101,
873-880.
26.Westerlund-Wikstrom, B., Tanskanen, J., Virkola,
R., Hacker, J., Lindberg, M., Skurnik, M., and Korhonen, T. K. (1997)
Functional expression of adhesive peptides as fusions to Escherichia
coli flagellin, Prot. Eng., 10, 1319-1326.
27.Lee, Y., Kim, J., Yun, D. S., Nam, Y. S.,
Shao-Horn, Y., and Belcher, A. M. (2012) Virus-templated Au and
Au–Pt core–shell nanowires and their electrocatalytic
activities for fuel cell applications, Energy Environ. Sci.,
5, 8328-8334.
28.Chen, P.-Y., Ladewski, R., Miller, R., Dang, X.,
Qi, J., Liau, F., Belcher, A. M., and Hammond, P. T. (2013)
Layer-by-layer assembled porous photoanodes for efficient electron
collection in dye-sensitized solar cells, J. Mater. Chem.
A, 1, 2217-2224.
29.Mao, C., Solis, D. J., Reiss, B. D., Kottmann, S.
T., Sweeney, R. Y., Hayhurst, A., Georgiou, G., Iverson, B., and
Belcher, A. M. (2004) Virus-based toolkit for the directed synthesis of
magnetic and semiconducting nanowires, Science, 303,
213-217.
30.Petrov, A., Lombardo, S., and Audette, G. F.
(2013) Fibril-mediated oligomerization of pilin-derived protein
nanotubes, J. Nanobiotechnol., 11, 24.
31.Rothemund, P. W. K. (2006) Folding DNA to create
nanoscale shapes and patterns, Nature, 440, 297-302.
32.Peng, L., Wu, C., You, M., Han, D., Chen, Y., Fu,
T., Ye, M., and Tan, W. (2013) Engineering and applications of
DNA-grafting polymer materials, Chem. Sci., 4,
1928-1938.
33.Yevdokimov, Y. M., and Sytchev, V. V. (2007)
Nanotechnology and nucleic acids, Open Nanosci. J., 1,
19-31.
34.Xiong, X., Wu, C., Zhou, C., Zhu, G., Chen, Z.,
and Tan, W. (2013) Responsive DNA-based hydrogels and their
applications, Macromol. Rapid. Commun., 34,
1271-1283.
35.Sleytr, U. B., Pum, D., and Sara, M. (1997)
Advances in S-layer nanotechnology and biomimetics, Adv.
Biophys., 34, 71-79.
36.Kovalenko, I., Zdyrko, B., Magasinski, A.,
Hertzberg, B., Milicev, Z., Burtovyy, R., Luzinov, I., and Yushin, G.
(2011) A major constituent of brown algae for use in high-capacity
Li-ion batteries, Science, 334, 75-79.
37.Mashaghi, S., Jadidi, T., Koenderink, G., and
Mashaghi, A. (2013) Lipid nanotechnology, Int. J. Mol. Sci.,
14, 4242-4282.
38.Beznosov, S. N., Pyatibratov, M. G., and Fedorov,
O. V. (2009) Archaeal flagella as matrices for new nanomaterials,
Nanotechnologies in Russia, 4, 373-378.
39.Tarasov, V. Yu., Kostyukova, A. S., Tiktopulo, E.
I., Pyatibratov, M. G., and Fedorov, O. V. (1995) Unfolding of tertiary
structure of Halobacterium halobium flagellins does not result
in flagella destruction, J. Prot. Chem., 14, 27-31.
40.Alam, M., and Oesterhelt, D. (1984) Morphology,
function and isolation of halobacterial flagella, J. Mol. Biol.,
176, 459-475.
41.Jarrell, K. F., Stark, M., Nair, D. B., and
Chong, J. P. (2011) Flagella and pili are both necessary for efficient
attachment of Methanococcus maripaludis to surfaces, FEMS
Microbiol. Lett., 319, 44-50.
42.Bardy, S. L., Mori, T., Komoriya, K., Aizawa, S., and Jarrell,
K. F. (2002) Identification and localization of flagellins FlaA and
FlaB3 within flagella of Methanococcus voltae, J.
Bacteriol., 184, 5223-5233.
43.Sumper, M. (1987) Halobacterial glycoprotein
biosynthesis, Biochem. Biophys. Acta, 906, 69-79.
44.Yaroslavtsev, A. B., Kulova, T. L., and Skundin,
A. M. (2015) Electrode nanomaterials for lithium-ion batteries,
Russ. Chem. Rev., 84, 826-852.
45.Beznosov, S. N., Pyatibratov, M. G., Fedorov, O.
V., Kulova, T. L., and Skundin, A. M. (2011) Electrochemical properties
of nanostructured material based on modified flagella of halophilic
archaea Halobacterium salinarum for negative electrode of
lithium-ion battery, Nanotechnologies in Russia, 6,
705-710.
46.Biasini, M., Bienert, S., Waterhouse, A., Arnold,
K., Studer, G., Schmidt, T., Kiefer, F., Cassarino, T. G., Bertoni, M.,
Bordoli, L., and Schwede, T. (2014) SWISS-MODEL: modelling protein
tertiary and quaternary structure using evolutionary information,
Nucleic Acids Res., 42, W252-W258.
47.Wu, C. H., Liu, I. J., Lu, R. M., and Wu, H. C.
(2016) Advancement and applications of peptide phage display technology
in biomedical science, J. Biomed. Sci., 23, 8.