REVIEW: Role of microRNA (miRNA) and Viroids in Lethal Diseases of Plants and Animals. Potential Contribution to Human Neurodegenerative Disorders
L. Cong1,2, Y. Zhao1,3, A. I. Pogue4, and W. J. Lukiw1,5,6,a*
1Neuroscience Center, Louisiana State University School of Medicine, Louisiana State University Health Sciences Center, New Orleans LA 70112-2272, USA2Department of Neurology, Shengjing Hospital, China Medical University, Heping District, Shenyang, Liaoning Province, China
3Department of Anatomy and Cell Biology, Louisiana State University School of Medicine, Louisiana State University Health Sciences Center, New Orleans LA 70112-2272, USA
4Alchem Biotech Research, Toronto ON M5S 1A8, Canada
5Department Neurology, Louisiana State University School of Medicine, New Orleans LA 70112-2272, USA
6Department Ophthalmology, Louisiana State University School of Medicine, New Orleans LA 70112-2272, USA
* To whom correspondence should be addressed.
Received April 19, 2018; Revision received May 25, 2018
Both plants and animals have adopted a common strategy of using ~18-25-nucleotide small non-coding RNAs (sncRNAs), known as microRNAs (miRNAs), to transmit DNA-based epigenetic information. miRNAs (i) shape the total transcriptional output of individual cells; (ii) regulate and fine-tune gene expression profiles of cell clusters, and (iii) modulate cell phenotype in response to environmental stimuli and stressors. These miRNAs, the smallest known carriers of gene-encoded post-transcriptional regulatory information, not only regulate cellular function in healthy cells but also act as important mediators in the development of plant and animal diseases. Plants possess their own specific miRNAs; at least 32 plant species have been found to carry infectious sncRNAs called viroids, whose mechanisms of generation and functions are strikingly similar to those of miRNAs. This review highlights recent remarkable and sometimes controversial findings in miRNA signaling in plants and animals. Special attention is given to the intriguing possibility that dietary miRNAs and/or sncRNAs can function as mobile epigenetic and/or evolutionary linkers between different species and contribute to both intra- and interkingdom signaling. Wherever possible, emphasis has been placed on the relevance of these miRNAs to the development of human neurodegenerative diseases, such as Alzheimer’s disease. Based on the current available data, we suggest that such xeno-miRNAs may (i) contribute to the beneficial properties of medicinal plants, (ii) contribute to the negative properties of disease-causing or poisonous plants, and (iii) provide cross-species communication between kingdoms of living organisms involving multiple epigenetic and/or potentially pathogenic mechanisms associated with the onset and pathogenesis of various diseases.
KEY WORDS: Alzheimer’s disease, inflammatory neurodegeneration, microRNA, plant miRNAs, small non-coding RNAs, viroidsDOI: 10.1134/S0006297918090031
Abbreviations: AD, Alzheimer’s disease; AMD, age-related macular degeneration; CNS, central nervous system; LPS, lipopolysaccharide; miRNA, microRNA; mRNA, messenger RNA; nt, nucleotide; sncRNA, small non-coding RNA; 3'-UTR, 3'-untranslated region; vsRNA, viroid-specific sncRNA.
COMMON FUNCTIONS of microRNA (miRNAs) IN PLANTS AND
ANIMALS
The existence of microRNAs (miRNAs) in plant and animal cells, their function, and structural variety were described 20 years ago. However, the first indications of the involvement of these miRNAs in the development of human neurological diseases was discovered almost 30 years ago [1-15]. It was originally found that both plant and animal miRNAs (i) are representative of small non-coding RNAs (sncRNAs) that widely occur in all eukaryotic cells; (ii) are derived from long double-stranded RNAs (dsRNAs) generated by RNA polymerases II and III (RNA Pol II and III) and then processed into smaller mature miRNAs 18-25 nucleotides (nt) long by nuclear ribonuclease III (RNase III) or RNase III-like processing enzymes; (iii) regulate post-transcriptional stability of messenger RNAs (mRNA) in both plants and animals; (iv) play a significant role in cell development, aging, the maintenance of cell health, disease development, and modulation of the cell transcriptome; (v) participate in molecular-genetic mechanisms involved in the development of various pathologies in animals, and especially, of lethal human diseases, such as age-related inflammation-associated neurodegeneration, cancer, cardiovascular disorders (e.g., retinal damage), and neurological diseases, including age-related macular degeneration (AMD) and Alzheimer’s disease (AD); (vi) modulate expression profiles by regulating gene expression at the level of mRNA complexity and stability and/or by inhibiting mRNA translation; and (vii) control gene expression in both plants and animals by downregulating target mRNAs (and therefore, preventing expression of genetic information encoded by these mRNAs) through upregulation of specific inducible miRNAs [1-15].
Plants have specific nucleic acid molecules similar to animal miRNAs, known as viroids, that act as pathogenic infectious agents in many plant species. Because viroids infect agriculturally important plants, they have attracted significant scientific, as well as socio-economic interest [12-21]. From an evolutionary point of view, miRNAs, including miRNA-155, miRNA-168, and the evolutionary ancient miRNA-854, are highly homologous and can be found in both plants and animals, which suggests that certain plant and animal miRNAs have a common origin and have remained similar over extraordinarily long periods of evolution (for example, Arabidopsis thaliana and Homo sapiens had diverged ~1.5 billion years ago) [1, 3, 17, 18]. This suggests the existence of interkingdom information exchange between animal and plants that involves sncRNAs and miRNAs. Plant miRNAs can enter an animal’s body via the diet and then perform various physiological and/or pathophysiological functions in the host organism [19-21]. Indeed, dietary miRNAs and/or sncRNAs may contribute to both intra- and interkingdom signaling, and by doing so, modulate molecular and genetic processes associated with human health and diseases. They might also be involved in the dietary, environmental, and molecular-genetic interactions of the host organism within the local ecosystem and entire biosphere of the earth [17-25].
miRNAs IN HEALTH AND DISEASE
The discovery of miRNAs has revolutionized our concepts of post-transcriptional gene control in aging, development, health, and disease in both plants and animals. miRNAs are produced from long RNA precursors (low-free-energy hairpin structures up to several hundred nucleotides (nt) long) generated by RNA Pol II and III from genes located on multiple chromosomes (all somatic and sex chromosomes in humans). microRNA precursors to (pre-miRNAs) are processed into small interfering RNAs and smaller mature miRNAs (~18-25 nt long) by nuclear RNase III or RNase III-like enzymes, such as the RNase III endoribonuclease Dicer [24-26]. Mature miRNAs guide the RNA-induced silencing complex (RISC) onto messenger RNAs (mRNAs); RISC has a catalytic endonuclease component Argonaute capable of rapid mRNA degradation [1, 4, 10, 24, 25].
It should be noted that several animal Argonaute proteins (e.g., human AGO1, AGO3, and AGO4) lack endonuclease activity and either decrease mRNA expression indirectly or inhibit its translation on ribosomes [1, 4, 10, 24, 25]. miRNAs are typically complementary to the 3′-untranslated regions (3′-UTRs) of multiple mRNAs (target mRNAs). miRNAs decrease the stability of target mRNAs, accelerate their degradation, or prevent their translation on the polyribosome complex [10, 24, 25]. For example, the 22-nt human pro-inflammatory miRNA-146a (MI0000477; 59% A+U, encoded on chr5q33.3; 5′-UGAGAACUGAAUUCCAUGGGUU-3′) is derived from the larger 98-nt pre-miRNA-146a hairpin precursor sequence 5′-GAUGUGUAUCCUCAGCUUUGAGAACUGAAUUCCAUGGGUUGUGUCAGUGUCAGACCUCUGAAAUUCAGUUCUUCAGCUGGGAUAUCUCUGUCAUCGU-3′ (55% A+U; mature miRNA sequence is underlined; also see below) [26-29]. Bioinformatics analysis has revealed that miRNA-146a is probably one of the most thoroughly selected and information-dense sncRNA [30-34]. For example, a 22-nt sncRNA consisting of four different ribonucleotides (adenine, A; cytosine, C; guanine, G; and uridine, U) can generate just a very few biologically useful miRNAs from over 1012 possible sequence combinations. Indeed, direct observations and analysis of the abundance, specificity, and complexity of miRNAs in the human central nervous system (CNS) revealed a relatively small number (~2650) of miRNAs in the entire human organism, and only ~40 of them are present in the CNS in amounts sufficient for their detection [31-35]. These results indicate that (i) unusually high evolutionary and selection pressure results in the utilization of only highly specific ribonucleotide sequences so they may function in highly selective biologically useful miRNA–mRNA interactions; (ii) negative charge along the length of an 18-25-nt miRNA provides a unique biophysical configuration based on the miRNA ribonucleotide composition and polyphosphate negative charge [34-36]; (iii) complementarity provides the basis for miRNA–mRNA interactions and, eventually, regulates transcriptional output, shapes the gene expression profiles, and determines the transcriptome composition during development, aging, and disease pathogenesis; and (iv) remarkably, only about one in 10 billion theoretically possible miRNAs is involved in actual miRNA–mRNA interactions and miRNA-mediated gene regulation and control of biological processes in human body, and less than one in ~200 billion possible miRNAs participate in the miRNA–mRNA-based regulation of gene expression profiles in the human nervous system.
In the following five sections we will briefly review and highlight some recent and remarkable discoveries including (i) infectious plant sncRNAs known as viroids; (ii) similarities in biological function, processing, structure, and molecular epigenetics of miRNAs and viroids; (iii) involvement of plant miRNAs in potential interkingdom and interspecies communication and disease development in humans; (iv) molecular genetics of inter- and intrakingdom signaling pathways involving specific miRNAs (e.g., miRNA-146a, miRNA-155, miRNA-168, and miRNA-854); and (v) existence of the same miRNAs in plants and animals that still causes controversy on the relevance of this fact and poses the question how these miRNAs can affect pathogenic mechanisms associated with cancer and neurological diseases, including inflammation-associated neurodegeneration [37-50].
Viroids are highly similar to miRNAs in their structure, processing, and biological functions. Viruses are small infectious agents typically consisting of a protein coat encasing a nucleic acid molecule that can only replicate in the cells of a host organism. The smallest known DNA virus is the single-stranded DNA (ssDNA) class II non-enveloped porcine circovirus (family Circoviridae) with an unsegmented circular genome of 1.7 × 103 nt; the smallest known RNA virus is the cancer-causing class VI enveloped Rous sarcoma virus (RSV; family Retroviridae) with a 3.5 × 104 nt sncRNA genome [47-51]. Viroids are a family of about 32 different non-coding un-encapsulated autonomously replicating circular infectious plant sncRNAs that are considerably smaller than any known ssDNA or sncRNA viruses. Similar to pre-miRNAs, viroid precursors range in size from ~246 to ~401 nt and possess the highest in vivo mutation rates among all known nucleic acids [51-58]. Interestingly, the range of viroid hosts appears to be currently expanding due to fast and continuing evolution of RNA sequences and structures that acquire novel biological functions [52, 54]. Viroids not only attract significant interest from the biological, evolutionary, scientific, and virologic points of view, but also cause serious agricultural and economic concerns, since viroid infections significantly reduce the yields of important food cultures including potatoes, citrus fruits, apples, avocados, eggplants, peaches, tomatoes, and coconuts [5, 52-56].
Viroids are replicated by the unidirectional rolling circle mechanism in the infected plant nucleus (family Pospiviroidae) or chloroplasts (family Avsunviroidae). After replication, viroid precursors exit the nucleus or chloroplast in a fashion similar to that of miRNA translocation – via the exportin 5-mediated or similar transport mechanisms [52-55]. Similar to miRNAs, viroid activity is associated with the emergence of small 21-24-nt viroid-specific sncRNAs (vsRNAs) processed from a dsRNA hairpin precursor by RNase III from the family of Dicer enzyme-like proteins (see above) [50-52, 56, 57]. Like miRNAs, pre-viroid sncRNAs and vsRNAs do not encode proteins, have no protein coat, do not reverse transcribe into DNA during replication, and are induced by external stressors and environmental factors [52, 55-58].
Mature vsRNAs are extraordinarily similar to miRNAs in their size, structure, action, processing, and capacity for cell-to-cell translocation. Indeed, both vsRNAs and miRNAs can participate in the transduction of sncRNA-mediated pathological signals to neighboring cells and tissues via passive diffusion and/or through the circulatory system and alter homeostatic gene expression profiles in the host cells [49-57]. miRNA-like vsRNAs can induce diseases in plants through similar genetic mechanisms. Like miRNAs, they can contribute to disease development in both plants and animals [52-58]. In contrast to viroids and other known structures, miRNAs do not replicate in vivo – they are probably too small to do so. In vitro experiments usually require nucleotide linkers to be added to miRNAs to enable their copying and replication by reverse transcription/polymerase chain reaction (RT-PCR). Interestingly, hairpin formation or circularization of pre-viroids, pre-miRNAs, and miRNAs, as well as formation of complex secondary and/or tertiary structures, may stabilize these unique molecules [59-64]. The possibility of cell-to-cell, tissue-to-tissue, and perhaps, species-to species transfer and spreading of signals mediated by miRNAs and viroids has a tremendous effect on our understanding of complex genetic interactions between diverse life forms in the plant and animal kingdoms and their capacity for the symbiotic exchange of highly selective biological information in the natural environment [13, 56-62].
Interkingdom communication – miRNAs and human diseases. Some of the earliest and most compelling evidence for miRNA participation in lethal human diseases came from the studies on the role of miRNA-15 and miRNA-16 in the development of chronic lymphocytic leukemia (CLL), a type of cancer in which bone marrow generates an excessive amount of lymphocytes [65-69] and from the discovery of the association between miRNA-146a and other pro-inflammatory miRNAs and lethal age-related inflammatory neurodegenerative disorders, such as Alzheimer’s disease (AD) [16, 30-33, 43, 49]. The discovery of extracellular miRNAs and their ability to cross biological and physiological barriers added to the idea that circulating miRNAs, such as the inducible pro-inflammatory miRNA-146a (see below), (i) can mediate both short- and long-range communications between various cell types and (ii) in doing so, can influence regulation of both physiological and pathological processes [46-50]. The signaling potential of multiple plant miRNAs and the intriguing possibility of “horizontal” interkingdom communication via orally ingested miRNAs that could influence gene expression in the host organism have been recently recognized by many independent research groups [6-9, 23, 51-73]. Currently, approximately ~872 miRNAs have been identified in ~71 individual plant species by genomic analysis and RNA sequencing; at least 325 miRNAs of the angiosperm plant Arabidopsis thaliana have been fully characterized [70-74]. Experiments on the transfer of dietary miRNAs, translocation analysis, cloning, RNA sequencing, and estimation of miRNA resistance to the strong oxidizing agent periodate have demonstrated that (i) about 5% of all miRNAs in the human blood serum are plant-derived; (ii) plant miRNAs can be 2′-O-methylated at the 3′-end, which makes them resistant to oxidation, depolymerization, and subsequent degradation; and (iii) miRNAs can be translocated, transported, or circulated as lipoprotein-attached ssRNAs, complexes with Argonaute, or while encapsulated in exosomes, microvesicles, or other types of membrane vesicles [29-36, 72-74]. It was found recently that 2′-O-methyl-protected plant miRNAs are able to efficiently cross the epithelial cell layer of the human gastrointestinal (GI) tract. These miRNAs can be transported by the circulatory system and access specific tissues/organs, including the CNS after passing through the blood–brain barrier (BBB) [35, 36, 73].
miRNAs from food have a significant potential for post-transcriptional gene regulation in the physiological compartments of a host organism, mediate gene-based communications inside the cells and between cells, tissues, and species, and perhaps contribute to cell dysfunction and neurodegenerative diseases [19, 20, 73, 75-78]. While some of these findings remain controversial and are still at the early stages of investigation, the ubiquity of miRNAs in food products of plant and animal origin suggests an interesting possibility that food is not only a source of fiber, probiotics, nutrients, and vitamins – it also carries highly specific information for post-transcriptional gene regulation [1, 77, 79, 80]. There are currently several highly illustrative examples of extracellular, intercellular, interspecific, and interkingdom communications via miRNAs (e.g., miRNA-146a, miRNA-155, miRNA-168, and miRNA-854). The data obtained by our group and other researchers have provided evidence that these “mobile” miRNAs that sometimes can be encapsulated into extracellular vesicles (i) are active in and common to plants and animals; (ii) can pass through intra- and inter-kingdom biological and physiological barriers; and (iii) can modulate essential biological or potentially pathological processes in plants and animals [77-81].
Specific miRNAs implicated in trans-species communication: miRNA-146a. This inducible NF-κB-regulated pro-inflammatory miRNA is probably one of the most studied miRNAs due to its role in innate immunity and inflammatory neurodegeneration in humans [35-40]. Human 22-nt miRNA-146a (hsa-miRNA-146a; MI0000477; chr5q33.3) was one of the first miRNAs found to be upregulated in brain cells, brain extracellular fluid (ECF), cerebral spinal fluid (CSF), and blood serum in affected individuals [81-84]. miRNA-146a is moderately abundant in diseased human brain and retina, but its amount can be significantly increased by pathology-related stress factors, including microbial components (e.g., lipopolysaccharide, LPS), neurotoxic metal sulfates, amyloid-beta (Aβ) peptides, and pro-inflammatory cytokines [35, 36, 40, 50]. miRNA-146a (i) was first characterized in 2006, in acute monocytic leukemia cell line THP-1 as an endotoxin-responsive gene and important regulatory component of mammalian innate and acquired immune responses [37-40]; (ii) is the first pro-inflammatory miRNA to be extensively characterized in AD brain in comparison to the same anatomical regions of healthy age-matched controls [38-40]; (iii) is the first miRNA that was found to be directly induced by the microbiome, more specifically, by extremely pro-inflammatory neurotoxic LPS secreted by the Gram-negative bacillus Bacteroides fragilis inhabiting the human GI tract [4, 14, 16, 40]; (iv) was the first pro-inflammatory miRNAs to be shown to “leak out” of cultured human neuronal glial (HNG) primary cells into the surrounding medium; (v) was detected in brain cells, ECF, CSF, and blood serum of AD patients [84-87]; and (vi) is upregulated in human prion diseases, including sporadic Creutzfeldt–Jakob disease (sCJD) and Gerstmann–Straussler–Scheinker (GSS) syndrome [60]. Interestingly, miRNA-146a is often induced in mammalian hosts infected with microbial pathogens, such as Listeria (Gram-positive, intracellular), Salmonella (Gram-negative, intracellular), Helicobacter (Gram-negative, extracellular/intracellular), Mycobacteria (intracellular), and common neurotrophic viruses, such as herpes simplex virus 1 (HSV-1) [40-43]. The role of miRNA-146a in the interspecific signaling in healthy and diseased plants and animals is under active investigation [34, 39, 41, 84-87].
miRNA-155. Human 24-nt A+U-rich miRNA-155 (hsa-miRNA-155; 5′-UUAAUGCUAAUCGUGAUAGGGGUU-3′; MI0000681; chr21q21.3) plays an important role in various physiological and pathological processes, such as blood and lung cancers, tumor growth, viral infection, immune and cardiovascular disorders, and inflammatory neurodegeneration including septic shock, AD, AMD and Down syndrome (trisomy 21) [88-101]. Inducible NF-κB-regulated miRNA-155a has been shown to modulate expression of cell regulatory factors (Bcl2, c-Fos, and STAT3) and transcription-regulating DNA-binding proteins that directly modify B cell activation and B cell transmembrane receptor signaling in chronic lymphocytic leukemia [67, 96-98]. Upregulated miRNA-155 expression in sporadic AD, AMD, and Down syndrome is associated with the downregulation of the soluble serum glycoprotein complement factor H (CFH) that stimulates inflammatory signaling and elicits an aberrant innate immune response, as well as reduced complement activation [31, 49, 68, 88-95]. miRNA-155 or miRNA-155-containing exosomes are involved in the T lymphocyte proliferation in septic shock [90] and in the T cell regulation [91]. Indeed, miRNA-155 plays an essential role in multiple autoimmune and innate immunity disorders, carcinogenesis, cardiovascular diseases, inflammation, and age-related neurodegeneration. The efficiency of anti-miRNA-155 therapy has been recently shown in clinical and experimental studies of acute myeloid leukemia [89], different types of lung cancer [92], experimental autoimmune myocarditis [94], and insect-borne transmissible infections, such as malaria, accompanied by nervous system damage and cognitive impairment [100, 101]. It is also interesting that miRNA-155 from animal fat tissue plays a key role in the development and activation of the immune system (including activation of regulatory T cells) and induction of pivotal epigenetic and immune regulatory signals that may promote the development of progressive inflammatory age-associated neurodegenerative diseases, e.g., AD [21, 76, 89, 95, 99, 101, 102].
Propolis is a mixture that honey bees (Apis mellifera) produce from the exudate of plant blossoms. It is a complex mixture of plant resins, waxes, essential oils, pollen, and natural pesticides, such as lipophilic acaricides, sesquiterpene quinones, coumarins, phenols, and other flavonoids and aromatic compounds [103, 104]. Propolis displays numerous remarkable regulatory, pharmacological, and anti-inflammatory properties, including immunomodulatory action on NF-κB and human miRNA-155 [85, 103-105]. Therefore, plants can exert interkingdom effects not only via direct interspecific or interkingdom transfer of miRNAs but also through the medicinal products of plant secretion that can target specific host miRNAs and promote their beneficial or detrimental activities. The existence of miRNA-155 homologs in plants and animals combined with the miRNA-155 capacity for intra- and interkingdom translocation may underlie an association between a high-fat cholesterol (HF-C) diet and inflammation-related pathologies, such as disorders of the cardiovascular system and CNS [46, 84, 105, 106]. In other words, plant extracts obtained through the diet can produce multiple beneficial effects on human cardiovascular and neurovascular systems via specific miRNAs, e.g., miRNA-155. This NF-κB-inducible miRNA was found in relatively high amounts in the human brain, retina, and blood. It is possible that it is directly involved in the development of cardiovascular and CNS disorders, including AD, BBB dysfunction, various types of malignancies, Down syndrome, and AMD [88, 99, 102, 106, 107].
miRNA-168a. miRNA-168 that belongs to a gene family of at least 49 related miRNA sequences, is one of the most extensively studied plant sncRNAs [108, 109]. miRNA-168 (21 nt, MIMAT0001726) from maize (Zea mays) represents an “archetypical plant miRNA”. At ~850 copies per maize cell, miRNA-168 is the most abundant miRNA that has a recognized ability for both interspecific and interkingdom communication [6, 73, 108]. Thus, after feeding domestic pigs (Sus domesticus) with fresh maize ad libitum for 7 days, at least 18 plant-derived miRNAs 2′-O-methylated at the 3′ end (including zma-miRNA-168a-5p) were detected in pig serum, brain, heart, and other organs. In vivo and in vitro experiments demonstrated that most maize miRNAs are capable of crossing the pig GI tract barriers to enter the bloodstream [73]. It was found that plant miRNAs were likely to specifically target endogenous porcine mRNAs and to influence gene expression in a fashion highly similar to that of mammalian miRNAs [10, 73, 108, 109]. Recent in silico study demonstrated that a small family of plant miRNAs, including miRNA-168a, is also present in a population of mammalian breast milk exosomes, although the significance of miRNA-168 presence in this unique secretory compartment is not well understood [19, 91]. Interestingly, (i) plant-derived miRNA-168 displays a high degree of complementarity with exon 4 of the mammalian low density lipoprotein receptor adapter protein 1 (LDLRAP1) mRNA [6, 73, 110]; (ii) LDLRAP1 mRNA encodes clathrin-associated sorting protein (CLASP) that predominantly localizes to the liver and facilitates removal of low density lipoprotein (LDL) from the human circulatory system [6, 20, 73]; (iii) miRNA-168 regulates expression of the AGO1 mRNA coding for the central component of the plant RNA silencing complex (RISC) that binds to a vast majority of plant miRNA undergoing processing [6, 20, 73, 110]. Our ongoing studies indicate that miRNA-168a is indeed abundant in several food plants and could be detected in the blood and CNS ([6]; unpublished data, 2018).
miRNA-854a. The primary sequences of miRNAs contain ribonucleotide fingerprints conserved across multiple miRNA species; these RNA sequence fingerprints include some of the most thoroughly studied, evolution-conserved sncRNA sequences ever discovered [4, 14, 34, 35, 58, 84, 107]. For example, using species- and genome-wide computational approaches, RNA sequencing, and comparison of miRNA structures and sequences, it was found that members of the miRNA-854 family are abundantly expressed in Arabidopsis thaliana, Caenorhabditis elegans, Mus musculus, and Homo sapiens ([2-4, 6, 9, 36]; unpublished results, 2018). In all these evolutionary distant species (Arabidopsis thaliana and Homo sapiens diverged about ~1.5 billion years ago) [16-18], the 21-nt G+A-enriched miRNA-854a (90.5% G+A; 5′-GAUGAGGAUAGGGAGGAGGAG-3′) plays a common regulatory role by targeting the 3′-UTR of the oligouridylate-binding protein 1b (UBP1b) mRNA. Interestingly, UBP1b encodes a highly conserved member of the family of heterogeneous nuclear RNA (hnRNA) binding proteins [2, 4, 6]. Hence, miRNA-854 is an important example of an interkingdom regulator of basal RNA-specific uridine-mediated transcriptional mechanism in both animals and plants that has existed over billions of years of evolution. In addition to the evolutionary conservation of primary nucleotide sequences, the secondary and tertiary structures of many miRNA precursors are also significantly conserved across multiple sncRNA species; many stem-loop configurations appear at equivalent positions in a significant number of pre-miRNA sequences [2, 3, 6, 14, 17, 35, 107].
The “ancient” miRNA-854 is significantly upregulated in AD-affected brain tissues; targeting of miRNA-854 to the UBP1b mRNA 3′-UTR has been predicted and actually observed in the AD brain. This may affect the availability of uridine (U) for its incorporation into newly synthesized RNA and is presumably related to the widely reported transcriptional impairments observed in AD tissues when compared to age- and gender-matched controls [6, 49, 72, 111-116].
Important questions. As in any newly emerging research area, many fundamental questions on the interspecific/interkingdom communications via miRNAs are still to be answered. Ultimate, basic, persistent, and recurring questions remain on the proportion of miRNAs participating in cell-to-cell or species-to-species communications and on the impact of miRNAs on cells, tissues, organs, and organisms. Which miRNAs are selectively secreted into the extracellular space? Which ones are not, and for what reason? Which miRNAs actually participate in cell-to-cell communication and/or interspecies signaling in health and disease? What are the mechanisms underlying selective export of miRNAs into the extracellular space and their trafficking to more distant locations? What have been the relationships between miRNAs and viroids over the course of evolution? Can a viroid mimic or alter the normal function of miRNA and contribute to health and/or disease? Are miRNAs and/or viroids able to penetrate through the GI tract and BBB of animals or though the plant cell walls? Do miRNAs of poisonous or neurotoxic plants contribute in any way to the toxicity usually associated with the known toxic components of these plants? And perhaps most importantly, can plant-derived miRNAs provide health benefits to humans who consume these plants, and is there any reciprocal action of animal miRNAs on plant miRNAs? All these questions still remain unanswered.
Interkingdom communication via miRNAs in health and disease: the controversy. Taken together, these multiple observations and recent findings suggest an existence of complex and highly interactive pathways governing miRNA biogenesis and horizontal transfer of miRNAs between plants and animals. As in any emerging research field, these findings are somewhat controversial. For example, recent reports on the structure and function of plant miRNAs provides evidence that plant miRNAs could have evolved independently from other miRNAs [62, 70], while bioinformatic analysis of the abundance, specificity, and diversity of cross-species miRNAs (xeno-miRNAs) has shown that xeno-miRNAs originate by processing of non-specific heterogeneous nuclear RNAs (hnRNAs) or sncRNAs rather than from dietary intake or other external miRNA sources [117]. Other research groups have proposed that sncRNAs, miRNA-like molecules, or miRNAs from medicinal plants can be used as novel bioactive components that might be beneficial for an animal organism [6, 80]. Similarly, poisonous or disease-causing plant species can damage animal’s health via their miRNAs [118]. Thus, deadly nightshade (Atropa belladonna), a perennial herbaceous plant of the Solanaceae family (a family consisting of about 98 genera and ~2700 species including tomato, potato, pepper, and tobacco) ranks as one of the most poisonous plants on Earth. Its leaves and berries contain extremely neurotoxic tropane alkaloids that include anisodine, atropine, hyoscyamine, methylecgonine, and scopolamine, whose action is associated with altered visual perception, ecstasy, hallucinations, and delirium and may contribute to adverse neurotoxic events and functional neurological deficits in the CNS resulting in irreversible neurodegeneration [119]. According to the unpublished results of microarray assay, leaves and berries of deadly nightshade also contain high amounts of miRNA-155, a pathogenic microRNA known to be upregulated in multiple autoimmune and innate immune disorders and in several progressive neurodegenerative disorders, including AD, AMD, and Down syndrome [33, 35, 45, 50, 88, 98, 107]. Therefore, in contrast to the health-promoting effects of certain miRNAs from medicinal plants, neurotoxic or pro-inflammatory miRNAs from poisonous or disease-inducing plants may contribute to the impairments in the CNS homeostasis through induction of novel pathogenic miRNA-mediated signaling pathways ([118-122], unpublished results).
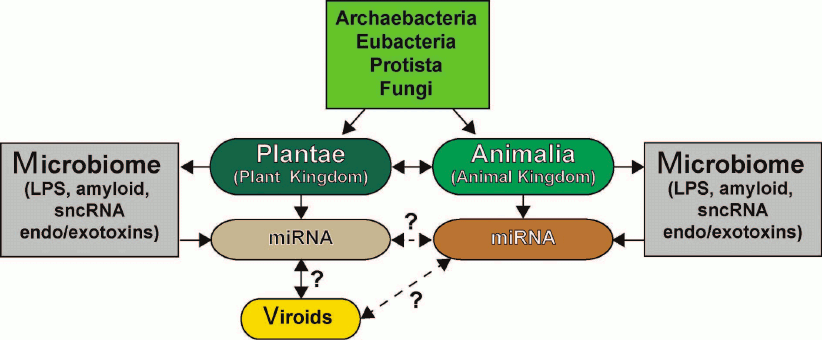
The similarities and differences in the sncRNA-based communication and information transfer between various organisms, the functional roles of sncRNAs in eukaryotic organisms, their evolutionary origin, potential for interaction with human microbiome, and role in proposed interspecific or interkingdom communications, as well as miRNA complexity, specialization, and abundance in plants and animals have been recently reviewed in [6, 16, 33, 84, 96, 118-122] (figure). The ongoing studies of the structure, activity, and biophysical properties of sncRNAs, miRNAs, and viroids in plants and animals will expand our understanding of sncRNA biology including multicomponent miRNA-based communication pathways and open up new frontiers with new and perhaps unexpected prospects.
Six kingdoms of life (Archaea, Eubacteria, Protista, Fungi, Plantae, and Animalia) and their potential and highly interactive contributions to sncRNA signaling via miRNAs and viroids [117-131]. Very little is known about sncRNAs and miRNAs of Archaea, Eubacteria, Protista, and Fungi and their interactions (if any) with plant and animal miRNAs. Both plants (Plantae) and animals (Animalia) make an extensive use of small ~18-25-nt non-coding single stranded RNAs (sncRNAs) known as microRNAs (miRNAs) that transmit critical DNA-based epigenetic information and impact the make-up of the cell transcriptome (see text). Researchers are just beginning to understand the relationship between plant and animal miRNAs; there is emerging evidence that miRNAs may be “epigenetic communication linkers” between different species of plants and animals. Different plant genera and species possess multiple forms of viroids, which are small miRNA-like sncRNAs generally associated with genetically inherited plant diseases. While there is recent evidence that plant miRNA may enter the human body via the diet, there are very few data on direct or reciprocal interactions between animal and plant miRNAs or between plant viroids and animal miRNAs (dashed line with question mark). While Archaea, Eubacteria, Protista, and Fungi appear to possess multiple forms of miRNA-like sncRNAs, interactions between these molecules and higher eukaryotic miRNAs have been acknowledged only recently [6, 14, 114, 115, 118, 123-130]. Microbiomes of plants and animals may act in part as “dynamic exchanges”, as “a reservoir”, or as “modulators” for miRNA-based communications between Plantae and Animalia (dashed arrow with question mark). Researchers have only started to understand in detail the effects of plant and animal microbiomes and their secretory products (e.g., LPS), bacterial amyloids, small non-coding RNAs (sncRNAs) and endotoxins/exotoxins (e.g., fragilysin) on miRNA abundance, complexity, and their role in interspecies communications [16, 119-134]. sncRNA- or miRNA-based signaling and intra- or interkingdom communications may explain in part social, cooperative, interdependent, mutualistic, and/or symbiotic relationships among the six kingdoms of life
Taken together, these recent studies have expanded our understanding of interspecific and interkingdom communications and strengthen the idea that the transfer of sncRNAs or miRNAs from one species to another is an intriguing direction for contemporary genetics research.
It is important to remember that (i) certain plant and animal sncRNAs (e.g., miRNAs) have preserved their size, mechanism of biogenesis, structure, and function as discrete information-carrying molecules through hundreds of millions of years of evolution; (ii) certain miRNAs have retained their essential role in the modulation of gene expression and may perform pathophysiological functions in healthy and diseased plants and animals; (iii) health-promoting or disease-linked miRNAs may provide a foundation for novel therapeutic approaches involving application of stabilized miRNAs or anti-miRNA agents; and (iv) perhaps most importantly, our food is not only an essential supply of nutrients for our bodies but may also be a source of sncRNA- or miRNA-based important genetic regulatory information that can either support our health and well-being or contribute to disease pathogenesis [6-9, 48, 118].
Based on 40 years of research into the structure and function of sncRNAs and miRNAs in the human CNS, we propose that (i) the reasons for the highly similar mode of action among plant and animal miRNAs is that these 18-25-nt molecules have originated from a common ancient RNA and then evolved separately as a result of evolutionary divergence between plants and animals; (ii) existence of sequence-specific sncRNAs in the same environment and their mechanistic similarity in plants and animals may represent an intriguing example of convergent evolution. Taken together, these recent findings support the following concepts: (i) our dietary habits may affect our physiological condition on a genetic level; (ii) nutrigenomics may be involved in the pathobiology of most prevalent age- and lifestyle-related human diseases, such as cancer, cardiovascular disorders, and inflammatory neurodegenerative diseases of the CNS (e.g., AD); (iii) externally sourced animal- or plant-derived sncRNAs and/or miRNAs may impact human health or disease; (iv) sncRNAs and/or miRNAs may be potential targets for genetic manipulation to increase stress resistance by consumption of certain animal and plant products; and (v) food-derived miRNAs and sncRNAs may provide us with another means to deliver necessary therapeutic agents or nutrients to improve and maintain human health or to treat diseases more efficiently. A number of excellent reviews on microRNAs, viroids, intra- and interkingdom communications, evolutionary aspects of miRNAs, their potential contributions to diseases, and novel bioinformatic tools to study these biological processes have been recently published [123-134].
Lastly, plant-derived sncRNAs, miRNAs, and viroids all have the capacity to rapidly evolve in response to numerous stressors that plants encounter during their growth cycle, such as competition with other species, drought, extreme temperatures, nutrient deprivation, salinity, exposure to heavy metals or other toxic species, etc. Transfer of these rapidly evolving sncRNAs to animals via dietary intake certainly occurs, but its relevance, if any, is not clearly understood. The rather unique 2′-O-methylation “blocking” of 3′-ends in plant miRNAs imparts a remarkable stability to these microRNAs in acidic domains of the human GI tract, and the influence of these and other sncRNAs on the gut microbiome is yet another interesting research area worthy of future investigation. Genomic interactions between plants and animals mediated by sncRNA and related molecules add another layer of genetic complexity to interspecific/interkingdom communications and to the relationships between different animal and plant species that make possible their successful coexistence in our biosphere.
Author Contributions
L. C., W. J. L., A. I. P., and Y. Z. discussed the genomic data and scientific implications of ideas presented in the review; A. P. and W. J. L. further researched and wrote this paper; the authors are sincerely grateful to colleagues and collaborators (including T. P. A. Kruck and the late J. M. Hill) for helpful discussion and sharing unpublished data.
Acknowledgements
This research work was presented in part at the Vavilov Institute of General Genetics autumn seminar series in Moscow, Russia, October 2016 and at the Society for Neuroscience (SFN) Annual Meeting, November 2017, Washington DC, USA. This paper is an updated and expanded version of the two previous reports entitled “Evolution of microRNA (miRNA) structure and function in plants and animals: relevance to aging and disease”, J. Aging Sci., 2 (2); pii: 119, 2014 and “Plant and animal microRNAs (miRNAs) and their potential for inter-kingdom communication” that appeared in Cell. Mol. Neurobiol., 38, 133-140; doi: 10.1007/s10571-017-0547-4, 2017 (see [4] and [6]).
We sincerely thank Dr. S. Bhattacharjee, Dr. T. P. A. Kruck (University of Toronto), and the late Dr. J. M. Hill (Louisiana State University) for helpful discussion of this controversial and emerging research area; P. N. Alexandrov, J. G. Cui, F. Culicchia, K. Navel, C. Hebel, and C. Eicken for kindly provided post mortem human and mammalian brain tissues and extracts, unpublished Western blot data, bioinformatic analysis, and data interpretation; and D. Guillot for expert technical assistance. We are grateful to many neuropathologists, physicians, and researchers in the USA, Canada, and Europe who have provided high-quality post mortem human CNS and tissue samples for our studies.
Funding
Studies on human and mouse miRNAs, sncRNAs, pro-inflammatory and pathogenic signaling, including innate immune response, neuroinflammation, and amyloidogenesis in AD, AMD, prion disease and other neurological disorders performed at the Lukiw laboratory was supported by the unlimited grant to the Louisiana State University from the Eye Center from Research to Prevent Blindness, Louisiana Biotechnology Research Network, and NIH (projects NEI EY006311, NIA AG18031 and NIA AG038834).
REFERENCES
1.Moran, Y., Agron, M., Praher, D., and Technau, U.
(2017) The evolutionary origin of plant and animal microRNAs, Nat.
Ecol. Evol., 1, 27.
2.Lukiw, W. J., Handley, P., Wong, L., and Crapper
McLachlan, D. R. (1992) BC200 RNA in normal human neocortex,
non-Alzheimer dementia (NAD), and senile dementia of the Alzheimer type
(AD), Neurochem. Res., 17, 591-597.
3.Axtell, M. J., Westholm, J. O., and Lai, E. C.
(2011) Vive la difference: biogenesis and evolution of microRNAs in
plants and animals, Genome Biol., 12, 221.
4.Pogue, A. I., Clement, C., Hill, J. M., and Lukiw,
W. J. (2014) Evolution of microRNA (miRNA) structure and function in
plants and animals: relevance to aging and disease, J. Aging
Sci., 2, 119.
5.Djami-Tchatchou, A. T., Sanan-Mishra, N., Ntushelo,
K., and Dubery, I. A. (2017) Functional roles of microRNAs in
agronomically important plants-potential as targets for crop
improvement and protection, Front. Plant Sci., 8,
378.
6.Zhao, Y., Cong, L., and Lukiw, W. J. (2018) Plant
and animal microRNAs (miRNAs) and their potential for inter-kingdom
communication, Cell. Mol. Neurobiol., 38, 133-140.
7.Reinhart, B. J., Weinstein, E. G., Rhoades, M. W.,
Bartel, B., and Bartel, D. P. (2002) MicroRNAs in plants, Genes
Dev., 16, 1616-1626.
8.Perge, P., Nagy, Z., Decmann, A., Igaz, I., and
Igaz, P. (2017) Potential relevance of microRNAs in inter-species
epigenetic communication, and implications for disease pathogenesis,
RNA Biol., 14, 391-401.
9.Pirro, S., Minutolo, A., Galgani, A., Potesta, M.,
Colizzi, V., and Montesano, C. (2016) Bioinformatics prediction and
experimental validation of microRNAs involved in cross-kingdom
interaction, J. Comput. Biol., 23, 976-989.
10.Guo, H., Ingolia, N. T., Weissman, J. S., and
Bartel, D. P. (2010) Mammalian microRNAs predominantly act to decrease
target mRNA levels, Nature, 466, 835-840.
11.Liang, H., Huang, L., Cao, J., Zen, K., Chen, X.,
and Zhang, C. Y. (2012) Regulation of mammalian gene expression by
exogenous microRNAs, Wiley Interdiscip. Rev. RNA, 3,
733-742.
12.Daros, J. A., Elena, S. F., and Flores, R. (2006)
Viroids: an Ariadne’s thread into the RNA labyrinth, EMBO
Rep., 7, 593-598.
13.Ding, B., and Wang, Y. (2009) Viroids: uniquely
simple and tractable models to elucidate regulation of cell-to-cell
trafficking of RNA, DNA Cell Biol., 28, 51-56.
14.Pogue, A. I., Hill, J. M., and Lukiw, W. J.
(2014) MicroRNA (miRNA): sequence and stability, viroid-like
properties, and disease association in the CNS, Brain Res.,
1584, 73-79.
15.Arteaga-Vazquez, M., Caballero-Perez, J., and
Vielle-Calzada, J. P. (2006) A family of microRNAs present in plants
and animals, Plant Cell, 18, 3355-3369.
16.Zhao, Y., and Lukiw, W. J. (2018)
Microbiome-mediated upregulation of microRNA-146a in sporadic
Alzheimer’s disease, Front. Neurol., 9, 145.
17.Marshall, M. (2018) Timeline: the evolution of
life, https://www.newscientist.com/article/dn17453-timeline-the-evolution-of-life/
(last accessed 11 June 2018).
18.Wang, D. Y., Kumar, S., and Hedges, S. B. (1999)
Divergence time estimates for the earl history of animal phyla and the
origin of plants, animals and fungi, Proc. Biol. Sci.,
266, 163-171.
19.Dato, S., Rose, G., Crocco, P., Monti, D.,
Garagnani, P., Franceschi, C., and Passarino, G. (2017) The genetics of
human longevity: an intricacy of genes, environment, culture and
microbiome, Mech. Ageing Dev., 165, 147-155.
20.Vaucheret, H., and Chupeau, Y. (2012) Ingested
plant miRNAs regulate gene expression in animals, Cell Res.,
22, 3-5.
21.Wagner, A. E., Piegholdt, S., Ferraro, M.,
Pallauf, K., and Rimbach, G. (2015) Food derived microRNAs, Food
Funct., 6, 714-718.
22.Lagos-Quintana, M., Rauhut, R., Lendeckel, W.,
and Tuschl, T. (2001) Identification of novel genes coding for small
expressed RNAs, Science, 294, 853-858.
23.Liang, H., Zen, K., Zhang, J., Zhang, C. Y., and
Chen, X. (2013) New roles for microRNAs in cross-species communication,
RNA Biol., 10, 367-370.
24.Bartel, D. P. (2009) MicroRNAs: target
recognition and regulatory functions, Cell, 136,
215-233.
25.Carthew, R. W., and Sontheimer, E. J. (2009)
Origins and mechanisms of miRNAs and siRNAs, Cell, 136,
642-655.
26.UniProt Database; http://www.uniprot.org/uniprot/Q9UPY3
(last accessed 11 June 2018).
27.GeneCards microRNA-146a; Human Gene Database;
microRNA-146a; Weitzmann Institute, Rehovot Israel; http://www.genecards.org/cgi-bin/carddisp.pl?gene=MIR146A
(last accessed 11 June 2018).
28.National Center for Biological Information
(NCBI); Bethesda MD, USA; Homo sapiens chromosome 5, GRCh38. p. 12.
Primary Assembly. https://www.ncbi.nlm.nih.gov/nuccore/NC_000005.10?strand=1&report=genbank&from=160485352&to=160485450
(last accessed 11 June 2018).
29.Idda, M. L., Munk, R., Abdelmohsen, K., and
Gorospe, M. (2018) Noncoding RNAs in Alzheimer’s disease,
Wiley Interdiscip. Rev. RNA, 9, doi:
10.1002/wrna.1463.
30.Miya Shaik, M., Tamargo, I. A., Abubakar, M. B.,
Kamal, M. A., Greig, N. H., and Gan, S. H. (2018) The role of microRNAs
in Alzheimer’s disease and their therapeutic potentials, Genes
(Basel), 9, E174.
31.Lukiw, W. J. (2007) MicroRNA speciation in fetal,
adult and Alzheimer’s disease hippocampus, Neuroreport,
18, 297-300.
32.Lukiw, W. J. (2012) Evolution and complexity of
microRNA in the human brain, Front. Genet., 3,
166-175.
33.Zhao, Y., Pogue, A. I., and Lukiw, W. J. (2015)
MicroRNA (miRNA) signaling in the human CNS in sporadic
Alzheimer’s disease – novel and unique pathological
features, Int. J. Mol. Sci., 16, 30105-30116.
34.Hill, J. M., and Lukiw, W. J. (2014) Comparing
miRNAs and viroids; highly conserved molecular mechanisms for the
transmission of genetic information, Front. Cell. Neurosci.,
8, 45.
35.Hill, J. M., and Lukiw, W. J. (2016) MicroRNA
(miRNA)-mediated pathogenetic signaling in Alzheimer’s disease
(AD), Neurochem. Res., 41, 96-100.
36.Lukiw, W. J. (2012) Evolution and complexity of
micro RNA in the human brain, Front. Genet., 3, 166.
37.Taganov, K. D., Boldin, M. P., Chang, K.-J., and
Baltimore, D. (2006) NF-κB-dependent induction of miRNA miR-146,
an inhibitor targeted to signaling proteins of innate immune responses,
Proc. Natl. Acad. Sci. USA, 103, 12481-12486.
38.Sethi, P., and Lukiw, W. J. (2009) Micro-RNA
abundance and stability in human brain: specific alterations in
Alzheimer’s disease temporal lobe neocortex, Neurosci.
Lett., 459, 100-104.
39.Mann, M., Mehta, A., Zhao, J. L., Lee, K.,
Marinov, G. K., Garcia-Flores, Y., and Baltimore, D. (2017) An
NF-κB-microRNA regulatory network tunes macrophage inflammatory
responses, Nat. Commun., 8, 851.
40.Zhao, Y., and Lukiw, W. J. (2018) Bacteroidetes
neurotoxins and inflammatory neurodegeneration, Mol. Neurobiol.,
doi: 10.1007/s12035-018-1015-y.
41.Maudet, C., Mano, M., and Eulalio, A. (2014)
MicroRNAs in the interaction between host and bacterial pathogens,
FEBS Lett., 588, 4140-4147.
42.Lukiw, W. J., Cui, J. G., Yuan, L. Y.,
Bhattacharjee, P. S., Corkern, M., Clement, C., Kammerman, E. M., Ball,
M. J., Zhao, Y., Sullivan, P. M., and Hill, J. M. (2010) Acyclovir or
Aβ42 peptides attenuate HSV-1-induced miRNA-146a levels in human
primary brain cells, Neuroreport, 21, 922-927.
43.Jaber, V., Zhao, Y., and Lukiw, W. J. (2017)
Alterations in micro RNA-messenger RNA (miRNA–mRNA) coupled
signaling networks in sporadic Alzheimer’s disease (AD)
hippocampal CA1, J. Alzheimers Dis. Parkinsonism, 7,
312.
44.Lau, N. C., Lim, L. P., Weinstein, E. G., and
Bartel, D. P. (2001) An abundant class of tiny RNAs with probable
regulatory roles in Caenorhabditis elegans, Science,
294, 858-862.
45.Lee, R. C., and Ambros, V. (2001) An extensive
class of small RNAs in Caenorhabditis elegans, Science,
294, 862-864.
46.Prasad, K. N. (2017) Oxidative stress and
pro-inflammatory cytokines may act as one of the signals for regulating
microRNAs expression in Alzheimer’s disease, Mech. Ageing
Dev., 162, 63-71.
47.Previdi, M. C., Carotenuto, P., Zito, D.,
Pandolfo, R., and Braconi, C. (2017) Noncoding RNAs as novel biomarkers
in pancreatic cancer: what do we know? Future Oncol., 13,
443-453.
48.Alural, B., Genc, S., and Haggarty, S. J. (2017)
Diagnostic and therapeutic potential of microRNAs in neuropsychiatric
disorders: past, present, and future, Prog. Neuropsychopharmacol.
Biol. Psychiatry, 73, 87-103.
49.Fabris, L., and Calin, G. A. (2016) Circulating
free xeno-microRNAs – the new kids on the block, Mol.
Oncol., 10, 503-508.
50.Ding, Y., Sun, X., and Shan, P. F. (2017)
MicroRNAs and cardiovascular disease, Biomed Res. Int., doi:
10.1155/2017/4080364.
51.Adams, M. J., and Carstens, E. B. (2012)
Ratification vote on taxonomic proposals to the International Committee
on Taxonomy of Viruses, Arch. Virol., 157, 1411-1422.
52.Diener, T. O. (2003) Discovering
viroids – a personal perspective, Nat. Rev.
Microbiol., 1, 75-80.
53.Navarro, B., Gisel, A., Rodio, M. E., Delgado,
S., Flores R., and Di Serio, F. (2012) Viroids: how to infect a host
and cause disease without encoding proteins, Biochimie,
94, 1474-1480.
54.Muller, S., and Appel, B. (2016) In vitro
circularization of RNA, RNA Biol., 14, 1018-1027.
55.Adams, M. J., Lefkowitz, E. J., King, A. M. Q.,
Harrach, B., Harrison, R. L., Knowles, N. J., Kropinski, A. M.,
Krupovic, M., Kuhn, J. H., Mushegian, A. R., Nibert, M., Sabanadzovic,
S., Sanfaçon, H., Siddell, S. G., Simmonds, P., Varsani, A.,
Zerbini, F. M., Gorbalenya, A. E., and Davison, A. J. (2017) Changes to
taxonomy and the International Code of Virus Classification and
Nomenclature ratified by the International Committee on Taxonomy of
Viruses, Arch Virol., 162, 2505-2538.
56.Fox, A., and Mumford, R. A. (2017) Plant viruses
and viroids in the United Kingdom: an analysis of first detections and
novel discoveries from 1980 to 2014, Virus Res., 241,
10-18.
57.Hammann, C., and Steger, G. (2012)
Viroid-specific small RNA in plant disease, RNA Biol.,
9, 809-819.
58.Hill, J. M., Zhao, Y., Bhattacharjee, S., and
Lukiw, W. J. (2014) miRNAs and viroids utilize common strategies in
genetic signal transfer, Front. Mol. Neurosci., 7,
10.
59.Zhang, B., Pan, X., Cobb, G. P., and Anderson, T.
A. (2006) Plant microRNA: a small regulatory molecule with big impact,
Dev. Biol., 289, 3-16.
60.Chen, X., Liang, H., Zhang, J., Zen, K., and
Zhang, C. Y. (2012) Secreted microRNAs: a new form of intercellular
communication, Trends Cell Biol., 22, 125-132.
61.Chellappan, P., Vanitharani, R., and Fauquet, C.
M. (2005) MicroRNA-binding viral protein interferes with
Arabidopsis development, Proc. Natl. Acad. Sci. USA,
102, 10381-10386.
62.Budak, H., and Akpinar, B. A. (2015) Plant
miRNAs: biogenesis, organization and origins, Funct. Integr.
Genomics, 15, 523-531.
63.Millar, A. A., and Waterhouse, P. M. (2005) Plant
and animal microRNAs: similarities and differences, Funct. Integr.
Genomics, 5, 129-135.
64.Perkel, J. M. (2013) Assume nothing: the tale of
circular RNA, Biotechniques, 55, 55-57.
65.Lukiw, W. J., and Bazan, N. G. (1997)
Cyclooxygenase 2 RNA message abundance, stability, and hypervariability
in sporadic Alzheimer neocortex, J. Neurosci. Res., 50,
937-945.
66.Calin, G. A., Dumitru, C. D., Shimizu, M., Bichi,
R., Zupo, S., Noch, E., Aldler, H., Rattan, S., Keating, M., Rai, K.,
Rassenti, L., Kipps, T., Negrini, M., Bullrich, F., and Croce, C. M.
(2002) Frequent deletions and down-regulation of micro-RNA genes
miRNA-15 and miRNA-16 at 13q14 in chronic lymphocytic leukemia,
Proc. Natl. Acad. Sci. USA, 99, 15524-15529.
67.Mirzaei, H., Fathullahzadeh, S., Khanmohammadi,
R., Darijani, M., Momeni, F., Masoudifar, A., Goodarzi, M., Mardanshah,
O., Stenvang, J., Jaafari, M. R., and Mirzaei, H. R. (2018) State of
the art in microRNA as diagnostic and therapeutic biomarkers in chronic
lymphocytic leukemia, J. Cell. Physiol., 233,
888-900.
68.Millan, M. J. (2017) Linking deregulation of
non-coding RNA to the core pathophysiology of Alzheimer’s
disease: an integrative review, Prog. Neurobiol.,
156, 1-68.
69.Recabarren, D., and Alarcon, M. (2017) Gene
networks in neurodegenerative disorders, Life Sci., 183,
83-97.
70.Budak, H., and Zhang, B. (2017) MicroRNAs in
model and complex organisms, Funct. Integr. Genomics, 17,
121-124.
71.Chen, X., Liang, H., Zhang, J., Zen, K., and
Zhang, C. Y. (2012) Horizontal transfer of microRNAs: molecular
mechanisms and clinical applications, Protein Cell, 3,
28-37.
72.Igaz, P., Nagy, Z., Vasarhelyi, B., Buzas, E.,
Falus, A., and Racz, K. (2012) Potential role for microRNAs in
inter-individual and inter-species communication, Orv. Hetil.,
153, 1647-1650.
73.Luo, Y., Wang, P., Wang, X., Wang, Y., Mu, Z.,
Li, Q., Fu, Y., Xiao, J., Li, G., Ma, Y., Gu, Y., Jin, L., Ma, J.,
Tang, Q., Jiang, A., Li, X., and Li, M. (2017) Detection of
dietetically absorbed maize-derived microRNAs in pigs, Sci.
Rep., 7, 645.
74.MirBASE release 21.0; microRNA database;
University of Manchester, Manchester UK; http://www.mirbase.org/cgi-bin/mirna_summary.pl?org=ath
(last accessed 11 June 2018).
75.Zhang, H., Li, Y., Liu, Y., Liu, H., Wang, H.,
Jin, W., Zhang, Y., Zhang, C., and Xu, D. (2016) Role of plant microRNA
in cross-species regulatory networks of humans, BMC Syst. Biol.,
10, 60.
76.Liang, H., Zhang, S., Fu, Z., Wang, Y., Wang, N.,
Liu, Y., Zhao, C., Wu, J., Hu, Y., Zhang, J., Chen, X., Zen, K., and
Zhang, C. Y. (2015) Effective detection and quantification of
dietetically absorbed plant microRNAs in human plasma, J. Nutr.
Biochem., 26, 505-512.
77.Walzer, K. A., and Chi, J. T. (2017)
Trans-kingdom small RNA transfer during host-pathogen interactions: the
case of P. falciparum and erythrocytes, RNA Biol.,
14, 442-449.
78.Hoy, A. M., and Buck, A. H. (2012) Extracellular
small RNAs: what, where, why? Biochem. Soc. Trans.,
40, 886-890.
79.Makarova, J. A., Shkurnikov, M. U., Wicklein, D.,
Lange, T., Samatov, T. R., Turchinovich, A. A., and Tonevitsky, A. G.
(2016) Intracellular and extracellular microRNA: an update on
localization and biological role, Prog. Histochem. Cytochem.,
51, 33-49.
80.Xie, W., Weng, A., and Melzig, M. F. (2016)
MicroRNAs as new bioactive components in medicinal plants, Planta
Med., 82, 1153-1162.
81.Turchinovich, A., Tonevitsky, A. G., and
Burwinkel, B. (2016) Extracellular miRNA: a collision of two paradigms,
Trends Biochem. Sci., 41, 883-892.
82.Rutter, B. D., and Innes, R. W. (2018)
Extracellular vesicles as key mediators of plant–microbe
interactions, Curr. Opin. Plant Biol., 44,
16-22.
83.Malloci, M., Perdomo, L., Veerasamy, M.,
Andriantsitohaina, R., Simard, G., and Martinez, M. C. (2018)
Extracellular vesicles: mechanisms in human health and disease,
Antioxid. Redox Signal., doi: 10.1089/ars.2017.7265.
84.Alexandrov, P. N., Dua, P., Hill, J. M.,
Bhattacharjee, S., Zhao, Y., and Lukiw, W. J. (2012) microRNA (miRNA)
speciation in Alzheimer’s disease (AD) cerebrospinal fluid (CSF)
and extracellular fluid (ECF), Int. J. Biochem. Mol. Biol.,
3, 365-373.
85.Andreeva, T. V., Lukiw, W. J., and Rogaev, E. I.
(2017) Biological basis for amyloidogenesis in Alzheimer’s
disease, Biochemistry (Moscow), 82, 122-139.
86.MiRBase; microRNA database; University of
Manchester, Manchester UK; http://www.mirbase.org/cgi-bin/mirna_entry.pl?acc=MI0000477
(last accessed 11 June 2018).
87.Genecards microRNA-146a; Weitzmann Institute,
Rehovot Israel; http://www.genecards.org/cgi-bin/carddisp.pl?gene=MIR146A
(last accessed 11 June 2018).
88.Li, Y. Y., Alexandrov, P. N., Pogue, A. I., Zhao,
Y., Bhattacharjee, S., and Lukiw, W. J. (2012) MiRNA-155 upregulation
and complement factor H deficits in Down’s syndrome,
Neuroreport, 23, 168-173.
89.Liang, H., Dong, Z., Liu, J. F., Chuang, W., Gao,
L. Z., and Ren, Y. G. (2017) Targeting miRNA-155 suppresses
proliferation and induces apoptosis of HL-60 cells by targeting
Slug/PUMA signal, Histol. Histopathol., 32,
899-907.
90.Chen, Y., Wang, G., Liu, Z., Wang, S., and Wang,
Y. (2016) Glucocorticoids regulate the proliferation of T cells via
miRNA-155 in septic shock, Exp. Ther. Med., 12,
3723-3728.
91.Melnik, B. C., John, S. M., and Schmitz, G.
(2014) Milk: an exosomal microRNA transmitter promoting thymic
regulatory T cell maturation preventing the development of atopy? J.
Transl. Med., 12, 43.
92.Shriram, V., Kumar, V., Devarumath, R. M., Khare,
T. S., and Wani, S. H. (2016) MicroRNAs as potential targets for
abiotic stress tolerance in plants, Front. Plant Sci., 7,
817.
93.Van Roosbroeck, K., Fanini, F., Setoyama, T.,
Ivan, C., Rodriguez-Aguayo, C., Fuentes-Mattei, E., Xiao, L., Vannini,
I., Redis, R. S., D’Abundo, L., Zhang, X., Nicoloso, M. S.,
Rossi, S., Gonzalez-Villasana, V., Rupaimoole, R., Ferracin, M.,
Morabito, F., Neri, A., Ruvolo, P. P., Ruvolo, V. R., Pecot, C. V.,
Amadori, D., Abruzzo, L., Calin, S., Wang, X., You, M. J., Ferrajoli,
A., Orlowski, R., Plunkett, W., Lichtenberg, T. M., Davuluri, R. V.,
Berindan-Neagoe, I., Negrini, M., Wistuba, I. I., Kantarjian, H. M.,
Sood, A. K., Lopez-Berestein, G., Keating, M. J., Fabbri, M., and
Calin, G. A. (2017) Combining anti-miRNA-155 with chemotherapy for the
treatment of lung cancers, Clin. Cancer Res., 23,
2891-2904.
94.Wong, L. L., Wang, J., Liew, O. W., Richards, A.
M., and Chen, Y. T. (2016) MicroRNA and heart failure, Int. J. Mol.
Sci., 17, 502.
95.Yan, L., Hu, F., Yan, X., Wei, Y., Ma, W., Wang,
Y., Lu, S., and Wang, Z. (2016) Inhibition of microRNA-155 ameliorates
experimental autoimmune myocarditis by modulating Th17/Treg immune
response, J. Mol. Med. (Berlin), 94, 1063-1079.
96.Zhao, Y., Pogue, A. I., and Lukiw, W. J. (2015)
MicroRNA (miRNA) signaling in the human CNS in sporadic
Alzheimer’s disease (AD) – novel and unique pathological
features, Int. J. Mol. Sci., 16, 30105-30116.
97.Zhao, Y., Jaber, V., Percy, M. E., and Lukiw, W.
J. (2017) A microRNA cluster (let-7c, miRNA-99a, miRNA-125b, miRNA-155
and miRNA-802) encoded at chr21q21.1-chr21q21.3 and the phenotypic
diversity of Down’s syndrome (DS; trisomy 21), J. Nat.
Sci., 3, e446.
98.Pogue, A. I., and Lukiw, W. J. (2018)
Up-regulated pro-inflammatory microRNAs (miRNAs) in Alzheimer’s
disease (AD) and age-related macular degeneration (AMD), Cell. Mol.
Neurobiol., 38, 1021-1031.
99.Barker, K. R., Lu, Z., Kim, H., Zheng, Y., Chen,
J., Conroy, A. L., Hawkes, M., Cheng, H. S., Njock, M. S., Fish, J. E.,
Harlan, J. M., Lopez, J. A., Liles, W. C., and Kain, K. C. (2017)
MiRNA-155 modifies inflammation, endothelial activation and blood-brain
barrier dysfunction in cerebral malaria, Mol. Med., 2,
23.
100.MiRBase; microRNA database; University of
Manchester, Manchester, UK; http://www.mirbase.org/cgi-bin/mirna_entry.pl?acc=MI0000477
(last accessed 11 June 2018).
101.Genecards microRNA-155; Weitzmann Institute,
Rehovot Israel; http://www.genecards.org/cgi-bin/carddisp.pl?gene=MIR155
(last accessed 11 June 2018).
102.Banerjee, N., Talcott, S., Safe, S., and
Mertens-Talcott, S. U. (2012) Cytotoxicity of pomegranate polyphenolics
in breast cancer cells in vitro and vivo: potential role
of miRNA-27a and miRNA-155 in cell survival and inflammation, Breast
Cancer Res. Treat., 136, 21-34.
103.De Figueiredo, S. M., de Freitas, M. C. D., de
Oliveira, D. T., de Miranda, M. B., Vieira-Filho, S. A., and
Caligiorne, R. B. (2017) Biological activities of red propolis: a
review, Recent Pat. Endocr. Metab. Immune Drug Discov.,
11, 3-12; doi: 10.2174/1872214812666180223120316.
104.Conti, B. J., Santiago, K. B., Cardoso, E. O.,
Freire, P. P., Carvalho, R. F., Golim, M. A., and Sforcin, J. M. (2016)
Propolis modulates miRNAs involved in TLR-4 pathway, NF-κB
activation, cytokine production and in the bactericidal activity of
human dendritic cells, J. Pharm. Pharmacol., 68,
1604-1612.
105.Alexandrov, P., Cui, J. G., Zhao, Y., and
Lukiw, W. J. (2005) 24S-hydroxycholesterol induces inflammatory gene
expression in primary human neural cells, Neuroreport,
16, 909-913.
106.Bagyinszky, E., Giau, V. V., Shim, K., Suk, K.,
An, S. S. A., and Kim, S. (2017) Role of inflammatory molecules in the
Alzheimer’s disease progression and diagnosis, J. Neurol.
Sci., 376, 242-254.
107.Hill, J. M., Pogue, A. I., and Lukiw, W. J.
(2015) Pathogenic microRNAs common to brain and retinal degeneration;
recent observations in Alzheimer’s disease and age-related
macular degeneration, Front. Neurol., 6, 232.
108.MiRBase; microRNA database; University of
Manchester; http://www.mirbase.org/cgi-bin/mirna_summary.pl?fam=MIPF0000081
(last accessed 11 June 2018).
109.Hu, H., Rashotte, A. M., Singh, N. K., Weaver,
D. B., Goertzen, L. R., Singh, S. R., and Locy, R. D. (2015) The
complexity of posttranscriptional small RNA regulatory networks
revealed by in silico analysis of Gossypium arboreum L.
leaf, flower and boll small regulatory RNAs, PLoS One,
10, e0127468.
110.Zhang, L., Hou, D., Chen, X., Li, D., Zhu, L.,
Zhang, Y., Li, J., Bian, Z., Liang, X., Cai , X., Yin, Y., Wang, C.,
Zhang, T., Zhu, D., Zhang, D., Xu, J., Chen, Q., Ba, Y., Liu, J., Wang,
Q., Chen, J., Wang, J., Wang, M., Zhang, Q., Zhang, J., Zen, K., and
Zhang, C. Y. (2012) Exogenous plant miRNA-168a specifically targets
mammalian LDLRAP1: evidence of cross-kingdom regulation by miRNA,
Cell Res., 22, 107-126.
111.New Scientist – Timeline: The
evolution of life; https://www.newscientist.com/article/dn17453-timeline-the-evolution-of-life/
(last accessed 11 June 2018).
112.Rosenberg, E., and Zilber-Rosenberg, I. (2016)
Microbes drive evolution of animals and plants: the hologenome concept,
MBio, 7, e01395.
113.Lukiw, W. J. (2004) Gene expression profiling
in fetal, aged, and Alzheimer hippocampus: a continuum of
stress-related signaling, Neurochem. Res., 29,
1287-1297.
114.Cui, J. G., Zhao, Y., and Lukiw, W. J. (2005)
Isolation of high spectral quality RNA using run-on gene transcription;
application to gene expression profiling of human brain, Cell. Mol.
Neurobiol., 25, 789-794.
115.Clement, C., Hill, J. M., Dua, P., Culicchia,
F., and Lukiw, W. J. (2016) Analysis of RNA from Alzheimer’s
disease post-mortem brain tissues, Mol. Neurobiol., 53,
1322-1328.
116.Barbash, S., and Sakmar, T. P. (2017)
Length-dependent gene mis-expression is associated with
Alzheimer’s disease progression, Sci. Rep.,
7, 190.
117.Kang, W., Bang-Berthelsen, C. H., Holm, A.,
Houben, A. J., Muller, A. H., Thymann, T., Pociot, F., Estivill, X.,
and Friedlander, M. R. (2017) Survey of 800+ data sets from human
tissue and body fluid reveals xenomiRs are likely artifacts,
RNA, 23, 433-445.
118.Lukasik, A., and Zielenkiewicz, P. (2016) Plant
microRNAs – novel players in natural medicine? Int. J.
Mol. Sci., 18, E9.
119.Kwakye, G. F., Jimenez, J., Jimenez, J. A., and
Aschner, M. (2018) Atropa belladonna neurotoxicity: implications
to neurological disorders, Food Chem. Toxicol.,
116, 346-353.
120.University of California Phylogeny Wing: The
Phylogeny of Life; http://www.ucmp.berkeley.edu/alllife/threedomains.html
(last accessed 11 June 2018).
121.Astrobiology at NASA – Life in the
Universe; The three domains of life; https://astrobiology.nasa.gov/news/the-three-domains-of-life/
(last accessed 11 June 2018).
122.Sciencing: Characteristics of the six kingdoms
of organisms; https://sciencing.com/characteristics-six-kingdoms-organisms-8242194.html)
(last accessed 11 June 2018).
123.Flores, R., Navarro, B., Kovalskaya, N.,
Hammond, R. W., and Di Serio, F. (2017) Engineering resistance against
viroids, Curr. Opin. Virol., 26, 1-7.
124.Lukasik, A., and Zielenkiewicz, P. (2014) In
silico identification of plant miRNAs in mammalian breast milk
exosomes – a small step forward? PLoS One, 9,
e99963.
125.Yu, Y., Jia, T., and Chen, X. (2017) The
“how” and “where” of plant microRNAs, New
Phytol., 216, 1002-1017.
126.Chen, X., Xie, D., Zhao, Q., and You, Z. H.
(2017) MicroRNAs and complex diseases: from experimental results to
computational models, Brief. Bioinform., doi:
10.1093/bib/bbx130.
127.Moran, Y., Agron, M., Praher, D., and Technau,
U. (2017) The evolutionary origin of plant and animal microRNAs,
Nat. Ecol. Evol., 1, 27.
128.Zhou, G., Zhou, Y., and Chen, X. (2017) New
insight into inter-kingdom communication: horizontal transfer of mobile
small RNAs, Front. Microbiol., 8, 768.
129.Singh, N. K. (2017) miRNAs target databases:
developmental methods and target identification techniques with
functional annotations, Cell. Mol. Life Sci., 74,
2239-2261.
130.Bhat, S. S., Jarmolowski, A., and
Szweykowska-Kulinska, Z. (2016) MicroRNA biogenesis: epigenetic
modifications as another layer of complexity in the microRNA expression
regulation, Acta Biochim. Pol., 63, 717-723.
131.Mal, C., Aftabuddin, M., and Kundu, S. (2018)
IIKmTA: inter- and intra-kingdom miRNA-target analyzer, Interdiscip.
Sci., doi: 10.1007/s12539-018-0291-6.
132.Mitchell, P. S., Parkin, R. K., Kroh, E. M.,
Fritz, B. R., Wyma, S. K., Pogosova-Agadjanyan, E. L., Peterson, A.,
Noteboom, J., O’Briant, K. C., Allen, A., Lin, D. W., Urban, N.,
Drescher, C. W., Knudsen, B. S., Stirewalt, D. L., Gentleman, R.,
Vessella, R. L., Nelson, P. S., Martin, D. B., and Tewari, M. (2008)
Circulating microRNAs as stable blood-based markers for cancer
detection, Proc. Natl. Acad. Sci. USA, 105,
10513-10518.
133.Arroyo, J. D., Chevillet, J. R., Kroh, E. M.,
Ruf, I. K., Pritchard, C. C., Gibson, D. F., Mitchell, P. S., Bennett,
C. F., Pogosova-Agadjanyan, E. L., Stirewalt, D. L., Tait, J. F., and
Tewari, M. (2011) Argonaute2 complexes carry a population of
circulating microRNAs independent of vesicles in human plasma, Proc.
Natl. Acad. Sci. USA, 108, 5003-5008.
134.Turchinovich, A., Weiz, L., Langheinz, A., and
Burwinkel, B. (2011) Characterization of extracellular circulating
microRNA, Nucleic Acids Res., 39, 7223-7233.