O-Antigens of Escherichia coli Strains O81 and HS3-104 Are Structurally and Genetically Related, Except O-Antigen Glucosylation in E. coli HS3-104
E. L. Zdorovenko1, Y. Wang2, A. S. Shashkov1, T. Chen2, O. G. Ovchinnikova1, B. Liu2,a, A. K. Golomidova3, V. V. Babenko4,b, A. V. Letarov3,c, and Y. A. Knirel1,d*
1Zelinsky Institute of Organic Chemistry, Russian Academy of Sciences, 119991 Moscow, Russia2TEDA Institute of Biological Sciences and Biotechnology, Nankai University, TEDA, 300457 Tianjin, China
3Winogradsky Institute of Microbiology, Research Center of Biotechnology, Russian Academy of Sciences, 117312 Moscow, Russia
4Federal Research and Clinical Centre of Physico-Chemical Medicine, 119435 Moscow, Russia
* To whom correspondence should be addressed.
Received October 8, 2017; Revision received November 22, 2017
Glycerophosphate-containing O-specific polysaccharides (OPSs) were obtained by mild acidic degradation of lipopolysaccharides isolated from Escherichia coli type strain O81 and E. coli strain HS3-104 from horse feces. The structures of both OPSs and of the oligosaccharide derived from the strain O81 OPS by treatment with 48% HF were studied by monosaccharide analysis and one- and two-dimensional 1H- and 13C-NMR spectroscopy. Both OPSs had similar structures and differed only in the presence of a side-chain glucose residue in the strain HS3-104 OPS. The genes and the organization of the O-antigen biosynthesis gene cluster in both strains are almost identical with the exception of the gtr gene cluster responsible for glucosylations in the strain HS3-104, which is located elsewhere in the genome.
KEY WORDS: lipopolysaccharide, O-specific polysaccharide, bacterial polysaccharide structure, O-antigen gene cluster, glucosylation, Escherichia coliDOI: 10.1134/S0006297918050061
Abbreviations: bp, base pair; COSY, correlation spectroscopy; GC, gas chromatography; Gro, glycerol; HMBC, heteronuclear multiple-bond correlation; HSQC, heteronuclear single quantum correlation; OPS, O-specific polysaccharide; ROESY, rotating-frame Overhauser effect spectroscopy; TOCSY, total correlation spectroscopy; Und-P, undecaprenyl phosphate.
Escherichia coli is a dominant facultative anaerobe of the gut
microbiome in many mammals, including humans. Gut microbiome also
contains numerous conditionally pathogenic strains [1]. The O-polysaccharide (O-antigen) is a component of
the cell wall outer membrane lipopolysaccharide (LPS) in Gram-negative
bacteria. It consists of oligosaccharide repeats (O-units) containing
two to eight residues of common or rarely occurring sugars and their
derivatives. The O-antigen is one of the most variable cell
constituents, with variation in the types of sugars present, their
arrangement within the O-unit, and the type of bonds in and between the
O-units, thus providing the basis for serotyping of bacteria [2]. Being exposed on the cell surface, the O-antigen
is a subject of intense selective pressure by the host immune system
and bacteriophages [3].
Genes involved in the O-antigen biosynthesis in E. coli and some related bacteria are usually clustered in a chromosomal locus located between two conserved genes, galF and gnd [4]. Most of these genes fall into one of the three major classes: genes for the synthesis of nucleotide-activated sugars, genes for glycosyl transferases involved in the formation of bonds between the sugars, and O-unit processing genes [5].
More than 180 O-serogroups of E. coli are internationally acknowledged (http://www.ssidiagnostica.com/e-coli-strains/). Their O-antigen gene clusters have been sequenced [6], and >140 O-polysaccharide (OPS) structures have been determined (http://nevyn.organ.su.se/ECODAB/; http://csdb.glycoscience.ru/bacterial/). The goal of our investigation is the development of a chemical basis for classification of E. coli strains and molecular diagnostics of infectious diseases caused by these bacteria. For this purpose, it is necessary to elucidate the structures of OPSs in type strains of all E. coli O-serogroups and to annotate genes in the O-antigen gene clusters of these strains, taking into account the OPS structures determined.
In this work, we deciphered the structures of the OPSs from the earlier unstudied E. coli serogroup O81 and of the structurally related OPS from the E. coli strain HS3-104. The O81-antigen gene cluster was present in both strains and was consistent with the E. coli O81 OPS structure also typical for the OPS main chain in the HS3-104 strain. OPS from the HS3-104 strain is modified by the side-chain glucosylation; enzymes catalyzing this process are located outside of the O-antigen gene cluster.
MATERIALS AND METHODS
Bacterial strains, cell cultivation, and isolation of lipopolysaccharides. Escherichia coli O81 type strain (laboratory stock G1217) was from the Institute of Medical and Veterinary Science, Adelaide, Australia. Escherichia coli HS3-104 strain was isolated from horse feces [7]. The bacterial cells were grown to the late log phase in 8 liters of Luria–Bertani broth (pH 7.0) with constant aeration at 37°C and then washed and dried as described in [8]. The cells were extracted by the phenol–water method [9]; LPS preparations were obtained as described earlier [10].
OPS isolation. LPSs were hydrolyzed with 2% acetic acid aqueous solution at 100°C for 2 h; precipitated lipids were removed by centrifugation (13,000g, 20 min). Carbohydrates were fractionated by gel filtration on a Sephadex G-50 Superfine column (56 × 2.6 cm; Amersham Biosciences, Sweden) in 0.05 M pyridinium acetate buffer (pH 4.5); elution was monitored with a differential refractometer (Knauer, Germany). The OPS yield was 5 and 6% of total LPS mass for E. coli strains O81 and HS3-104, respectively.
Dephosphorylation. OPS from strain O81 (10 mg) was treated with aqueous 48% HF at 4°C for 24 h. HF was removed by a stream of air; the residue was dried over P2O5 and fractionated by gel filtration on a Fractogel TSK HW 40S column (85 × 1.6 cm) in 1% acetic acid to obtain oligosaccharide 1 (1.4 mg).
Monosaccharide analysis. Alditol acetates [11] were obtained by OPS hydrolysis with 2 M trifluoroacetic acid (120°C, 2 h) and analyzed by gas chromatography (GC) on an HP-5 capillary column with an Agilent 7820 GC system using a temperature gradient from 160°C (1 min) to 290°C at 7°C/min.
NMR spectroscopy. Samples were lyophilized from 99.9% D2O and examined as solutions in 99.95% D2O. 13C NMR spectra were recorded with a Bruker DRX-500 instrument; other spectra were recorded with a Bruker Avance II 600 MHz spectrometer at 30°C (oligosaccharide 1 and O81 OPS) or at 60°C (HS3-104 OPS) using sodium 3-(trimethylsilyl)propanoate-2,2,3,3-d4 (δH 0, δC —1.6) as an internal standard for calibration. Two-dimensional NMR spectra were obtained using standard Bruker software and the Bruker TopSpin 2.1 program for acquiring and processing of NMR data. The spin-lock time of 60 ms and the mixing time of 150 ms were used in the two-dimensional 1H,1H TOCSY and 1H,1H ROESY experiments, respectively. A 60-ms delay was used for evolution of long-range coupling to optimize 1H,13C HMBC spectroscopy experiments.
Sequencing and analysis of genes. Genomic DNA from E. coli HS3-104 cells was isolated with a Wizard® Genomic DNA Purification Kit (Promega, USA) according to the manufacturer’s protocol. Extracted DNA (100 ng) was disrupted into 200-300-bp fragments by sonication using a Covaris S220 System (Covaris, USA). The shotgun library was prepared with an Ion Xpress™ Plus Fragment Library Kit (Life Technologies, USA). Emulsion PCR was performed using an Ion PGM™ Template OT2 200 Kit (Life Technologies). DNA sequencing was performed using an Ion Torrent PGM (Life Technologies) with an Ion 318 chip using Ion PGM™ Sequencing 200 Kit v2 (Life Technologies). The genome sequence was assembled using the Newbler v.3.0 program with standard parameters. To correct Ion Torrent homopolymer errors that could result in assembly errors [12] and artificial frame shifts in coding sequences, the HomoHomo tool was applied (www.github.com/paraslonic/HomoHomo) [13]. The corrected draft of the genome sequence was annotated using the PROKKA 1.7 program [14]. Sequences of the contigs obtained by the whole-genome sequencing were deposited at the DDBJ/ENA/GenBank under accession No. PHFI00000000.
Suggested gene functions were assigned using the NCBI BLAST program to screen homologous sequences in the GenBank database. Conserved domains in each protein were annotated by comparing them to conserved protein motifs from the Pfam database (pfam.sanger.ac.uk). The TMHMM analysis program v.2.0 (http://www.cbs.dtu.dk/services/TMHMM-2.0/) was used to identify potential transmembrane domains. The functions of glycosyl transferase genes were predicted by comparing to genes of other E. coli O-serogroups with the same glycoside bonds in their OPSs.
RESULTS AND DISCUSSION
Structure of O-specific polysaccharides. OPSs were obtained by mild acidic degradation of LPSs isolated from bacterial cells by phenol–water extraction and then separated from low-molecular-mass components by gel filtration on a Sephadex G-50 column. GC analysis of alditol acetates derived after full acidic hydrolysis of the O81 and HS3-104 OPSs identified Glc, Gal and GalNAc at the ratios of 1.5 : 1.0 : 1.3 and 2.5 : 1.0 : 1.3, respectively.
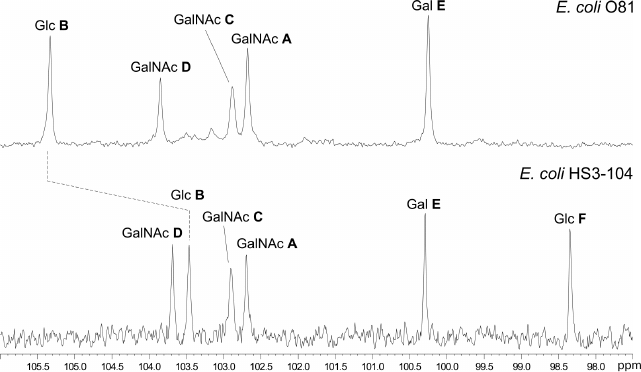
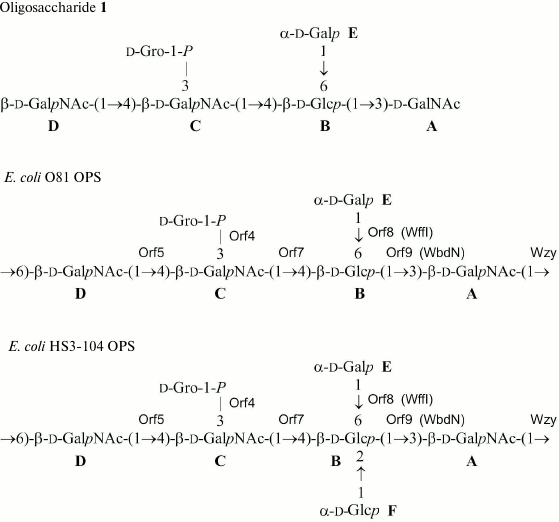
The 31P NMR spectrum of the O81 OPS contained a signal for a monophosphate group at δ 0.23. Assignment of the 1H NMR and 13C NMR spectra of the O81 OPS (Fig. 1) was complicated, most likely, due to non-stoichiometric phosphorylation. For this reason, the OPS was dephosphorylated by treatment with 48% aqueous HF. Dephosphorylation was accompanied by cleavage of the β-GalpNAc-(1→6)-GalpNAc bond (Fig. 2). The resulting oligosaccharide 1 was studied by one- and two-dimensional NMR spectroscopies (for 1H and 13C NMR signal assignment in oligosaccharide 1 see Table 1). It was found that oligosaccharide 1 contained the same monosaccharides as the initial OPS. The structure of oligosaccharide 1 (Fig. 2) was established using two-dimensional 1H,1H ROESY and 1H,13C HMBC spectroscopy (Table 2).
Fig. 1. Region of anomeric carbons in the 13C NMR spectra of OPSs from E. coli strains O81 (top) and HS3-104 (bottom).
Fig. 2. Structures of E. coli OPSs and oligosaccharide 1. Escherichia coli O81 transferases are indicated next to the bonds they were assigned to. Homologs of Orf8 and Orf9 with known functions are given in parentheses.
Table 1. 1H and 13C
NMR chemical shifts (δ, ppm) measured at 30°C
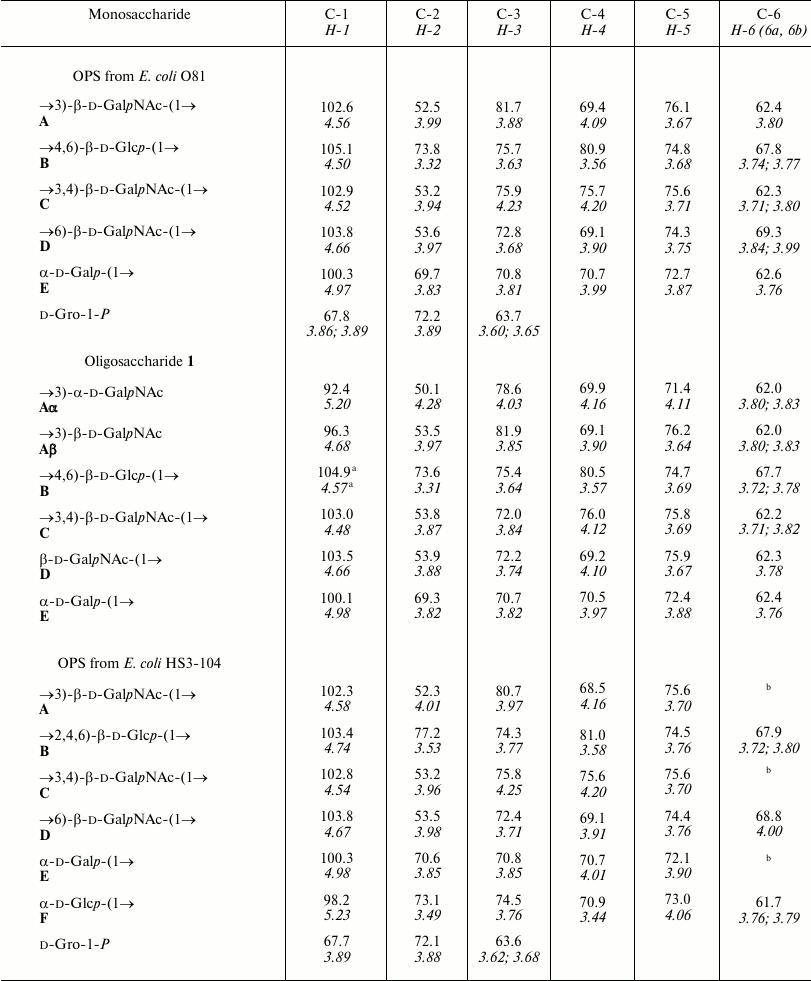
Note: 1H NMR chemical shifts are italicized. Chemical shifts
for N-acetyl groups: δH 2.03-2.08; δC
23.7-23.9 (Me), 175.2-176.4 (CO).
a When linked to Bα; δC = 105.1
and δH = 4.51 when linked to Bβ.
b Chemical shifts are δC 62.3-62.5 and
δH 3.74-3.88.
Table 2. Correlations for H-1 and C-1 in the
two-dimensional 1H,1H ROESY and
1H,13C HMBC spectra of oligosaccharide 1
from E. coli O81
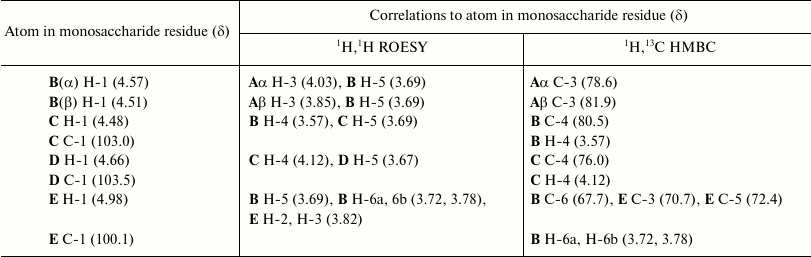
The data obtained for oligosaccharide 1 enabled assignment of the major series of signals in the 1H and 13C NMR spectra of the O81 OPS (Table 1). Comparison of 13C NMR chemical shifts for these two compounds showed that the GalNAc residue (A) at the reducing end of oligosaccharide 1 is β-linked in the OPS. The correlation between C-1 of unit A and H-6b of another GalNAc residue (D) at δ 102.6/3.99 indicated that the repeating units are connected by the β-GalpNAc-(1→6)-GalpNAc (A→D) bonds.
Beside monosaccharides, the OPS also contained glycerol 1-phosphate bound to the third GalNAc residue (unit C) at position 3, as follows from the correlation between the phosphate group signal and the GalNAc (unit C) H-3 and glycerol H-1 at δ 0.23/4.23 and 0.23/3.87, respectively, in the two-dimensional 1H,31P HMBC spectrum. Therefore, the O81 OPS has the structure shown in Fig. 2. Most likely, the minor series of signals in the OPS O81 spectrum belongs to the nonphosphorylated polysaccharide, but assignment of its NMR signals was complicated by multiple coincidences with signals of the major series.
Similar analysis of the NMR spectra allowed elucidation of the structure of the HS3-104 strain OPS (for assigned 1H NMR and 13C NMR (Fig. 1) signals see Table 1). The HS3-104 OPS structure is similar to that of the O81 OPS and differs only in the presence of an additional side-chain α-Glcp residue (unit F). The two-dimensional ROESY spectrum showed a correlation between the unit F H-1 and unit B H-2; therefore Glc (unit F) is attached to Glc (unit B) at position 2. The sequence of other constituent monosaccharides and the bonds between them were the same as in the O81 OPS; hence, the HS3-104 OPS has the structure shown in Fig. 2.
The OPSs studied are distinguished in the presence of a side-chain glycerol 1-phosphate. Phosphorylated OPSs are not uncommon for E. coli, including those containing glycerol 1-phosphate that can be either attached as a side chain (serogroups O37, O81, O100) or incorporated into the main chain (serogroups O28ab, O28ac, O29, O42) (http://nevyn.organ.su.se/ECODAB/; http://csdb.glycoscience.ru/bacterial/). Glycerol 2-phosphate or d-glyceric acid 2-phosphate are present in the OPSs of E. coli O130 and O82, respectively. In some other E. coli OPSs, phosphate links ribitol and a monosaccharide (serogroups O118 and O151) or two monosaccharides in the main chain (serogroups O84, O152, O160, O172, O173, O181) (http://nevyn.organ.su.se/ECODAB/; http://csdb.glycoscience.ru/bacterial/).
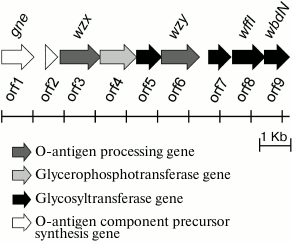
Characterization of the O81 antigen gene cluster. The O-antigen gene cluster of E. coli O81 has been sequenced earlier [6]. It contains nine genes transcribed in the direction from galF to gnd (Fig. 3). Descriptions of all open reading frames (orfs) and their homologs found by bioinformatic analysis are given in Table 3. No genes for the synthesis of nucleotide sugar precursors of Glc and Gal were found between galF and gnd in E. coli O81, as they are usually located outside the O-antigen gene cluster [15].
Fig. 3. Organization of E. coli O81 O-antigen gene cluster (adapted on the basis of Iguchi et al. data [6]). Homologs of orf8 and orf9 with known functions are designated.
Table 3. Characteristics of orfs in
the O81 antigen gene cluster
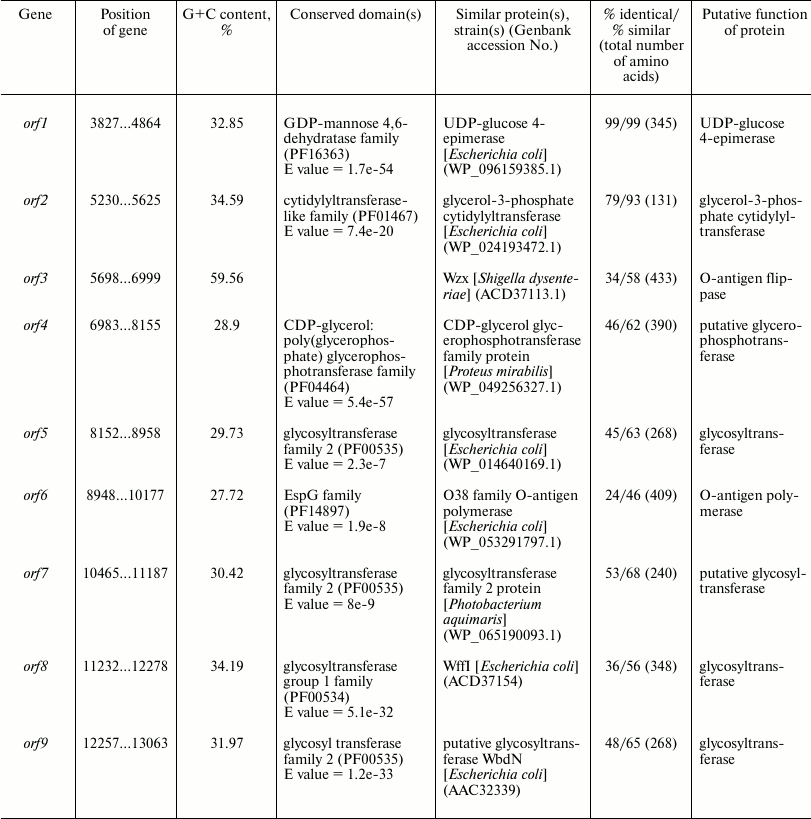
Either GlcNAc or GalNAc is the first sugar in the O-unit of most E. coli strains. The wecA (rfe) gene is responsible for the transfer of d-GlcNAc-P from UDP-d-GlcNAc to undecaprenyl phosphate (Und-P) with the formation of UndPP-d-GlcNAc. This gene is located in the cluster responsible for the synthesis of enterobacterial common antigen [16]. When the first sugar is GalNAc (as in E. coli O81), 4-epimerase Gnu converts UndPP-d-GlcNAc into UndPP-d-GalNAc [17], and the gnu gene is located immediately upstream of the galF gene [18]. In E. coli O81, this gene has been identified and named gne [6]. However, the name Gne has been already used for another 4-epimerase that converts UDP-d-GlcNAc to UDP-d-GalNAc [18]; therefore, we recommend renaming the gene responsible for the synthesis of UndPP-d-GalNAc as gnu. There are two more d-GalNAc residues in the O81-O unit. orf1 was identified as a gene coding for 4-epimerase involved in the synthesis of UDP-d-GalNAc and named gne accordingly.
orf2 was annotated as glycerol-3-phosphate cytidyltransferase gene; we presume that its protein product might be involved in the formation of d-Gro-1-P. However, this suggestion requires experimental confirmation.
There are five transferase genes in the O81 O-antigen gene cluster, that were annotated taking into account the established OPS structure. orf4 was identified as a glycerophosphotransferase gene that encodes the enzyme that supposedly transfers d-Gro-1-P to one of the d-GalNAc residues.
orfs 5, 7, 8, and 9 were annotated as glycosyltransferase genes (Figs. 3 and 4). WbdN, a UDP-Glc:GalNAc-diphosphate-lipid β1,3-Glc-transferase, has been found to catalyze formation of the β-d-Glcp-(1→3)-d-GalpNAc bond in E. coli O157 [19]. Since Orf9 of E. coli O81 shares 48% identity with WbdN from E. coli O157 and O-antigens of these two strains have the β-d-Glcp-(1→3)-d-GalpNAc disaccharide fragment in common, we propose that Orf9 is a homolog of WbdN responsible for the synthesis of this fragment. α-d-Galp-(1→6)-d-Glcp is the only identical bond in the O-antigens of E. coli strains O130 [20] and O81. Orf8 of E. coli O81 is 36% identical to WffI of E. coli O130 [15]; therefore, these two enzymes might have the same role in the formation of the α-d-Galp-(1→6)-d-Glcp bond. Hence, orf5 and orf7 are β-d-GalpNAc-transferases that catalyze formation of the β-d-GalpNAc-(1→4)-d-GalpNAc and β-d-GalpNAc-(1→6)-d-Glcp bonds. No detailed assignment has been done for these orfs due to the lack of their homologs with known functions in available databases.
orf3 and orf6 were identified as wzx and wzy genes for the O-unit flippase and O-antigen polymerase, respectively. The presence of these genes indicated that synthesis and translocation of the O81 O-antigen occurs by the Wzx/Wzy-dependent mechanism that includes assembly of the O-unit on a lipid carrier at the inner membrane cytoplasmic side followed by the O-unit translocation to the periplasmic side.
The genome of E. coli HS3-104 has been sequenced. Its O-antigen gene cluster is essentially identical to that of E. coli O81 (>99% nucleotide identity). Side-chain glucosylation in the OPS of E. coli HS3-104 is evidently driven by the gtrABC genes found on a 3145-bp contig, for which such function has been well established [21, 22]. Encoded GtrA, GtrB, and GtrC are identical to the corresponding proteins of E. coli KCJK4935 (GenBank accession numbers PBU28851, PBU28852, and PBU28853) by 98, 100, and 99%, respectively. Undecaprenol glucosyltransferase GtrB (synthesis of UndP-d-Glc from UDP-d-Glc and UndP) and flippase GtrA (translocation of UndP-d-Glc through the cytoplasmic membrane) are highly conserved proteins, whereas glucosyltransferase GtrC, that links a glucosyl group to the O-antigen backbone, is serotype-specific. The gtr gene cluster is present in many enteric bacteria and usually has a bacteriophage origin [21-23].
In conclusion, the content and assigned functions of the genes involved in the O-antigen synthesis in the studied E. coli strains are consistent with the OPS structures established.
Acknowledgments
This work was supported by the Russian Science Foundation (project 14-14-01042-P) and the National Natural Science Foundation of China General Program (projects 81471904, 31470194, 31371259; participation in characterization of O-antigen gene clusters).
REFERENCES
1.Kaper, J. B., Nataro, J. P., and Mobley, H. L.
(2004) Pathogenic Escherichia coli, Nat. Rev. Microbiol.,
2, 123-140.
2.Reeves, P. (1995) Role of O-antigen variation in
the immune response, Trends Microbiol., 3, 381-386.
3.Reeves, P., and Wang, L. (2002) Genomic
organization of LPS-specific loci, in Pathogenicity Islands and the
Evolution of Pathogenic Microbes, Springer, pp. 109-135.
4.Bastin, D. A., and Reeves, P. R. (1995) Sequence
and analysis of the O antigen gene (rfb) cluster of
Escherichia coli O111, Gene, 164, 17-23.
5.Feng, L., Senchenkova, S. N., Yang, J., Shashkov,
A. S., Tao, J., Guo, H., Cheng, J., Ren, Y., Knirel, Y. A., and Reeves,
P. R. (2004) Synthesis of the heteropolysaccharide O-antigen of
Escherichia coli O52 requires an ABC transporter: structural and
genetic evidences, J. Bacteriol., 186, 4510-4519.
6.Iguchi, A., Iyoda, S., Kikuchi, T., Ogura, Y.,
Katsura, K., Ohnishi, M., Hayashi, T., and Thomson, N. R. (2015) A
complete view of the genetic diversity of the Escherichia coli
O-antigen biosynthesis gene cluster, DNA Res., 22,
101-107.
7.Golomidova, A. K., Kulikov, E. E., Prokhorov, N.
S., Guerrero-Ferreira, R. C., Knirel, Y. A., Kostryukova, E. S.,
Tarasyan, K. K., and Letarov, A. V. (2016) Branched lateral tail fiber
organization in T5-like bacteriophages DT57C and DT571/2 as revealed by
genetic and functional analysis, Viruses, 8, 26.
8.Robbins, P. W., and Uchida, T. (1962) Studies on
the chemical basis of the phage conversion of O-antigens in the E-group
Salmonellae, Biochemistry, 1, 323-335.
9.Westphal, O., and Jann, K. (1965) Bacterial
lipopolysaccharides. Extraction with phenol–water and further
applications of the procedure, Methods Carbohydr. Chem.,
5, 83-91.
10.Senchenkova, S. N., Guo, X., Naumenko, O. I.,
Shashkov, A. S., Perepelov, A. V., Liu, B., and Knirel, Y. A. (2016)
Structure and genetics of the O-antigens of Escherichia coli
O182-O187, Carbohydr. Res., 435, 58-67.
11.Sawardeker, J. S., Sloneker, J. H., and Jeanes,
A. (1965) Quantitative determination of monosaccharides as their
alditol acetates by gas liquid chromatography, Anal. Chem.,
37, 1602-1603.
12.Bragg, L. M., Stone, G., Butler, M. K.,
Hugenholtz, P., and Tyson, G. W. (2013) Shining a light on dark
sequencing: characterising errors in ion torrent PGM data, PLoS
Comput. Biol., 9, e1003031.
13.Rakitina, D. V., Manolov, A. I., Kanygina, A. V.,
Garushyants, S. K., Baikova, J. P., Alexeev, D. G., Ladygina, V. G.,
Kostryukova, E. S., Larin, A. K., Semashko, T. A., Karpova, I. Y.,
Babenko, V. V., Ismagilova, R. K., Malanin, S. Y., Gelfand, M. S.,
Ilina, E. N., Gorodnichev, R. B., Lisitsyna, E. S., Aleshkin, G. I.,
Scherbakov, P. L., Khalif, I. L., Shapina, M. V., Maev, I. V., Andreev,
D. N., and Govorun, V. M. (2017) Genome analysis of E. coli
isolated from Crohn’s disease patients, BMC Genomics,
18, 544.
14.Seemann, T. (2014) Prokka: rapid prokaryotic
genome annotation, Bioinformatics, 30, 2068-2069.
15.Liu, B., Knirel, Y. A., Feng, L., Perepelov, A.
V., Senchenkova, S. N., Wang, Q., Reeves, P. R., and Wang, L.
(2008) Structure and genetics of Shigella O antigens,
FEMS Microbiol. Rev., 32, 627-653.
16.Meier-Dieter, U., Barr, K., Starman, R., Hatch,
L., and Rick, P. D. (1992) Nucleotide sequence of the Escherichia
coli rfe gene involved in the synthesis of enterobacterial common
antigen. Molecular cloning of the rfe-rff gene cluster, J.
Biol. Chem., 267, 746-753.
17.Rush, J. S., Alaimo, C., Robbiani, R., Wacker,
M., and Waechter, C. J. (2010) A novel epimerase that converts
GlcNAc-P-P-undecaprenol to GalNAc-P-P-undecaprenol in Escherichia
coli O157, J. Biol. Chem., 285, 1671-1680.
18.Cunneen, M. M., Liu, B., Wang, L., and Reeves, P.
R. (2013) Biosynthesis of UDP-GlcNAc, UndPP-GlcNAc and UDP-GlcNAcA
involves three easily distinguished 4-epimerase enzymes, Gne, Gnu and
GnaB, PLoS One, 14, e67646.
19.Gao, Y., Liu, B., Strum, S., Schutzbach, J. S.,
Druzhinina, T. N., Utkina, N. S., Torgov, V. I., Danilov, L. L.,
Veselovsky, V. V., Vlahakis, J. Z., Szarek, W. A., Wang, L., and
Brockhausen, I. (2012) Biochemical characterization of WbdN, a
β1,3-glucosyltransferase involved in O-antigen synthesis in
enterohemorrhagic Escherichia coli O157, Glycobiology,
22, 1092-1102.
20.Perepelov, A. V., Liu, B., Senchenkova, S. N.,
Shevelev, S. D., Wang, V., Shashkov, A. S., Feng, L., Wang, L., and
Knirel, Y. A. (2007) The structure of the glycerophosphate-containing
O-specific polysaccharide from Escherichia coli O130, Russ.
J. Bioorg. Chem., 33, 57-60.
21.Allison, G. E., and Verma, N. K. (2000)
Serotype-converting bacteriophages and O-antigen modification in
Shigella flexneri, Trends Microbiol., 8,
17-23.
22.Wang, W., Perepelov, A. V., Feng, L., Shevelev,
S. D., Wang, Q., Senchenkova, S. N., Han, W., Li, Y., Shashkov, A. S.,
Knirel, Y. A., Reeves, P. R., and Wang, L. (2007) A group of
Escherichia coli and Salmonella enterica O antigens
sharing a common backbone structure, Microbiology, 153,
2159-2167.
23.Knirel, Y. A., Sun, Q., Senchenkova, S. N.,
Perepelov, A. V., and Xu, J. (2015) O-Antigen modifications providing
the antigenic diversity of Shigella flexneri and underlying
genetic mechanisms, Biochemistry (Moscow), 80,
901-915.