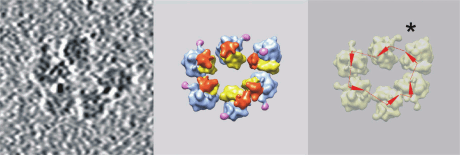REVIEW: Three-Dimensional Organization of Polyribosomes – A Modern Approach
Z. A. Afonina and V. A. Shirokov*
Institute of Protein Research, Russian Academy of Sciences, 142290 Pushchino, Moscow Region, Russia; E-mail: shirokov@vega.protres.ru* To whom correspondence should be addressed.
Received June 28, 2017; Revision received August 10, 2017
Polyribosomes in cells usually have a certain structural organization whose significance has not yet been elucidated. The development of cryo electron tomography has provided a new approach to study polyribosome structure. New data confirm or correct observations made earlier by classical techniques of electron microscopy. The existence of circular and linear (zigzag) topology of polyribosomes was confirmed, and their relationship with the frequently observed two-row forms was clarified. Contacts between ribosomes have been identified in densely packed three-dimensional helical polyribosomes. At the same time, modern cell-free translation systems have opened the possibility of investigating polyribosomes on mRNA of a given structure to elucidate the mechanism of polyribosome structure formation, especially of circular polyribosomes. There is an increasing amount of data supporting the idea of interdependence between polyribosome structure and their translational activity. Moreover, participation of polyribosomes in mRNA transport and localization of protein synthesis in the cell has been shown. Improvement of the resolution and the development of the cryo electron tomography technique for the analysis of polyribosomes in situ will enable further progress in understanding the process of protein synthesis in cells.
KEY WORDS: cryo electron tomography, polyribosome structure, circular polyribosomes, helical polyribosomesDOI: 10.1134/S0006297918140055
Abbreviations: cryo-EM, cryo electron microscopy; cryo-ET, cryo electron tomography; eIF, eukaryotic initiation factor; EM, electron microscopy; nt, nucleotide; PABP, poly(A)-binding protein; UTR, untranslated region of mRNA.
A polyribosome (polysome) is a group of several ribosomes bound to one
mRNA molecule and translating it in one direction [1-6]. The classical scheme of the
polyribosome depicts it in the form of “beads” of ribosomes
on a “thread” of mRNA. However, already the earliest
electron microscopy (EM) studies showed that ribosomes in a polysome
are specifically ordered. Visualization of polyribosomes by classical
EM techniques (ultrathin cell sections, negative and positive staining,
metal shadowing) revealed eukaryotic polyribosomes as double rows,
helices, and circles [7-14]
(Fig. 1).
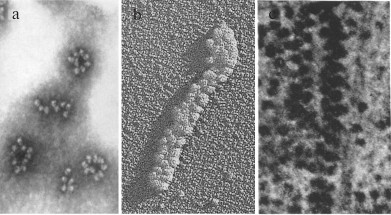
Fig. 1. Different forms of eukaryotic polyribosomes visualized by classical EM (adapted with some changes from several sources [8, 20, 26]). a) Negatively stained sample of polyribosomes purified from mouse tumor cells [8]. b) Platinum shadowed sample of polyribosomes formed in a wheat germ cell-free translation system [20]. c) Section of the cultured cell [26].
Ring-shaped polyribosomes, first described by Rich and coauthors in 1963 [1], have always been of particular interest to researchers. This form was shown both for free cytoplasmic and membrane-bound polyribosomes in eukaryotes [8-11]. It is considered that 5′- and 3′-terminal regions of mRNA in circular polyribosomes are kept close to each other through the interaction of proteins bound to the 5′- and 3′-untranslated regions (UTR), ensuring more efficient functioning of the polyribosome because of the reinitiation of ribosomes on the same mRNA after translation termination [15-17]. This model of the process of circular translation in eukaryotic polyribosomes, called the “closed-loop model”, is widely accepted today [16]. The interaction of mRNA termini through the complex of eIF4E–eIF4G–PABP proteins binding 5′-terminal cap and 3′ poly(A)-tail was demonstrated by atomic force microscopy of mRNA–protein complexes not bound to ribosomes [18].
Another form of polyribosomes often detected by traditional EM is a double-row. Double rows of polysomal ribosomes were often interpreted as collapsed circles with topologically circular (noncovalently closed) mRNA whose 5′- and 3′-ends were located close to each other on one of the extremities of a polyribosome [12, 19, 20]. However, 2-D EM images left open the possibility of an alternative linear configuration of some double-row polyribosomes, with a zigzag path of mRNA (mRNA termini located at opposite ends of a polyribosome). Such a configuration was well traced in double-row linear polyribosomes of prokaryotes [21].
One more form of polyribosome organization, revealed by traditional EM methods in cells under specific or suboptimal growth conditions, is long densely packed polyribosomes [22-25]. Such polyribosomes were often interpreted as 3-dimentional helices. In the model [26] based on EM images of polyribosomes of different length oriented at different angles in ultrathin cell sections, the polyribosome was represented as a left-handed helix with four ribosomes per turn; this model was confirmed recently with cryo-EM. The assumption of low translational activity of this type of polyribosomes was expressed by a number of authors [25, 27], who observed the accumulation of helical polyribosomes during viral infection in eukaryotic cells, accompanying a decrease in the rate of protein synthesis and the accumulation of viral particles [25]. Thus, the main ideas about the spatial organization of eukaryotic polyribosomes and their possible functional role were formulated in early studies.
It should be noted, however, that fixation, dehydration, staining, and drying of a sample when preparing the polyribosome specimen on an EM-grid can lead to distortion of the real shape of polyribosomes. In recent decades, a rapidly evolving method of cryo electron microscopy (cryo-EM) has been widely used for structural studies of the translation machinery of pro- and eukaryotes. Sample preparation for cryo-EM omits steps of drying and staining; on the contrary, macromolecules in a buffer with suitable salt composition are flash frozen and analyzed in a layer of amorphous ice. Localization of mRNA, tRNAs, and translation factors on a ribosome is studied by a single-particle analysis approach, now allowing near atomic resolution. The structure of such complexes is solved by assembling in 3-D space a large number (hundreds of thousands) of 2-D cryo-EM images of identical complexes registered in different projections. Another approach in cryo-EM – cryo electron tomography (cryo-ET) – is used for analysis of macromolecular complexes having the unique structures, e.g. polyribosomes. Cryo-ET is now the single method allowing determination of the structure of polyribosomes with resolution of 25-50 Å in a close-to-native environment [28-37]. With such resolution, the orientation of individual ribosomes in a polysome can be determined, which makes it possible to trace the path of the mRNA in polyribosomes and, thereby determine their topology.
CRYO ELECTRON TOMOGRAPHY OF POLYRIBOSOMES
Polyribosomes were studied by cryo electron tomography (cryo-ET) in samples obtained from a cell-free translation system (in vitro) [28, 30-33], in preparations purified from cells [28, 34], in cell sections [36], or even in the whole cells (in situ) [29]. For the analysis, a specimen of polyribosomes is flash frozen at ultralow temperature, and its images are acquired in the cryo electron microscope at different tilt angles ranging from –70° to +70°. Series of projections are then used to reconstruct a 3-D density (tomogram) [38]. Cryo electron tomography of polyribosomes is often used in combination with subtomogram analysis – a set of data-processing methods necessary to extract from tomograms information about localization and orientation of ribosomes. If polyribosomes are studied in vivo, surrounded by other macromolecules, visual identification of ribosomes in a tomogram can be complicated. In this case, a template-matching method [39, 40] is used, which finds a correlation between the known structure of a ribosome and the ribosome density in a tomogram [41].
Because biological macromolecules are very sensitive to irradiation by electrons, the cumulative electron dose used for acquisition of a tilt series usually should not exceed 60-100 e/Å2. The use of such a low dose of electrons results in a rather low signal from macromolecules in a tomogram. Subtomogram averaging [42] can be used to increase the signal-to-noise ratio and, as a result, the resolution. Averaging of subtomograms containing individual polysomal ribosomes is done either using an external model (a known ribosome structure), or without a model using a maximum likelihood algorithm [42, 43]. The resulting averaged ribosome (with 25-50 Å resolution) is fitted into the initial tomogram according to the rotation and shift parameters found during the averaging process for each of polysomal ribosome, which allows to interpret the structure of polyribosomes. Knowing the arrangement and mutual orientation of polysomal ribosomes, as well as the mRNA entrance and exit sites in a ribosome [44, 45], it is possible to determine the putative path of an mRNA chain in each polyribosome. Such an analysis allows a conclusion about the linear or circular topology of mRNA in a polysome.
STRUCTURE OF PROCARYOTIC POLYRIBOSOMES
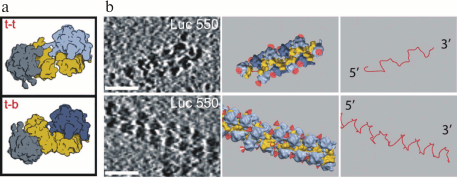
For the first time cryo-ET analysis of the polyribosome structure was applied to the Escherichia coli polysomes formed in vivo and in vitro [28]. In a cell-free translation system, mRNA coding for full-length or shortened luciferase without the stop codon was used. On such mRNAs, long polyribosomes visible on 2-D images as double-row structures were formed. The cryo-ET analysis of polyribosomes with subsequent subtomogram averaging showed that polyribosomes generally have either “sinusoidal” (zigzag) or 3-D helical path of the mRNA chain (Fig. 2). The ribosomes are mainly contacting each other through their small subunits. In case of the “sinusoidal” mRNA path, an alternation of two types of contacts between 40S subunits can be observed – the “top-to-top” and the “top-to-bottom” (Fig. 2a). In the case of the 3-D-helical path of the mRNA, only one type of contacts – the “top-to-top” – is observed. The structural analysis of polyribosomes from E. coli spheroplasts revealed inter-ribosomal contacts similar to those described above for polyribosomes from the cell-free translation system. Thus, in studied prokaryotic systems polyribosomes always had linear topology of mRNA. These data are consistent with the fact that the 5′- and 3′-ends of mRNA in prokaryotes are spatially separated: while on the 5′-end of mRNA the formation of polyribosomes is taking place, its 3′-end is still being synthesized by the RNA-polymerase [46, 47].
Fig. 2. Structural organization of prokaryotic polyribosomes according to cryo-ET data. a) Types of inter-ribosomal contacts (t-t, top-to-top; t-b, top-to-bottom). b) Examples of planar zigzag (upper row) and 3-D helical polyribosome (lower row); left panel, tomogram sections (scale bar, 50 nm); middle panel, structure of polysomes (the 40S subunit is shown in yellow, the 60S in blue, red color designates peptide exit site from the ribosomal tunnel); right panel, schematic representation of putative mRNA path in corresponding polyribosomes. Adapted with changes from the work [28].
STRUCTURE OF EUKARYOTIC POLYRIBOSOMES
The structural organization of eukaryotic polyribosomes was studied by cryo-ET in situ (in a cell) [29], in preparations of polyribosomes purified from cells [34], and in samples of polyribosomes formed in vitro – in cell-free translation systems [30-33].
Studying polyribosomes in situ. Analysis of the polysome structure in an intact cell is of the greatest interest. Cellular polyribosomes in situ were studied in protrusions of human glioma cells, which had a thickness up to 200-400 nm [29]. For this, glioma cells were cultivated directly on EM grids and instantly frozen for visualization in cryo-EM and acquisition of tilt series. Visual detection of polyribosomes in the tomograms was complicated by the presence of neighboring monosomes. Therefore, specific contacts between adjacent ribosomes were analyzed to identify the polysomal ribosomes. As a result, four frequent types of contacts were revealed that were specific to 25-30% of all polysomal ribosomes. Two of these four types were similar to “top-to-top” contacts described for prokaryotic polyribosomes [28]. Computer modeling showed that multiple replicating of these ribosomal contacts in a polyribosome model could result in the formation of several polysome folding variants leading either to a three-dimensional helix or to a flat spiral configuration. It was confirmed that these types of polyribosome organization are actually observed in tomograms. In this way the densely packed 3-D helices as well as planar double-row polyribosomes with variable packing and linear topology of mRNA were found. Thus, in situ analysis showed the existence of only topologically linear eukaryotic polyribosomes [29]. However, it is possible that circular polyribosomes were not detected in the cells because of the absence of tight inter-ribosomal contacts, since the identification of polysomes was based on them.
The structural organization of polyribosomes in samples purified from MCF-7 cells by centrifugation through a sucrose gradient was analyzed by cryo-ET and atomic force microscopy [34]. In these samples, not only the linear, but also the ring-shaped polyribosomes were found.
Structure of polyribosomes in vitro. During translation of mRNA in eukaryotic cell-free systems, polysomes similar to cellular polyribosomes are formed [19, 20]. The use of such systems allows, unlike the cellular situation, to study the polyribosomes formed on specific mRNAs added to the translation system. It gives the possibility to investigate the relationship between the structure of polyribosomes and the construction of the mRNA used, primarily, the properties of its 5′- and 3′-UTRs.
Circular eukaryotic polyribosomes. The classical mechanism of mRNA circularization in polyribosomes involves the interaction (via the initiation factor eIF4G) of initiation factor eIF4E bound to the 5′-cap with PABP bound to the 3′-poly(A) tail [16, 17]. Formation of a circular structure by free (not within a polysome) capped polyadenylated mRNA in the presence of these factors was demonstrated by atomic force microscopy [18]. Besides, functional data showing a synergistic effect of the 5′-cap of mRNA and its 3′-poly(A) tail on translation also suggested interaction of these elements in polyribosomes [48-52]. For long time, however, there was no direct structural evidence supporting the idea that the complex of proteins eIF4E–eIF4G–PABP triggers the formation of topologically circular polyribosomes. The first direct probing of the “closed loop” model was attempted recently by cross-linking of eIF4E/4G and PAB1 with opposing ends of mRNAs in living cells [53]. Indeed, an increased crosslink yield with the distant mRNA ends was found for both proteins indicating their participation in the interaction of 5´ and 3´ ends. Remarkably, the yield of eIF4E/4G–3´ end cross-link was relatively higher. That may reflect the transient character of the interaction and/or the existence of other protein partners (besides PAB1) at the 3´ end.
Topologically circular polyribosomes were found and first studied by cryo-ET in a wheat germ cell-free translation system [30, 31, 33]. The wheat germ translation system has an important advantage, which is the absence of endogenous mRNA and polyribosomes. Using this system, one can follow for a long period of time the formation of polyribosomes on a specific mRNA [33]. To check the hypothesis that interaction of cap with poly(A)-tail is involved in polyribosome circularization, the structure of polyribosomes formed on mRNAs with different UTRs was analyzed [31]. In case of capped polyadenylated mRNA, a large portion (about 40%) of polysomal ribosomes was found in polyribosomes with circular topology. It turned out, however, that topologically circular polyribosomes are successfully formed on a capped mRNA without 3′-poly(A) tail (Fig. 3) in amount comparable to that on the capped polyadenylated mRNA. Moreover, similar content of circular polyribosomes was also found in the translation system containing mRNA with nonhomologous 5′- and 3′-UTRs containing no cap and no poly(A)-tail. It was concluded that the interaction of cap and poly(A)-tail mediated by the eIF4E–eIF4G–PABP complex is probably not a unique way of circularization of polysomal mRNA. The authors suggested that the interaction of some components of initiation (probably eIF3 and eIF4F) and termination complexes can play a role in circularization of mRNA in polyribosomes [31].
Fig. 3. An example of a circular polyribosome formed in a wheat germ cell-free translation system on capped mRNA without poly(A) tail. Left panel, tomogram section; middle panel, structure of the polyribosome (the head of the 40S subunit is shown in red, the body of the 40S subunit in yellow, the 60S subunit in blue, the P-stalk in pink); right panel, schematic representation of the putative mRNA path in the polyribosome. The orientation of the mRNA strand in each ribosome is shown with an arrow (from the entry of mRNA into the ribosome towards the exit); the putative mRNA path through ribosomes is shown as a dashed line. The asterisk shows a possible site of interaction of mRNA ends. Adapted from the article [31].
Structure of double-row polyribosomes. For a long time, it was considered that eukaryotic polyribosomes detected on 2-D EM images as double rows of ribosomes are circles in which the opposite sides were collapsed and have antiparallel packing (circular topology of mRNA) [12, 19, 20]. However, one could assume that the mRNA follows a zigzag path through a double row of ribosomes (topologically linear) and its ends are located at opposite extremities of the polyribosome. Recent cryo-ET studies of pro- and eukaryotic polyribosome structures finally resolved the question about the topology of mRNA in double-row polyribosomes. The first study proved the existence of double-row polyribosomes with linear topology of mRNA: such polyribosomes formed in a E. coli cell-free translation system have a sinusoidal (zigzag) path of mRNA [28].
In later studies, cryo-ET structural analysis of polyribosomes formed in a wheat germ cell-free translation system [30, 33] showed that double-row eukaryotic polyribosomes can have different topology. In the same sample of polyribosomes, both circular and double-row polyribosomes were found in approximately equal ratio, as well as helical polyribosomes. It turned out that among double-row polyribosomes topologically circular polyribosomes (collapsed circles) represented only about 10%; all other polyribosomes had linear (zigzag) topology of mRNA [30]. This demonstrates that in reality (in a frozen sample of polyribosomes) circular polysomes are quite rarely collapsed. At the same time, in classical EM most circles can collapse during adsorption on EM support and take the form of a double-row, as they are usually represented in these cases.
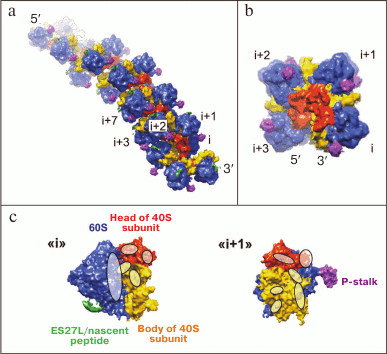
Three-dimensional helical polyribosomes. 3-D helical polyribosomes were found by cryo-ET both in pro- [28] and eukaryotic systems [29, 32]. Pro- and eukaryotic 3-D polyribosomes are quite similar, and they form densely packed left-handed helices with four ribosomes per turn, with 40S subunits oriented inwards and 60S subunits exposed outside (Fig. 4a). The mutual positioning of adjacent ribosomes in such polysomes is described as the “head to head” orientation, described for the first time for prokaryotic polysomes [28]. The arrangement of ribosomes in a turn of a helix in axial projection resembles the positioning of ribosomes in a crystal [45], however they are packed more tightly (Fig. 4b).
To study in detail the contacts between ribosomes in a 3-D helical polyribosome, the crystal structure of the yeast ribosome was fitted into the cryo-ET structure of a helical polyribosome formed in the wheat germ cell-free translation system [32]. As a result, ribosomal proteins and elements of rRNA forming the inter-ribosomal contacts in a helical polyribosome were identified. The main interaction takes place between the heads and the bodies of 40S subunits of adjacent ribosomes, with the L1-stalk of one of the two ribosomes is also involved (Fig. 4c). The first region of contact includes the interaction of helix h16 (18S rRNA) from the 40S subunit of the (i+1) ribosome (located closer to the 5′-end) with the L1 protein of the 60S subunit, and also with proteins S1e, S11, and S26e of the 40S subunit of the (i) ribosome (located closer to the 3′-end). The second region of contact is formed by the interaction of the heads of the 40S subunits of adjacent ribosomes: one contact region comprises proteins S10e and S12e of the (i+1) ribosome and S7, S19e and S25e proteins with rRNA extension segment ES9S of the (i) ribosome, and the second contact region comprises S10 protein of the (i+1) ribosome and RACK1 protein of the (i) ribosome. In addition to the interaction of neighboring ribosomes, some interactions between ribosomes in adjacent turns of a helix might also exist. Thus, it can be seen from the structure that a density corresponding to the beginning of the extension segment ES27L of the ribosome in one turn is directed towards the P-stalk of the ribosome located one turn upstream of the helix (towards the 5′-end of the mRNA) [32].
Fig. 4. Structure of the densely packed 3-D helical polyribosome. Side view (a) and the view on the helical axis (b) (scaled up 2-fold); (i+n) – the number of a polysomal ribosome in the direction from 3′- to 5′-end of mRNA. c) Regions of contacts between adjacent ribosomes (i) and (i+1) of a helical polyribosome presented on panel (b). Adapted with modifications from article [32].
The presence of different structural forms of polyribosomes in a cell could reflect a variety of translated mRNAs. During translation in a cell-free system, however, formation of various forms of polyribosomes happens on the same mRNA, which requires assuming the possibility of structural transformation of polysomes. Therefore, the ratio of various forms of polyribosomes depending on the incubation time of the system was investigated [33]. The maximal number of circular and zigzag polyribosomes was found after short time of incubation in the translation system (the first several rounds of translation); longer incubation time led to the formation of many densely packed 3-D helical polyribosomes. A model of the process of eukaryotic polyribosomes formation, based on the statistical ratio of structural forms of polyribosomes and their loading with ribosomes after different times of translation, has been proposed: (1) short (≥100 nucleotides (nt) of mRNA per ribosome) circular and zigzag polyribosomes are formed initially, (2) increase in the loading of polyribosomes with ribosomes (≤100 nt of mRNA per ribosome) leads to the formation of small structural elements of the 3-D helical organization, resulting in the de-circularization of circular polyribosomes and their transformation into linear zigzag polyribosomes, (3) upon further loading of zigzag polyribosomes with ribosomes they are transformed into the densely packed 3-D helices (30-60 nt of mRNA per ribosome). Such a tight ribosome packing is likely to cause the low translational activity of helical polyribosomes [33].
STRUCTURE OF MEMBRANE-BOUND POLYRIBOSOMES
Unlike cytoplasmic polyribosomes, polyribosomes bound to membrane structures are localized in a 2-dimentional space of the membrane surface that restricts the possible ways of inter-ribosomal contacts in them. The structure of membrane-bound polysomes is defined especially by the interaction of the nascent peptide with the translocon. The structure of membrane-bound polyribosomes has been studied by cryo-ET in preparation of microsomes from dog pancreas [35] and in thin lamellas produced from HeLa cells by a cryo focused ion beam technique (cryo-FIB) [36]. As expected, the ribosomes were found universally directed towards the membrane by the side where the exit from the ribosomal tunnel is located. The extension segments ES27L and ES7L of 28S rRNA are involved in contacts of the ribosome with the membrane. Each next ribosome is usually turned relative to the previous one, and the ribosomes are arranged along the mRNA in a round-shape configuration, so that the mRNA entry and exit sites of adjacent ribosomes form a smooth pathway [35]. In general, the mutual positioning of ribosomes resembles the shape of a planar spiral, which agrees with data of the early works. A similar organization of membrane-bound polyribosomes was also found in yeast mitochondria. Interestingly, in this case one of the contacts of the ribosome with the membrane is also formed by the extension segment 96-ES1 of the 21S rRNA [37].
CONCLUDING REMARKS
The use of cryo-ET for studying the structure of polyribosomes made it possible to determine the topology of mRNA in circular, double-row, and helical polyribosomes and to describe the details of the interaction between ribosomes in polysomes. The results obtained also allowed to make essential functional conclusions and assumptions.
It turned out that the cap-structure and the poly(A)-tail of mRNA are not necessary for the formation of circular polyribosomes, in contrast to what was assumed earlier within the conventional model of “circular translation”. This leads to the existence of another, perhaps more general, mechanism of circularization. For example, the mechanism might be based on the interaction of the components of initiation and termination or post-termination complexes. A possible candidate for such interaction is the ABCE1 protein, which participates in ribosome recycling, interact with terminated 80S ribosome [54] and, probably, has affinity to initiation factors eIF2, eIF3, eIF5 [55, 56]. Moreover, the possibility cannot be excluded that specific positioning of mRNA entry and exit sites on the ribosome, inducing the turn of each downstream ribosome by a certain angle in a polyribosome (as found in the structure of membrane-bound and cytoplasmic polysomes) [29, 35, 36], can by itself promote the formation of a circular configuration. In general, all three mechanisms can operate at the same time, providing the dynamic circular state of a polyribosome necessary for effective reinitiation and circular translation.
3-D helical polyribosomes are a type of organization revealed by cryo-ET in both prokaryotes and eukaryotes. The dense packing of ribosomes in such polysomes can determine their reduced translational activity, which can serve as an element of regulation of translation rate or, if necessary, its complete stopping for conservation and a subsequent fast restart. “Stopped” polyribosomes were found recently in sarcoma cells using fluorescence microscopy [57]. The temporary arrest of translating polyribosomes can be important for transferring the polyribosomes to a specific cell compartment where they can be reactivated. Such mechanism was suggested for polyribosomes translating synaptic mRNAs in neurons. The advantage of this mechanism is in the fast local synthesis of proteins involved in synaptic signal transduction [58]. In addition, a similar model can be realized in polyribosomes translating dendrite-specific mRNAs. It was shown that the transport of mRNA with 3′-UTR of the Arc gene, which defines its dendrite-specific localization, occurs only after polyribosomes have formed on it [59]. Formation of the 3-D helical polyribosomes was previously observed during viral infection, linking their formation with a decrease in the level of translation in the infected cells [25].
The polyribosome, as an assembly of several translating ribosomes, can provide cotranslational formation of oligomeric protein complexes with polypeptide chains being synthesized by adjacent ribosomes. Such a mechanism is assumed to be involved in the formation of the β-galactosidase tetramer [60] and in the formation of so-called “vault particles” consisting of 78 copies of MVP protein (95.8 kDa) [61].
The modern development of cryo-ET technology provides new technical solutions to increase the resolution (phase plate, direct electron detection cameras, new software for analyzing structural data). New techniques become available to study cellular structures in situ (cryo-focused ion beam) and combine fluorescence and electron microscopy in one instrument (correlative microscopy). This will undoubtedly ensure rapid progress in the study of the mechanisms of the formation of three-dimensional organization of polyribosomes and of their contribution to the regulation and localization of protein synthesis in a cell.
Acknowledgments
This publication was supported by the Russian Foundation for Basic Research (projects Nos. 16-34-60148 and 15-04-08649) and the program “Molecular and Cell Biology” of the Russian Academy of Sciences Presidium.
REFERENCES
1.Warner, J. R., Rich, A., and Hall, C. E. (1962)
Electron microscope studies of ribosomal clusters synthesizing
hemoglobin, Science, 138, 1399-1403.
2.Warner, J. R., Knopf, P. M., and Rich, A. (1963) A
multiple ribosomal structure in protein synthesis, Proc. Natl. Acad.
Sci. USA, 49, 122-129.
3.Rich, A., Warner, J. R., and Goodman, H. M. (1963)
The structure and function of polyribosomes, Cold Spring Harb. Symp.
Quant. Biol., 28, 269-285.
4.Gierer, A. (1963) Function of aggregated
reticulocyte ribosomes in protein synthesis, J. Mol. Biol.,
6, 148-157.
5.Wettstein, F. O., Staehelin, T., and Noll, H.
(1963) Ribosomal aggregate engaged in protein synthesis:
characterization of the ergosome, Nature, 197,
430-435.
6.Penman, S., Scherrer, K., Becker, Y., and Darnell,
J. E. (1963) Polyribosomes in normal and poliovirus-infected HeLa cells
and their relationship to messenger RNA, Proc. Natl. Acad. Sci.
USA, 49, 654-662.
7.Palade, G. E. (1955) A small particular component
of the cytoplasm, J. Biophys. Biochem. Cytol., 1,
59-68.
8.Shelton, E., and Kuff, E. L. (1966) Substructure
and configuration of ribosomes isolated from mammalian cells, J.
Mol. Biol., 22, 23-31.
9.Mathias, A. P., Williamson, R., Huxley, H. E., and
Page, S. (1964) Occurrence and function of polysomes in rabbit
reticulocytes, J. Mol. Biol., 9, 154-167.
10.Yazaki, K., Yoshida, T., Wakiyama, M., and Miura,
K. (2000) Polysomes of eukaryotic cells observed by electron
microscopy, J. Electron Microsc. (Japan), 49,
663-668.
11.Christensen, A. K., Kahn, L. E., and Bourne, C.
M. (1987) Circular polysomes predominate on the rough endoplasmic
reticulum of somatotropes and mammotropes in the rat anterior
pituitary, Am. J. Anat., 178, 1-10.
12.Christensen, A. K., and Bourne, C. M. (1999)
Shape of large bound polysomes in cultured fibroblasts and thyroid
epithelial cells, Anat. Rec., 255, 116-129.
13.Behnke, O. (1963) Helical arrangement of
ribosomes in the cytoplasm of differentiating cells of the small
intestine of rat fetuses, Exp. Cell Res., 30,
597-598.
14.Waddington, C. H., and Perry, M. M. (1963)
Helical arrangement of ribosomes in differentiating muscle cells,
Exp. Cell Res., 30, 599-600.
15.Philipps, G. R. (1965) Haemoglobin synthesis and
polysomes in intact reticulocytes, Nature, 205,
567-570.
16.Jacobson, A. (1996) Poly(A) metabolism and
translation: the closed-loop model, in Translational Control
(Hershey, J. W. B., Mathews, M. B., and Sonenberg, N., eds.) Cold
Spring Harbor Laboratory Press, Cold Spring Harbor, N. Y., pp.
451-480.
17.Preiss, T., and Hentze, M. W. (1999) From factors
to mechanisms: translation and translational control in eukaryotes,
Curr. Opin. Genet. Dev., 9, 515-521.
18.Wells, S. E., Hillner, P. E., Vale, R. D., and
Sachs, A. B. (1998) Circularization of mRNA by eukaryotic translation
initiation factors, Mol. Cell, 2, 135-140.
19.Madin, K., Sawasaki, T., Kamura, N., Takai, K.,
Ogasawara, T., Yazaki, K., Toshiaki, T., Takei, T., Miura, K., and
Endo, Y. (2004) Formation of circular polyribosomes in wheat germ
cell-free protein synthesis system, FEBS Lett., 562,
155-159.
20.Kopeina, G. S., Afonina, Z. A.,
Gromova, K. V., Shirokov, V. A., Vasiliev, V. D.,
and Spirin, A. S. (2008) Step-wise formation of eukaryotic double-row
polyribosomes and circular translation of polysomal mRNA, Nucleic
Acids Res., 36, 2476-2488.
21.Slayter, H., Kiho, Y., Hall, C., and Rich, A.
(1968) An electron microscopic study of large bacterial polyribosomes,
J. Cell Biol., 37, 583-590.
22.Rosenbaum, R. M., and Wittner, M. (1970)
Ultrastructure of bacterized and axenic trophozoites of
Entamoeba histolytica with particular reference to
helical bodies, J. Cell Biol., 45, 367-382.
23.Kusamrarn, T., Sobhon, P., and Bailey, G. B.
(1975) The mechanism of formation of inhibitor-induced ribosome helices
in Entamoeba invadens, J. Cell Biol., 65,
529-539.
24.Jensen, W. A. (1968) Cotton embryogenesis.
Polysome formation in the zygote, J. Cell Biol., 36,
403-406.
25.Djaczenko, W., Benedetto, A., and Pezzi, R.
(1970) Formation of helical polyribosomes in poliovirus-infected cells
of the 37 RC line, J. Cell Biol., 45, 173-177.
26.Weiss, P., and Grover, N. B. (1968) Helical array
of polyribosomes, Proc. Natl. Acad. Sci. USA, 59,
763-768.
27.Wooding, F. B. (1968) Ribosome helices in mature
cells, J. Ultrastruct. Res., 24, 157-164.
28.Brandt, F., Etchells, S. A., Ortiz, J. O.,
Elcock, A. H., Hartl, F. U., and Baumeister, W. (2009) The native 3D
organization of bacterial polysomes, Cell, 136,
261-271.
29.Brandt, F., Carlson, L. A., Hartl, F. U.,
Baumeister, W., and Grunewald, K. (2010) The three-dimensional
organization of polyribosomes in intact human cells, Mol. Cell,
39, 560-569.
30.Afonina, Zh. A., Myasnikov, A. G., Khabibullina,
N. F., Belorusova, A. Y., Menetret, J.-F., Vasiliev, V. D., Klaholz, B.
P., Shirokov, V. A., and Spirin, A. S. (2013) Topology of mRNA chain in
isolated eukaryotic double-row polyribosomes, Biochemistry
(Moscow), 78, 445-454.
31.Afonina, Z. A., Myasnikov, A. G., Shirokov, V.
A., Klaholz, B. P., and Spirin, A. S. (2014) Formation of circular
polyribosomes on eukaryotic mRNA without cap-structure and
poly(A)-tail: a cryo electron tomography study, Nucleic Acids
Res., 42, 9461-9469.
32.Myasnikov, A. G., Afonina, Z. A., Menetret,
J.-F., Shirokov, V. A., Spirin, A. S., and Klaholz, B. P. (2014) The
molecular structure of the left-handed supra-molecular helix of
eukaryotic polyribosomes, Nat. Commun., 5, 5294.
33.Afonina, Z. A., Myasnikov, A. G., Shirokov, V.
A., Klaholz, B. P., and Spirin, A. S. (2015) Conformation changes of
eukaryotic polyribosomes during multi-round translation, Nucleic
Acids Res., 43, 618-628.
34.Viero, G., Lunelli, L., Passerini, A., Bianchini,
P., Gilbert, R. J., Bernabo, P., Tebaldi, T., Diaspro, A., Pederzolli,
C., and Quattrone, A. (2015) Three distinct ribosome assemblies
modulated by translation are the building blocks of polysomes, J.
Cell Biol., 208, 581-596.
35.Pfeffer, S., Brandt, F., Hrabe, T.,
Lang, S., Eibauer, M., Zimmermann, R., and
Forster, F. (2012) Structure and 3D arrangement of endoplasmic
reticulum membrane-associated ribosomes, Structure, 20,
1508-1518.
36.Mahamid, J., Pfeffer, S., Schaffer,
M., Villa, E., Danev, R., Cuellar, L. K.,
Forster, F., Hyman, A. A., Plitzko, J. M., and
Baumeister, W. (2016) Visualizing the molecular sociology at the HeLa
cell nuclear periphery, Science, 351, 969-972.
37.Pfeffer, S., Woellhaf, M. W., Herrmann, J. M.,
and Forster, F. (2015) Organization of the mitochondrial translation
machinery studied in situ by cryoelectron tomography, Nat.
Commun., 26, 6019.
38.Lucic, V., Forster, F., and Baumeister, W. (2005)
Structural studies by electron tomography: from cells to molecules,
Annu. Rev. Biochem., 74, 833-865.
39.Bohm, J., Frangakis, A. S., Hegerl, R., Nickell,
S., Typke, D., and Baumeister, W. (2000) Toward detecting and
identifying macromolecules in a cellular context: template matching
applied to electron tomograms, Proc. Natl. Acad. Sci. USA,
97, 14245-14250.
40.Frangakis, A. S., Bohm, J., Forster, F., Nickell,
S., Nicastro, D., Typke, D., Hegerl, R., and Baumeister, W. (2002)
Identification of macromolecular complexes in cryoelectron tomograms of
phantom cells, Proc. Natl. Acad. Sci. USA, 99,
14153-14158.
41.Ortiz, J. O., Forster, F., Kurner, J.,
Linaroudis, A. A., and Baumeister, W. (2006) Mapping 70S ribosomes in
intact cells by cryoelectron tomography and pattern recognition, J.
Struct. Biol., 156, 334-341.
42.Wan, W., and Briggs, J. A. (2016)
Cryo-electron tomography and subtomogram averaging, Methods
Enzymol., 579, 329-367.
43.Scheres, S. H., Melero, R., Valle,
M., and Carazo, J. M. (2009) Averaging of electron subtomograms
and random conical tilt reconstructions through likelihood
optimization, Structure, 17, 1563-1572.
44.Evstafieva, A. G., Shatsky, I. N., Bogdanov, A.
A., Semenkov, Y. P., and Vasiliev, V. D. (1983) Localization of
5′- and 3′-ends of the ribosome-bound segment of template
polynucleotides by immune electron microscopy, EMBO J.,
2, 799-804.
45.Yusupova, G. Z., Yusupov, M. M., Cate, J. H., and
Noller, H. F. (2001) The path of messenger RNA through the ribosome,
Cell, 106, 233-241.
46.Miller, O. L., Jr., Hamkalo, B. A.,
and Thomas, C. A., Jr. (1970) Visualization of bacterial genes in
action, Science, 169, 392-395.
47.Kohler, R., Mooney, R. A., Mills, D. J., Landick,
R., and Cramer, P. (2017) Architecture of a transcribing-translating
expressome, Science, 356, 194-197.
48.Gallie, D. R. (1991) The cap and poly(A) tail
function synergistically to regulate mRNA translational efficiency,
Genes Dev., 5, 2108-2116.
49.Le, H., Tanguay, R. L., Balasta, M. L., Wei, C.
C., Browning, K. S., Metz, A. M., Goss, D. J., and Gallie, D. R. (1997)
Translation initiation factors eIF-iso4G and eIF-4B interact with the
poly(A)-binding protein and increase its RNA binding activity, J.
Biol. Chem., 272, 16247-16255.
50.Tarun, S. Z. J., and Sachs, A. B. (1996)
Association of the yeast poly(A) tail binding protein with translation
initiation factor eIF-4G, EMBO J., 15, 7168-7177.
51.Imataka, H., Gradi, A., and Sonenberg, N. (1998)
A newly identified N-terminal amino acid sequence of human eIF4G binds
poly(A)-binding protein and functions in poly(A)-dependent translation,
EMBO J., 17, 7480-7489.
52.Borman, A. M., Michel, Y. M., Malnou, C. E., and
Kean, K. M. (2002) Free poly(A) stimulates capped mRNA translation
in vitro through the eIF4G-poly(A)-binding protein interaction,
J. Biol. Chem., 277, 36818-36824.
53.Archer, S. K., Shirokikh, N. E., Hallwirth, C.
V., Beilharz, T. H., and Preiss, T. (2015) Probing the closed-loop
model of mRNA translation in living cells, RNA Biol., 12,
248-254.
54.Pisarev, A. V., Skabkin, M. A., Pisareva, V. P.,
Skabkina, O. V., Rakotondrafara, A. M., Hentze, M. W., Hellen, C. U.,
and Pestova, T. V. (2010) The role of ABCE1 in eukaryotic
posttermination ribosomal recycling, Mol. Cell, 37,
196-210.
55.Dong, J., Lai, R., Nielsen, K., Fekete, C. A.,
Qiu, H., and Hinnebusch, A. G. (2004) The essential ATP-binding
cassette protein RLI1 functions in translation by promoting
preinitiation complex assembly, J. Biol. Chem., 279,
42157-42168.
56.Chen, Z. Q., Dong, J., Ishimura, A., Daar, I.,
Hinnebusch, A. G., and Dean, M. (2006) The essential vertebrate ABCE1
protein interacts with eukaryotic initiation factors, J. Biol.
Chem., 281, 7452-7457.
57.Yan, X., Hoek, T. A., Vale, R. D., and Tanenbaum,
M. E. (2016) Dynamics of translation of single mRNA molecules in
vivo, Cell, 165, 976-89.
58.Graber, T. E., Hebert-Seropian, S.,
Khoutorsky, A., David, A., Yewdell, J. W., Lacaille, J. C., and
Sossin, W. S. (2013) Reactivation of stalled polyribosomes in synaptic
plasticity, Proc. Natl. Acad. Sci. USA, 110,
16205-16210.
59.Wang, C., Han, B., Zhou, R., and Zhuang, X.
(2016) Real-time imaging of translation on single mRNA transcripts in
live cells, Cell, 165, 990-1001.
60.Kiho, Y., and Rich, A. (1964) Induced enzyme
formed on bacterial polyribosomes, Proc. Natl. Acad. Sci. USA,
51, 111-118.
61.Mrazek, J., Toso, D., Ryazantsev, S., Zhang, X.,
Zhou, Z. H., Fernandez, B. C., Kickhoefer, V. A., and Rome, L. H.
(2014) Polyribosomes are molecular 3D nanoprinters that orchestrate the
assembly of vault particles, ACS Nano, 8,
11552-11559.