Downregulation of LINC00894-002 Contributes to Tamoxifen Resistance by Enhancing the TGF-β Signaling Pathway
Xiulei Zhang1, Meiting Wang2,3, Huihui Sun1, Tao Zhu1, and Xiangting Wang1,4,a*
1Department of Cell and Developmental Biology, School of Life Sciences, University of Science and Technology of China, 230026 Hefei, China2College of Liren, Yanshan University, 066004 Qinhuangdao, China
3Department of Neurobiology and Biophysics, School of Life Sciences, University of Science and Technology of China, 230026 Hefei, China
4CAS Center for Excellence in Molecular Cell Science, Chinese Academy of Sciences, 230026 Hefei, China
* To whom correspondence should be addressed.
Received September 4, 2017; Revision received February 9, 2018
Tamoxifen is a widely used personalized medicine for estrogen receptor (ER)-positive breast cancer, but approximately 30% of patients receiving the treatment relapse due to tamoxifen resistance (TamR). Recently, several reports have linked lncRNAs to cancer drug resistance. However, the role of lncRNAs in TamR is unclear. To identify TamR-related lncRNAs, we first used a bioinformatic approach to predict whether they have connection with known TamR-associated genes by starBase v2.0 and divided them into two groups. Group A contains lncRNAs that connect with known TamR genes and group B contains lncRNAs that show no predicted interaction. Among the 12 lncRNAs in group A, 58.3% of them are either up- or downregulated in MCF-7/TamR cells compared to the sensitive cells. In contrast, the expression levels of all group B lncRNAs are not changed in MCF-7/TamR cells. LINC00894-002 exhibits the most sophisticated network pattern and is the most downregulated lncRNA in MCF-7/TamR cells. Moreover, we find that LINC00894-002 is directly upregulated by ERα. Knocking down LINC00894-002 downregulates expression of miR-200a-3p and miR-200b-3p, upregulates the expression of TGF-β2 and ZEB1, and finally contributes to TamR. Herein, we report the first case of an inhibitory lncRNA against TamR through the miR-200-TGF-β2-ZEB1 signaling pathway.
KEY WORDS: tamoxifen resistance, estrogen receptor, breast cancer, LINC00894-002, miR-200-TGF-β2-ZEB1DOI: 10.1134/S0006297918050139
Abbreviations: ASO, antisense oligonucleotide; ChIP, chromatin immunoprecipitation; ER, estrogen receptor; lncRNAs, long noncoding RNAs; SRB, sulforhodamine B; TamR, tamoxifen resistance.
Breast cancer is of high incidence and is the leading cause of cancer
death among females, with an estimated 1.7 million cases and 521,900
deaths worldwide in 2012 [1]. Tamoxifen, a
selective estrogen receptor (ER) modulator, is the gold standard for
treating ER-positive breast cancer. However, roughly 75% of all breast
cancer cases are ER-positive and up to 30% of ER-positive breast cancer
patients receiving tamoxifen treatment relapse due to tamoxifen
resistance (TamR) [2]. Multiple mechanisms have
been shown to contribute to TamR. Patients with higher levels of ER
correlate with better response to tamoxifen therapy, and lack of ER
expression contributes to TamR [3]. The TGF-β
signaling pathway can promote various kinds of chemoresistance through
enhancing cell proliferation, migration, and invasion by inducing
epithelial–mesenchymal transition [4].
MicroRNAs (miRNAs) are also involved in TamR. For example, miR-200 can
sensitize tamoxifen response through directly targeting multiple
TGF-β signaling molecules [5].
Long noncoding RNAs (lncRNAs) are a class of newly identified noncoding transcripts that are usually composed of more than 200 nucleotides [6]. Recently, lncRNAs have been shown to be a novel class of regulatory molecules in drug resistance. HOX antisense intergenic RNA (HOTAIR) is transcribed from the antisense strand of the HOXC locus and is a most upregulated lncRNA in breast cancer. HOTAIR has been found to be able to promote multiple drug resistance events. In human lung adenocarcinoma cells, HOTAIR contributes to cisplatin resistance via downregulation of p21WAF1/CIP1 expression [7]. During the preparation of this manuscript, HOTAIR was reported as a promoter for TamR through enhancing ER signaling in breast cancer [8]. Besides, breast cancer antiestrogen resistance 4 (BCAR4), urothelial carcinoma-associated 1 (UCA1), lncRNA-ROR (ROR, regulator of reprogramming), colon cancer associated transcript 2 (CCAT2), and DSCAM-AS1 are also reported to enhance TamR [9-13]. However, the overall roles of lncRNAs in TamR remain to be discovered.
In this work, we first used a bioinformatic approach to establish the connection of lncRNA with TamR-related mRNAs and miRNAs by starBase v2.0 [14]. Among the 16 tested lncRNAs, LINC00894-002 (ENST00000444489), a lncRNA derived from X chromosome, shows the most sophisticated pattern in network predication and is the most downregulated lncRNA in tamoxifen-resistant MCF-7 cells (MCF-7/TamR) versus sensitive MCF-7/WT cells. Further studies showed that LINC00894-002 posessed an inhibitory effect on TamR. Moreover, LINC00894-002 is directly upregulated by ERα and positively regulates the expression levels of miR-200a-3p and miR-200b-3p, which inhibits the downstream TGF-β2-ZEB1 signaling pathway. To the best of our knowledge, this is the first reported inhibitory lncRNA against TamR. Our data suggest that the inhibitory role of LINC00894-002 may be achieved via the miR-200-TGF-β2-ZEB1 signaling pathway.
MATERIALS AND METHODS
Cell lines, cell culture, and reagents. The authentication of MCF-7/WT and MCF-7/TamR cells by STR profile analysis was performed by a third party (Genewiz, China) (data are available upon request). MCF-7/TamR cell line was derived from its parental cell line MCF-7/WT continuously exposed to tamoxifen (1 μM) (Sigma, USA) [15]. Both cell lines were cultured in RPMI 1640 (Invitrogen, USA) medium with 2 g/liter NaHCO3 (Sigma), 10% fetal bovine serum (FBS) (Invitrogen), and 1% penicillin-streptomycin (WISENT, China). To maintain the resistance of MCF-7/TamR, it was continuously cultured with 1 μM tamoxifen.
Sulforhodamine B (SRB) assay. The SRB assay was modified according to the method described by Pauwels et al. [16]. Briefly, 3000 cells/well were seeded in 96-well plates for 24 h before receiving vehicle or tamoxifen treatment. After 96 h, the culture medium was aspirated, and the remaining cells were fixed with 10% trichloroacetic acid (TCA) at 4°C (200 µl/well) for 1 h. The cells were then washed five times with 100 µl deionized water and left to air-dry at room temperature. The cells were then stained with 100 µl 0.4% SRB (Sigma) at 37°C for 30 min and subsequently washed five times with 1% acetic acid to remove unbound stain. Lastly, the plates were dried at 37°C, and the bound protein stain was solubilized with 100 µl of 10 mM unbuffered Tris-base on a shaker for 30 min. Afterwards, the 96-well plates were transferred to a CLARIOstar microplate reader (BMG LABTECH, Germany) to detect optical density (OD) at 515 nm. The formula below was used to calculate the cell growth rate [17]:
cell growth rate (%) = [(mean ODtamoxifen – mean OD24h)/(mean ODvehicle – mean OD24h)] × 100%.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR). Total RNA was extracted from cultured cells using TRIzol reagent (Invitrogen) followed with DNase I (Thermo Scientific, USA) treatment. The RNAs were then reverse-transcribed with HiScript II 1st Strand cDNA Synthesis Kit (Vazyme, China), followed by qPCR on a LightCycler® 96 System (Roche, Switzerland) using SYBR Green master mix (Vazyme) and specific primers (GENERAL BIOSYSTEMS, China) (primers are listed in Table S1; see Supplement to this paper on the site of the journal http://protein.bio.msu.ru/biokhimiya and Springer site Link.springer.com). MiRNAs were reverse-transcribed and detected by the All-in-OneTM miRNA qRT-PCR Detection System (GeneCopoeia, USA) following the manufacturer’s instructions.
Nucleus–cytoplasm segregation experiment. MCF-7/TamR cells were lysed in 500 µl RSB-100 buffer (100 mM Tris-HCl, 100 mM NaCl, 2.5 mM MgCl2, 40 μg/ml digitonin, pH 7.4) on ice for 8 min and then centrifuged at 2000g for 8 min. The supernatant fraction was collected as the cytoplasm fraction. The pellet was then resuspended in 300 µl RSB-100T (RSB-100 with 0.5% Triton X-100) on ice for 8 min and centrifuged at 2000g for 8 min; the precipitate was collected as the nucleus fraction. The cytoplasm and nucleus fractions were used for RNA extraction following procedures described above.
Western blotting. Proteins were extracted from cultured cells using TRIzol reagent following the manufacturer’s instructions. Equal (by protein content) aliquots from each sample were separated by SDS-PAGE and transferred to PVDF membranes (Merck, Germany). Subsequently, the membranes were blocked in 5% milk/1× TBS for 1 h and then incubated with the primary antibody against GAPDH (Proteintech, USA) overnight. After a thorough wash, the membranes were incubated with rabbit anti-mouse horseradish peroxidase-conjugated secondary antibody (Proteintech) for 1 h. Finally, after washing, bands were visualized by ECL detection reagent (Thermo Scientific) on a UVP device (Analytik Jena AG, Germany).
Antisense oligonucleotide (ASO) transfection. MCF-7/TamR cells (3·105 per well) were seeded in 6-well plates for 12 h and then starved for 12 h before transfection. The ASO specifically targeting LINC00894-002, lnc-ATB, or the negative control (Ribobio, China) were transfected at a final concentration of 50 nM by Lipofectamine® 3000 (Invitrogen) following the manufacturer’s instructions. The target sequences of ASO against LINC00894-002 and lnc-ATB for transfection were, respectively, GAAGTGTGTGTGGCTGGAAG and TATGGCCTAGATTACCTTTCCA.
Chromatin immunoprecipitation (ChIP). First, 107 cells were cross-linked with 1% formaldehyde at room temperature (~25°C) for 10 min and neutralized with 0.125 M glycine for 5 min. After sonication, soluble chromatin was incubated with 2 μg ERα antibody (Abcam, UK) at 4°C overnight. Immunoprecipitated complexes were collected using Protein A/G PLUS-Agarose (Santa Cruz Biotechnology, USA). Subsequently, immuno-complexes were washed, and DNA was extracted and purified using an EasyPure Genomic DNA Kit (TransGene Biotech, China). The extracted DNA was finally detected by RT-qPCR.
Establishment of interaction network (see scheme below).
(1) Building tamoxifen resistance related factor pool. First, we used “tamoxifen resistance” as a keyword to search abstracts from the Pubmed website (http://www.ncbi.nlm.nih.gov/pubmed/) and downloaded them as tamoxifen resistance (TamR) related factor pool. Then, we analyzed sentences of each abstracts and saved the demonstrated TamR-related mRNAs and miRNAs.
(2) Downloading starBase v2.0 interaction pairs list. We downloaded miRNA–lncRNA and mRNA–miRNA interaction list for building an interaction network from the starBase v2.0 website (http://starbase.sysu.edu.cn/index.php).
(3) mRNA–miRNA–lncRNA interaction network. We selected 16 misregulated lncRNAs in TamR from our in-house microarray analysis, analyzed their potentially related miRNAs from the miRNA–lncRNA list, and used the TamR-related miRNAs to filter the lncRNAs list. The other filter was TamR-related mRNAs and their potentially related miRNAs from the mRNA–miRNA list, and we used this filter to establish an mRNA–miRNA–lncRNA interaction network. In the end, we merged the results from the above two filters and used the Cytoscape software to draw the interaction network.
Scheme
Statistical analysis. Data were expressed as mean ± SEM and qPCR data were normalized to 18S. All statistical analyses were performed using the GraphPad Prism 6 software (GraphPad Software Inc, USA). For comparisons, Student’s t-test (two-tailed) was performed as indicated; * p < 0.05 was considered significant. The IC50 (half-maximal inhibitory concentration) was calculated by SPSS Statistics 22 (IBM, USA).
RESULTS
Established interaction network between lncRNA and known TamR-related genes. Numerous lncRNAs were either up- or downregulated in MCF-7/TamR cells when compared with MCF-7/WT cells by microarray analysis [8]. To narrow the targeted lncRNAs for experimental test, we designed a bioinformatic approach. First, we generated a TamR-related genes list including 80 mRNAs and 11 miRNAs from published literature (see Table S2 in Supplement) and then downloaded the lncRNA–miRNA and miRNA–mRNA interaction list from starBase v2.0 [14]. Next, we randomly selected 16 lncRNAs from our in-house microarray analysis (data not shown) to establish lncRNA–miRNA–mRNA interaction network. We found 12 lncRNAs were involved in the TamR network, namely AC005076.5, AC016747.3, CTD-2201E18.3, HOTAIRM1, LINC00263, LINC00894-002, RP11-37B2.1, RP11-65F13.2, RP11-267N12.3, RP11-457M11.5, RP11-845C23.2, and RUSC1-AS1. These 12 lncRNAs were referred as group A in the following connection network (see Fig. S1 in Supplement). The other four lncRNAs – AC068491.1, RSBN1L-AS1, RP11-319G9.5 and MIR22HG – were not involved in the TamR network and were referred to as group B in the following context.
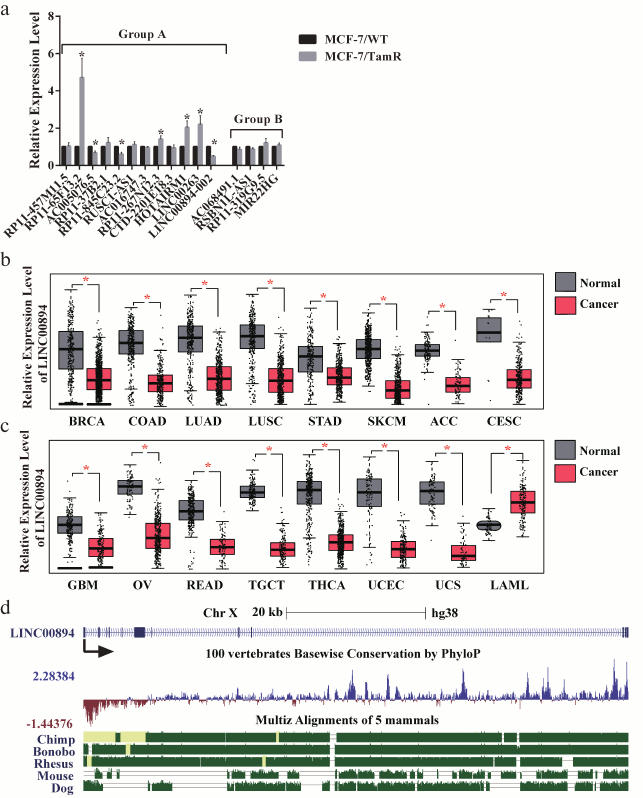
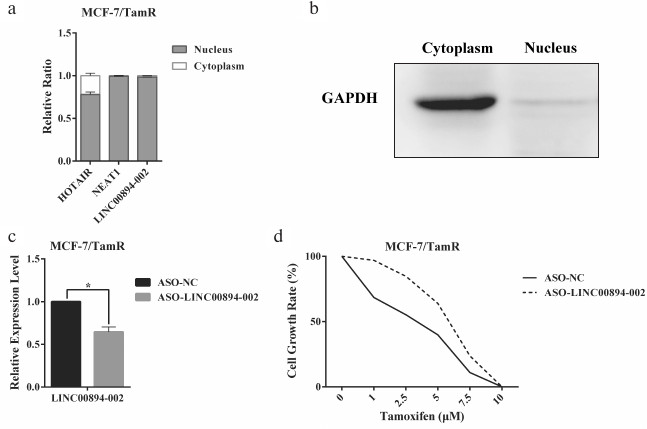
Identification of LINC00894-002 as an inhibitory lncRNA against TamR. The expression level changes of the 16 lncRNAs in MCF-7/WT and MCF-7/TamR cells were detected by RT-qPCR. Among the 12 lncRNAs in group A, seven lncRNAs were shown to be misregulated in TamR cells, including four upregulated and three downregulated lncRNAs (Fig. 1a). In contrast, none of the four negative lncRNAs in group B were changed (Fig. 1a). LINC00894-002 was shown to be the most downregulated lncRNA (Fig. 1a). Interestingly, LINC00894-002 also exhibited the most sophisticated pattern in terms of interaction network (Fig. S1, panel (l)). Coding potential analysis by the LNCipedia website demonstrated its noncoding feature [18]. We mined data from the GEPIA website and found LINC00894 was significantly downregulated in 15 kinds of cancer tissues including breast invasive cancer (BRCA), except that it was significantly upregulated in LAML (acute myeloid leukemia) (Fig. 1, b and c) [19], which indicated that it may act as a tumor-suppressor gene. The sequence of LINC00894-002 orthologs were obviously conserved in primates but less well conserved in other animals such as mouse and dog (Fig. 1d) (data from UCSC Genome browser). Further subcellular fractionation assay showed that LINC00894-002 was located in the nucleus (Fig. 2, a and b). For the functional verification and working mechanism identification, we designed the specific ASO for the knocking down experiments in MCF-7/TamR cells [20]. To test whether LINC00894-002 could affect TamR, we performed sulforhodamine B (SRB) cell proliferation assay after knocking it down. The results showed that the IC50 of transfected cells increased from 2.435 μM in the presence of ASO-NC to 5.361 μM when transfected with ASO-LINC00894-002 (Fig. 2, c and d). Our data suggested that knocking down of LINC00894-002 enhanced TamR. LINC00894-002 was an inhibitory factor against TamR in breast cancer.
Fig. 1. Characteristics of LINC00894-002. a) Expression level changes of 16 tested lncRNAs in MCF-7/WT and MCF-7/TamR cells. Sixteen lncRNAs were analyzed for their interaction network with miRNA/TamR and mRNA/TamR, and their expression levels in MCF-7/WT and MCF-7/TamR cells were measured by RT-qPCR (n = 5). Group A, lncRNAs that positively interacted with examined miRNA/TamR and mRNA/TamR (Fig. S1, a-l); group B, lncRNAs that showed no interaction. b, c) Relative expression levels of LINC00894 in different normal and cancer tissues. LINC00894 was significantly downregulated in breast invasive cancer (BRCA), colon adenocarcinoma (COAD), lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), stomach adenocarcinoma (STAD), skin cutaneous melanoma (SKCM), adrenocortical carcinoma (ACC), cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC), glioblastoma multiforme (GBM), ovarian serous cystadenocarcinoma (OV), rectum adenocarcinoma (READ), testicular germ cell tumors (TGCT), thyroid carcinoma (THCA), uterine corpus endometrial carcinoma (UCEC), and uterine carcinosarcoma (UCS), but significantly upregulated in acute myeloid leukemia (LAML) (data mined from GEPIA website). d) UCSC genome browser view of LINC00894 locus.
Fig. 2. LINC00894-002 acts as an inhibitory lncRNA against TamR in MCF-7/TamR cells. a, b) Subcellular location of LINC00894-002. Cytoplasm and nucleus were segregated by subcellular fractioning, and the expression levels of indicated lncRNAs in separate fractions of MCF-7/TamR cells were measured by RT-qPCR (n = 3). a) HOTAIR and NEAT1 were used as nucleus-associated RNA control. b) The expression level of GAPDH in cytoplasm and nucleus fractions was detected by Western blot, and it was used as cytoplasm-associated protein. c) Knocking down efficiency of LINC00894-002 ASO. After transfection of MCF-7/TamR cells with ASO-LINC00894-002 and ASO-NC, the expression level of LINC00894-002 was measured by RT-qPCR. d) Representative data of the cell growth rate of MCF-7/TamR cells. The cell growth rate of MCF-7/TamR cells transfected with ASO-LINC00894-002 and ASO-NC was determined by SRB assays.
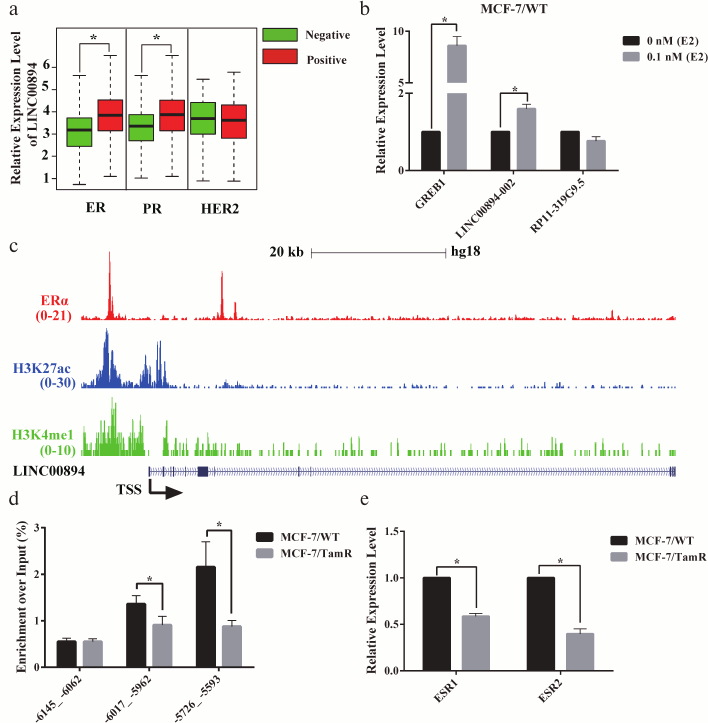
LINC00894-002 is directly up-regulated by estrogen receptor (ER). HOTAIR and DSCAM-AS1 were two lncRNAs that were directly regulated by ERα and contributed to TamR [8, 13]. We therefore hypothesized whether ERα may be also an upstream signal for LINC00894-002. To test our hypothesis, we analyzed the correlation between the expression level of LINC00894 and receptors status of breast cancer from TANRIC website and found LINC00894 was positively related with ER and progesterone receptor (PR), but no relation with human epithelial growth factor receptor 2 (HER2) (Fig. 3a) [21]. To further test our hypothesis, we treated MCF-7/WT cells with β-estradiol (E2) to detect whether LINC00894-002 could be induced by E2. The following RT-qPCR results showed that GREB1, a well-known E2-responsive gene, and LINC00894-002 were up-regulated after E2 induction, while RP13-319G9.5, a lncRNA in group B, was not induced (Fig. 3b). To verify whether E2 up-regulated expression of LINC00894-002 through direct ERα binding on its regulatory elements, we analyzed a published ERα, H3K27ac and H3K4me1 ChIP-seq data (data from GSE45822) and found ERα indeed occupied the regulatory region of the LINC00894 gene. Moreover, this region was strongly occupied by H3K27ac and H3K4me1, suggesting LINC00894 being in an active status (Fig. 3c and Fig. S2 in Supplement). Furthermore, our ERα ChIP-qPCR results also showed LINC00894-002 was directly regulated by ERα on the –6 kb ~ –5.5 kb genomic region upstream to the transcription start site of it (Fig. 3d). It has been reported that ERα expression was down-regulated in certain TamR cell lines [3]. Our RT-qPCR results demonstrated that ESR1 and ESR2 were down-regulated in MCF-7/TamR cells (Fig. 3e), and our ERα ChIP-qPCR results showed ERα occupancy was significantly reduced in MCF-7/TamR cells when compared with parental MCF-7/WT cells (Fig. 3d), which explained the low expression level of LINC00894-002 (Fig. 1a). These data indicated that LINC00894-002 was directly up-regulated by ERα, and the low expression level of LINC00894-002 in TamR cells was likely due to the down-regulation of ESR1.
Fig. 3. LINC00894-002 is directly up-regulated by estrogen receptor (ER). a) The correlation between LINC00894 and receptor status of breast cancer (data mined from TANRIC website and presented as boxplot). b) LINC00894-002 was up-regulated by β-estradiol (E2). MCF-7/WT cells were hormone-starved for 3 days, followed with 0.1 nM β-estradiol treatment for 6 h. Then the expression changes of GREB1 (E2-response marker), LINC00894-002 and RP11-319G9.5 (a lncRNA showed no interaction with miRNA/TamR and mRNA/TamR) were measured by RT-qPCR (n = 3). c) UCSC Genome Browser view of ERα (red), H3K27ac (blue) and H3K4me1 (green) binding events at the genomic region of the LINC00894 (data from GSE45822; the arrow showed the transcriptional direction). d) ERα ChIP-qPCR experiment was performed on MCF-7/WT and MCF-7/TamR cells, and qPCR primer pairs were designed from the ERα binding peak regions (–6017_ –5962, –5726_ –5593) and no binding peak region (–6145_ –6062) (n = 3). e) The expression levels of ESR1 and ESR2 in MCF-7/WT and MCF-7/TamR cells were measured by RT-qPCR (n = 4).
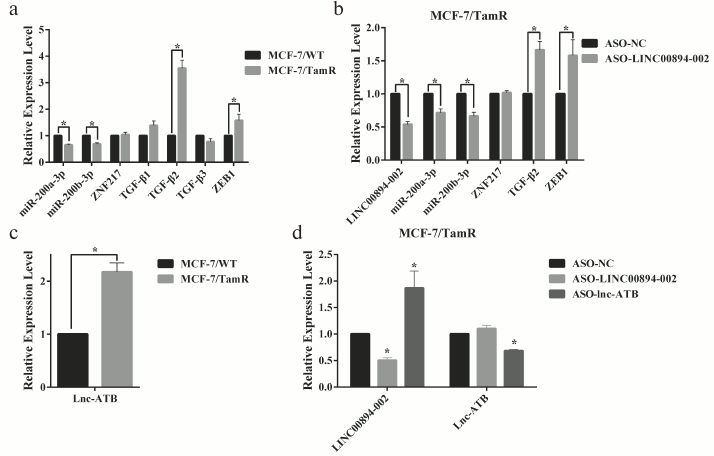
Downregulation of LINC00894-002 contributes to TamR via miR-200-TGF-β2-ZEB1 signaling pathway. The miR-200-TGF-β2-ZEB1 pathway was reported to participate in TamR [5, 22, 23]. Our interaction network of LINC00894-002 also indicated a predicted connection of this lncRNA with miR-200, TGF-β2, and ZEB1 (Fig. S1, panel (l)). Moreover, in our MCF-7/WT and TamR cells, LINC00894-002 positively correlated with miR-200a-3p and miR-200b-3p, but negatively correlated with TGF-β2 and ZEB1 (Fig. 4a). To test whether LINC894-002 was an upstream signal for the miR-200-TGF-β2-ZEB1 pathway, we performed RT-qPCR assay after knocking down the LINC00894-002. Our results showed that LINC00894-002 ASO downregulated miR-200a-3p and miR-200b-3p, whereas it upregulated TGF-β2 and ZEB1 (Fig. 4b). Lnc-ATB, a TGF-β-signaling upregulated lncRNA, could enhance TGF-β-ZEB1 signaling pathway by competitively binding the miR-200 family [24]. Interestingly, we found lnc-ATB was upregulated in TamR cells (Fig. 4c) and could inhibit the expression of LINC00894-002 but was not affected by downregulation of LINC00894-002 (Fig. 4d), which suggested that lnc-ATB acted as an inhibitory upstream signal for LINC00894-002. These results indicated downregulation of LINC00894-002 contributed to TamR via miR-200-TGF-β2-ZEB1 signaling pathway.
Fig. 4. LINC00894-002 inhibits TamR via miR-200-TGF-β2-ZEB1 signaling pathway. a) The expression levels of involved RNAs in miR-200-TGF-β2-ZEB1 signaling pathway in MCF-7/WT and MCF-7/TamR cells were measured by RT-qPCR (n = 4). b) The expression level changes of involved RNAs in miR-200-TGF-β2-ZEB1 signaling pathway in MCF-7/TamR cells after transfection with ASO-LINC00894-002 and ASO-NC (n = 4). c) RT-qPCR analysis of expression level of lnc-ATB in TamR cells (n = 3). d) The expression level changes of LINC00894-002 and lnc-ATB after MCF-7/TamR cells respectively transfected with LINC00894-002 ASO and lnc-ATB ASO (n = 3).
DISCUSSION
LncRNAs play critical roles in numerous physiological and pathophysiological processes [25]. However, the discovery and study of lncRNAs remain hampered by the lack of an effective prediction method from large scale screening data. In this study, we used the starBase v2.0 to facilitate the selection of most possible TamR-related lncRNAs by establishing lncRNA–miRNA–mRNA interaction network [14]. This bioinformatic prediction allows us to identify a lncRNA, LINC00894-002, which acts as an inhibitory lncRNA against TamR. Thus, our method is instructive to establish interaction network between lncRNAs from a large-scale dataset with known genes in the desired function for further investigation.
In the study of the function and working mechanism of lncRNAs in breast cancer, only GAS5, XIST and NKILA exert tumor suppressive function [26]. LINC00894 is a significantly downregulated gene in many cancer tissues, thus it may act as a tumor suppressor. In the study of TamR, tamoxifen inhibits the expression of LINC00894-002, and in turn LINC00894-002 attenuates TamR. HOTAIR, UCA1, lncRNA-ROR, CCAT2 and DSCAM-AS1 are reported to enhance TamR [8-13], and LINC00894-002 is the first inhibitory lncRNA against TamR. The study of lncRNA on cancer and TamR brings a new therapeutic concept – lncRNA medicine, which may further promote the treatment of disease.
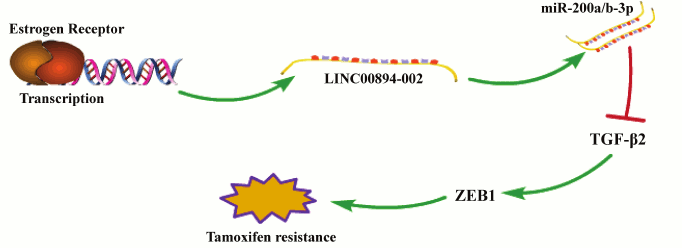
ER and TGF-β signaling pathways are two crucial pathways in the development of TamR and can act in crosstalk. For instance, ER signaling can attenuate TGF-β signaling by suppression of transcriptional activity of Smad3, whereas TGF-β signaling can activate ER signaling [27]. In this study, we identified an ER-enhancing lncRNA that can mediate TamR through negatively regulating the TGF-β2-ZEB1 signaling pathway (Fig. 5). Our findings indicate that LINC00894-002 may act as a bridge for connecting the inhibitory crosstalk between ER and TGF-β signaling pathway in chemoresistance. In our study, we demonstrate that LINC00894-002 positively regulates the expression level of miR-200a-3p and miR-200b-3p, and for the location of LINC00894-002 in the nucleus, we hypothesize that it may participate in the maturation process of miRNA. Our further results show that downregulation of LINC00894-002 downregulates the expression of DROSHA and DICER1, which are key factors in miRNA maturation (data not shown) [28]. It would be worthwhile to further study LINC00894-002, for it might be a novel TamR biomarker and potential therapeutic target for breast cancer.
Fig. 5. A schematic model of the working mechanism of LINC00894-002 in TamR. Estrogen receptor (ER) directly promotes the transcription of LINC00894-002, and LINC00894-002 inhibits TamR by the miR-200-TGF-β2-ZEB1 signaling pathway.
Acknowledgments
We appreciate the bioinformatic instruction from Dr. Ao Li (University of Science and Technology of China) and technical support from the Core Facility Center for Life Sciences (University of Science and Technology of China).
This work was supported by the National Natural Science Foundation of China (31471226, 91440108) and the Fundamental Research Funds for The Central Universities (WK2070000044, WK2070000034).
REFERENCES
1.Torre, L. A., Bray, F., Siegel, R. L., Ferlay, J.,
Lortet-Tieulent, J., and Jemal, A. (2015) Global cancer statistics,
2012, CA Cancer J. Clin., 65, 87-108.
2.Criscitiello, C., Fumagalli, D., Saini, K. S., and
Loi, S. (2010) Tamoxifen in early-stage estrogen receptor-positive
breast cancer: overview of clinical use and molecular biomarkers for
patient selection, Onco Targets Ther., 4, 1-11.
3.Garcia-Becerra, R., Santos, N., Diaz, L., and
Camacho, J. (2012) Mechanisms of resistance to endocrine therapy in
breast cancer: focus on signaling pathways, miRNAs and genetically
based resistance, Int. J. Mol. Sci., 14, 108-145.
4.Xia, H., and Hui, K. M. (2014) Mechanism of cancer
drug resistance and the involvement of noncoding RNAs, Curr. Med.
Chem., 21, 3029-3041.
5.Manavalan, T. T., Teng, Y., Litchfield, L. M.,
Muluhngwi, P., Al-Rayyan, N., and Klinge, C. M. (2013) Reduced
expression of miR-200 family members contributes to antiestrogen
resistance in LY2 human breast cancer cells, PLoS One.,
8, e62334.
6.Gibb, E. A., Brown, C. J., and Lam, W. L. (2011)
The functional role of long non-coding RNA in human carcinomas, Mol.
Cancer, 10, 1.
7.Liu, Z., Sun, M., Lu, K., Liu, J., Zhang, M., Wu,
W., De, W., Wang, Z., and Wang, R. (2013) The long noncoding RNA HOTAIR
contributes to cisplatin resistance of human lung adenocarcinoma cells
via downregualtion of p21 WAF1/CIP1 expression, PloS One,
8, e77293.
8.Xue, X., Yang, Y. A., Zhang, A., Fong, K. W., Kim,
J., Song, B., Li, S., Zhao, J. C., and Yu, J. (2016) LncRNA HOTAIR
enhances ER signaling and confers tamoxifen resistance in breast
cancer, Oncogene, 35, 2746-2755.
9.Godinho, M., Meijer, D., Setyono-Han, B., Dorssers,
L. C., and van Agthoven, T. (2011) Characterization of BCAR4, a novel
oncogene causing endocrine resistance in human breast cancer cells,
J. Cell. Physiol., 226, 1741-1749.
10.Li, X., Wu, Y., Liu, A., and Tang, X. (2016) Long
non-coding RNA UCA1 enhances tamoxifen resistance in breast cancer
cells through a miR-18a-HIF1alpha feedback regulatory loop, Tumour
Biol., 37, 14733-14743.
11.Zhang, H. Y., Liang, F., Zhang, J. W., Wang, F.,
Wang, L., and Kang, X. G. (2017) Effects of long noncoding
RNA–ROR on tamoxifen resistance of breast cancer cells by
regulating microRNA-205, Cancer Chemother. Pharmacol.,
79, 327-337.
12.Caia, Y., He, J., and Zhang, D. (2016)
Suppression of long non-coding RNA CCAT2 improves tamoxifen-resistant
breast cancer cells’ response to tamoxifen, Mol. Biol.,
50, 725-730.
13.Niknafs, Y. S., Han, S., Ma, T., Speers, C.,
Zhang, C., Wilder-Romans, K., Iyer, M. K., Pitchiaya, S., Malik, R.,
Hosono, Y., Prensner, J. R., Poliakov, A., Singhal, U., Xiao, L.,
Kregel, S., Siebenaler, R. F., Zhao, S. G., Uhl, M., Gawronski, A.,
Hayes, D. F., Pierce, L. J., Cao, X., Collins, C., Backofen, R.,
Sahinalp, C. S., Rae, J. M., Chinnaiyan, A. M., and Feng, F. Y. (2016)
The lncRNA landscape of breast cancer reveals a role for DSCAM-AS1 in
breast cancer progression, Nat. Commun., 7, 12791.
14.Li, J. H., Liu, S., Zhou, H., Qu, L. H., and
Yang, J. H. (2014) starBase v2.0: decoding miRNA–ceRNA,
miRNA–ncRNA and protein–RNA interaction networks from
large-scale CLIP-Seq data, Nucleic Acids Res., 42,
D92-97.
15.Lu, M., Ding, K., Zhang, G., Yin, M., Yao, G.,
Tian, H., Lian, J., Liu, L., Liang, M., Zhu, T., and Sun, F. (2015)
MicroRNA-320a sensitizes tamoxifen-resistant breast cancer cells to
tamoxifen by targeting ARPP-19 and ERRgamma, Sci. Rep.,
5, 8735.
16.Pauwels, B., Korst, A. E., de Pooter, C. M.,
Pattyn, G. G., Lambrechts, H. A., Baay, M. F., Lardon, F., and
Vermorken, J. B. (2003) Comparison of the sulforhodamine B assay and
the clonogenic assay for in vitro chemoradiation studies,
Cancer Chemother. Pharmacol., 51, 221-226.
17.Vichai, V., and Kirtikara, K. (2006)
Sulforhodamine B colorimetric assay for cytotoxicity screening, Nat.
Protoc., 1, 1112-1116.
18.Volders, P.-J., Helsens, K., Wang, X., Menten,
B., Martens, L., Gevaert, K., Vandesompele, J., and Mestdagh, P. (2013)
LNCipedia: a database for annotated human lncRNA transcript sequences
and structures, Nucleic Acids Res., 41, D246-D251.
19.Tang, Z., Li, C., Kang, B., Gao, G., Li, C., and
Zhang, Z. (2017) GEPIA: a web server for cancer and normal gene
expression profiling and interactive analyses, Nucleic Acids
Res., doi: 10.1093/nar/gkx247 [Epub ahead of print].
20.Dias, N., and Stein, C. (2002) Antisense
oligonucleotides: basic concepts and mechanisms, Mol. Canc.
Ther., 1, 347-355.
21.Li, J., Han, L., Roebuck, P., Diao, L., Liu, L.,
Yuan, Y., Weinstein, J. N., and Liang, H. (2015) TANRIC: an interactive
open platform to explore the function of lncRNAs in cancer, Cancer
Res., 75, 3728-3737.
22.MacCallum, J., Keen, J., Bartlett, J., Thompson,
A., Dixon, J., and Miller, W. (1996) Changes in expression of
transforming growth factor beta mRNA isoforms in patients undergoing
tamoxifen therapy, Br. J. Cancer, 74, 474.
23.Arteaga, C. L., Koli, K. M., Dugger, T. C., and
Clarke, R. (1999) Reversal of tamoxifen resistance of human breast
carcinomas in vivo by neutralizing antibodies to transforming
growth factor-β, J. Natl. Cancer Inst., 91,
46-53.
24.Yuan, J-h, Yang, F., Wang, F., Ma, J-z, Guo, Y-j,
Tao, Q-f, Liu, F., Pan, W., Wang, T-t, and Zhou, C-c. (2014) A long
noncoding RNA activated by TGF-β promotes the invasion-metastasis
cascade in hepatocellular carcinoma, Cancer Cell, 25,
666-681.
25.Wilusz, J. E., Sunwoo, H., and Spector, D. L.
(2009) Long noncoding RNAs: functional surprises from the RNA world,
Genes Dev., 23, 1494-1504.
26.Wang, J., Ye, C., Xiong, H., Shen, Y., Lu, Y.,
Zhou, J., and Wang, L. (2016) Dysregulation of long non-coding RNA in
breast cancer: an overview of mechanism and clinical implication,
Oncotarget, 8, 5508-5522.
27.Buck, M. B., and Knabbe, C. (2006) TGF-beta
signaling in breast cancer, Ann. N. Y. Acad. Sci., 1089,
119-126.
28.Lin, S., and Gregory, R. I. (2015) MicroRNA
biogenesis pathways in cancer, Nat. Rev. Cancer,
15, 321-333.
Supplementary Figures and Tables (PDF)