REVIEW: The Role of Chromosome–Nuclear Envelope Attachments in 3D Genome Organization
I. V. Sharakhov1,2*, S. M. Bondarenko2, G. N. Artemov2, and A. V. Onufriev3,4
1Virginia Polytechnic Institute and State University, Fralin Life Science Institute, Department of Entomology, 24061 Blacksburg, VA, USA; E-mail: igor@vt.edu2Tomsk State University, Laboratory of Ecology, Genetics and Environmental Protection, Tomsk, Russia
3Virginia Polytechnic Institute and State University, Department of Physics, 24060 Blacksburg, VA, USA
4Virginia Polytechnic Institute and State University, Department of Computer Science, 24061 Blacksburg, VA, USA
* To whom correspondence should be addressed.
Received October 20, 2017; Revision received December 11, 2017
Chromosomes are intricately folded and packaged in the cell nucleus and interact with the nuclear envelope. This complex nuclear architecture has a profound effect on how the genome works and how the cells function. The main goal of review is to highlight recent studies on the effect of chromosome–nuclear envelope interactions on chromatin folding and function in the nucleus. The data obtained suggest that chromosome–nuclear envelope attachments are important for the organization of nuclear architecture in various organisms. A combination of experimental cell biology methods with computational modeling offers a unique opportunity to explore the fundamental relationships between different aspects of 3D genome organization in greater details. This powerful interdisciplinary approach could reveal how the organization and function of the genome in the nuclear space is affected by the chromosome–nuclear envelope attachments and will enable the development of novel approaches to regulate gene expression.
KEY WORDS: chromosomes, Drosophila, nuclear architecture, nuclear envelope, mosquitoes, modelingDOI: 10.1134/S0006297918040065
Abbreviations: Chr, chromosome; DamID, DNA adenine methyltransferase identification; FISH, fluorescence in situ hybridization; Hi-C, chromosome conformation capture; LAD, lamina-associated domain; NE, nuclear envelope; TAD, topologically associated domain.
Non-random 3D organization of the genome plays a crucial role in the
regulation of gene expression, DNA double-strand break repair, and DNA
replication [1-4]. Deep
understanding of cell physiology will require elucidation of the
structure-function relationships in the nucleus, which is a complex,
interacting, and dynamic biological system. In eukaryotes, the genomic
material –chromatin fiber – is packed in a complex,
hierarchical manner [5, 6].
Unlike most proteins that adopt the same unique compact 3D shape in all
cells, the conformational states of chromatin fibers are not nearly as
compact or ordered. It is likely that these states are governed by a
set of principles most of which are still poorly understood. In
contrast to the unique native (folded) state of proteins, the state of
folded chromatin in the cell nucleus is expected to depend on its
interaction with the nuclear envelope (NE) [7, 8]. Experimental studies of polytene and regular
diploid (non-polytene) nuclei in Drosophila and other eukaryotes
have revealed at least two types of interactions in the nucleus:
chromosome–NE (Chr–NE) attachments and
chromosome–chromosome (Chr–Chr) contacts.
Organization and functional significance of Chr–NE attachments. Statistically significant high-frequency attachments exist between certain chromosomal loci and the NE. Several lines of evidence, including microscopy and genome-wide mapping studies using DamID (DNA adenine methyltransferase identification), show that in Drosophila and human nuclei, gene-poor and transcriptionally silent regions tend to form high-frequency contacts with NE [9-12]. In D. melanogaster, lamina-associated domains (LADs) usually span from 7 to 700 kb (average size, 90 kb) and bear on average seven genes per LAD [13]. Lamin binding is linked to a combination of several features including late replication, large intergenic region size, low gene expression level, and the lack of active histone marks [9, 12]. Depletion of lamin B (Dm0) in Drosophila cells causes visible detachment of chromosomal loci from the NE [14, 15]. In contrast, removal of essentially all lamins in mouse embryonic stem cells does not have any detectable effect on the Chr–NE attachments [16] indicating that lamins are not required for the formation of LADs in every cell type of every organism.
Many Chr–NE attachments are tissue-specific and, hence, may be responsible for the tissue-specific gene expression [1, 17, 18]. Chr–NE attachments regulate gene expression by repositioning a gene closer to the NE, i.e., to the region where transcription is usually repressed [1, 9, 14]. For example, in D. melanogaster somatic cells, testis-specific genes tend to be organized in clusters that are embedded in repressive nuclear peripheral compartments. Depletion of lamin B (Dm0) results in the derepression of these genes in somatic tissues together with their detachment from the NE [14]. Another consequence of lamin B depletion in the nucleus is misexpression of key developmental genes. Thus, detachment of the hunchback gene from the NE in D. melanogaster coincides with the reduction of its silencing and extension of the time window for neuroblast competence [15]. Moreover, the loss of lamin B in Drosophila fat body during aging leads to systemic inflammation and disruption of tissue homeostasis [19].
Organization of Chr–Chr interactions. Chromosomes fold in a characteristic way within a limited volume of the interphase nucleus thereby creating chromosome territories [11, 20-22]. Spatial chromosome organization is manifested in short-range and long-range intra-chromosomal interactions, as well as inter-chromosomal interactions. Long-range interactions between polytene chromosomes have been microscopically observed in Drosophila [23]. More recently, Hi-C (chromosome conformation capture) techniques [24-27] have been used to identify intra- and inter-chromosomal interactions in nonpolytenized nuclei. The Hi-C studies have led to the discovery of 3D structures that favor short-range, rather than long-range, chromosome interactions. These chromatin structures were named topologically associating domains (TADs) [28]. TADs are delimited by sharp boundaries defined by housekeeping genes and insulator-binding sites [29]; they correspond to chromosomal domains [30] and bands in polytene chromosomes [26, 31].
Functional significance of short-range Chr–Chr contacts. The functions of TADs depend on their structural organization that can be classified based on the chromatin type [2, 32]. Accordingly, several types of TADs can be recognized in Drosophila.
1) Active TADs belong to the most heterogeneous RED and YELLOW chromatin types. RED and YELLOW chromatin types represent housekeeping and tissue-specific genes, respectively, of the euchromatin. Active TADs have binding sites for many chromatin factors and are marked by multiple histone modifications, such as H3K4me, H3K36ac, H3K79ac [33]. They represent a highly accessible environment and participate in long-range interactions that favor coregulation of distant genes by increasing the local concentration of factors required for their transcription [34]. Active TADs are located predominately in the inner part of the nucleus and preferentially contact other active TADs, rather than inactive ones [2].
2) Polycomb-repressed TADs correspond to the BLUE chromatin, which is marked by the Polycomb group (PcG) proteins and H3K27me3. Polycomb-repressed TADs form long-range intra- and inter-chromosome contacts that occur in regions of high Polycomb concentration, the so-called Polycomb foci visible under a microscope [35]. Depletion of CTCF in D. melanogaster affects long-range interactions between homeotic complexes located in different Polycomb-repressed TADs, without affecting their local clustering [36].
3) Lamin-repressed TADs correspond to the BLACK chromatin (intercalary heterochromatin), marked by lamin and SUUR (suppressor of under-replication). These TADs might colocalize with lamin-associated domains (LADs). More than 5000 genes are located inside LADs and interact with lamin in Drosophila [13]. Interestingly, these genes have a very low transcriptional activity and are not associated with any active histone or heterochromatin marks. Repression at the nuclear lamina appears to be weaker compared to more specialized silencing mechanisms, such as in the Polycomb-repressed TADs [14, 37, 38]. This correlates well with the fact that H3K27me3 histone-associated Drosophila LADs contain less genes than Polycomb-repressed domains [32].
4) GREEN chromatin includes pericentromeric heterochromatin and can contain heterochromatic TADs. GREEN chromatin is marked by histone lysine methyltransferase SU(VAR)3-9, H3K9me2/3, and heterochromatin protein 1 (HP1) [32]. Average transcriptional activity is lower in heterochromatic domains than in the active euchromatic TADs but higher than in Polycomb- and lamin-repressed TADs. Long-range interactions between heterochromatic foci and euchromatic genes can drive tissue-specific gene silencing due to the proximity effect [39].
Regions between TADs (inter-TADs) correspond to decompacted interbands of polytene chromosomes in Drosophila; they consist of active chromatin and constitutively transcribed (housekeeping) genes [26].
Functional significance of long-range Chr–Chr contacts. Most active or potentially active euchromatic TADs are located in the inner part of the nucleus where they form long-range contacts with other active or potentially active TADs, resulting in the formation of larger active transcriptional factories [2]. Transcriptional factories represent a highly accessible environment and are formed by the interactions that favor coregulation of distant genes by increasing the local amounts of factors required for their transcription [34]. In contrast, PcG-associated TADs participate in long-range intra- and inter-chromosomal contacts and form strongly repressed larger hubs with high PcG concentration [40]. These large PcG hubs are visible under a light microscope [35]. Such long-range Chr–Chr interactions are usually tissue-specific because they occur between genes sharing either PcG-mediated repression [41, 42] or activation by transcription factors [43, 44] specifically in cell types where the regulation is mediated.
The interplay between Chr–NE attachments and Chr–Chr contacts. The driving forces that bring together long-range chromosomal contacts are not well-understood. It is possible that several active TADs form transcription factories, because transcriptional complexes cooperatively bring chromosomes toward a more internal position in the nucleus. In contrast, chromosome loci depleted in active TADs would be pushed to a more peripheral location. However, recent data indicate that chromosomal loci have the peripheral location because of specific interactions with the NE [1, 9, 13, 45]. Therefore, the internal positioning of active chromatin could be due to a passive force, resulting in preferential positioning of specific chromosomal loci at the nuclear periphery. Indeed, recent modeling results indicate that the number of Chr–NE attachments may affect the frequency of Chr–Chr interactions in D. melanogaster. Introduction of Chr–NE attachments decreases the overall probability of Chr–Chr contacts in trans configuration but increases the frequency of Chr–Chr interactions in cis configuration [46]. Attachment to the NE could constrain chromatin condensation and prevent chromosome territories from freely diffusing and mixing in the entire nuclear volume, which would be critical for maintaining the delicate 3D genome architecture. Experimental studies demonstrated the role of Chr–NE attachments in the maintenance of nuclear architecture and separation of chromosome territories [47, 48], as well as in the reduction of recombination between distant genomic loci and lowering the chances of non-allelic recombination and double-strand break repair [1, 49]. It is possible that Chr–NE attachments regulate expression of genes located not only at the nuclear periphery but also genes involved in long-range Chr–Chr interactions. Moreover, spatial networks of inter-connected genes may be a result of chromosome folding influenced by Chr–NE attachments. The functional significance of such configurations is that the long-range contact between two loci increases concentrations of common regulatory factors. When a factor dissociates, it is more likely to be retrapped by the cluster formed by long-range contacts than to diffuse away to another single TAD [50]. Also, interacting TADs may ensure robust gene regulation by constraining genes to the areas inaccessible for long-range interactions [51].
Studies of Chr–NE attachments using polytene chromosomes. The link between Chr–NE attachments and 3D genome organization can be uncovered with the help of giant (polytene) chromosomes in Drosophila and mosquitoes. Each of these chromosomes contain 1024 bundled replicas of the corresponding “regular” interphase chromosome and, thus, the entire chromosome set of a single nucleus becomes readily visible under a light microscope [22, 52, 53]. Drosophila polytene chromosomes have the same banding patterns as nonpolytene chromosomes, as it was found by analyzing the data on localization of interband- and band-specific proteins (modENCODE) and genomic positions of interbands and bands [54-57]. Recent Hi-C studies have revealed that TADs, and the regions between them (which are conserved between polytene and diploid cells) correspond to the bands and interbands of polytene chromosomes in Drosophila [26, 31]. Moreover, polytene chromosomes and nonpolytene chromosomes have similar sets of Chr–NE attachments, as seen by comparing DamID data obtained in embryonic cells [9] with light microscopy data obtained in salivary glands [11, 17, 58]. Belyaeva et al. [55] compared distribution of LADs [13] and localization of 60 regions of intercalary heterochromatin in polytene chromosomal arms and found complete overlap for four regions of intercalary heterochromatin. Most of the intercalary heterochromatin regions have shown partial overlap with LADs, six regions have shown no overlap with any of the LADs, and one region of the intercalary heterochromatin encompassed five separate LADs. A microscopic study of polytene nuclei offers important methodological advantages. The large size of a polytene nucleus (30-35 µm) facilitates discrimination between the peripheral position of a chromosomal locus and its actual attachment to the NE. Characteristic chromatin fibers that physically penetrate the NE, as well as tight bends and strong twists, can often be seen where a chromosomal locus attaches to the periphery [11, 23, 59, 60]. Spatial positions of multiple regions in a nucleus can be determined by reading their banding pattern with the help of oligopaint probes [61]. Microscopy data can be converted into R(x, y, z) curves corresponding to each chromosome, and the corresponding nuclear architecture can be computationally simulated. This level of structural completeness is currently not possible in the studies of regular nonpolytene chromosomes.
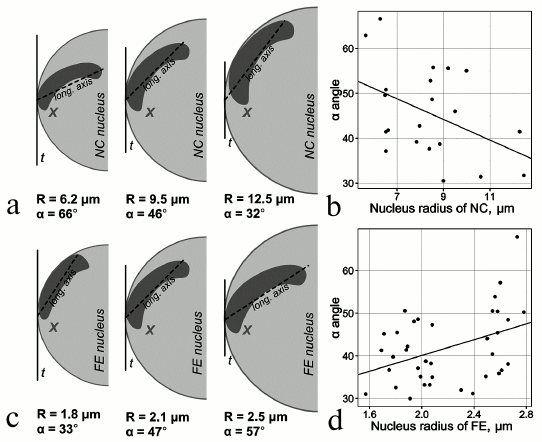
Studies of 3D organization of chromosomes in mosquitoes. Previous studies have shown tissue-specific features of chromosome attachments to the NE in various organisms including malaria mosquitoes [62-65]. Early works on mosquitoes were among the first to demonstrate substantial differences in the spatial organization of polytene chromosomes in the nuclei of salivary gland cells, malpighian tubules, and ovarian nurse cells in malaria mosquitoes of the Anopheles maculipennis complex [64, 66]. The chromosomes had typical Rabl organization in salivary glands and malpighian tubules. In contrast, the ovarian nurse cells had no single chromocenter (and Rabl organization), and some of their chromosomes attached to the NE with the pericentromeric beta-heterochromatin or specific loci of intercalary heterochromatin [67]. Another study focused on tissue-specific features of chromosome architecture in closely related malaria mosquitoes [63]. This study showed that the X chromosome–NE attachment is stronger in nurse cells then in salivary glands of An. messeae. Microdissected DNA probes corresponding to the regions of the X chromosome (2BC) and 3R chromosome (32D) attachment to the NE were hybridized with intact nuclei of nurse cells, salivary gland cells, follicle epithelium cells, and imaginal discs cells in 3D-FISH (3D fluorescence in situ hybridization) experiments. Only salivary gland cells and follicle epithelium cells had no statistical differences in the interposition of 2BC and 32D. On average, the X chromosome and chromosome 3R were located closer to each other in somatic tissue cell than in nurse cells. In the nuclei of imaginal disc cells, the distance between the X and 3R chromosomes was intermediate between those observed in the nurse cells and somatic cells. A more recent study conducted a thorough analysis of tissue-specific dynamics of the X chromosome in follicular epithelium and nurse cells of An. atroparvus ovaries [65]. A fluorescently labeled X chromosome painting probe was hybridized with formaldehyde-fixed ovaries of mosquitoes using the 3D-FISH method, and the results were processed with the TANGO software for chromosome spatial organization analysis [68]. It was shown that the volume and position of the X chromosome are tissue-specific. Unlike nurse cell nuclei, the growth of follicular epithelium nuclei was not accompanied with the proportional growth of the X chromosome. The dynamics of the X chromosome–NE attachment regions was also tissue-specific (Fig. 1). During the nucleus growth in nurse cells, the polytene X chromosome moved toward the periphery, whereas in follicular epithelium, the X chromosome telomere moved from the nuclear envelope to the intranuclear space [65]. It is possible that Chr–NE attachments facilitate or prevent tissue-specific Chr–Chr contacts, where actively transcribed genes colocalize and exchange transcription sites.
Fig. 1. Tissue-specific dynamics of the X chromosome location during nuclear growth. a, b) Decrease in the X chromosome longitudinal axis related to the nucleus growth in nurse cells. c, d) Increase in the X chromosome longitudinal axis related to the nucleus growth in follicular epithelium [65].
In addition to tissue-specific differences, essential differences in the nuclear architecture among seven species of the An. maculipennis complex have been found [69]. These closely related species of malaria mosquitoes, some of which are homosequential (e.g., An. maculipennis and An. subalpinus), had different patterns of Chr–NE attachments. Spatial organization of chromosomes in ovarian nurse cell of experimental F1 hybrids (An. maculipennis × An. subalpinus and An. sacharovi × An. matrinius) confirmed species-specific chromosome organization in the parental species. However, despite species-specific Chr–NE attachments, no differences in the 3D organization of nurse cell chromosomes between An. messeae and An. atroparvus have been observed [63]. More detailed studies are needed to address species-specific aspects of the nuclear architecture. Such studies will be essential for assessing the possibility of using 3D genome organization as a phylogenetic marker for understanding the process of mosquito speciation.
Possible role of NE–chromosome attachments in genome rearrangements. Cytogenetic and genomic studies in various organisms have provided evidence for nonuniform distribution of chromosomal rearrangements and breakpoints [70-72]. Chromosomal rearrangements, which are believed to be affected by nuclear architecture, are linked to cancer [73]. Unequal chances of chromosome arms and lineages in generating and fixing inversions have serious consequences for differential adaptive and evolutionary plasticity of organisms. Therefore, it is important to understand the forces that govern the structural malleability of genomes. Analysis of nuclear architecture in diverse systems revealed a general principle: because of nonrandom nuclear organization, certain loci are closer together than other loci and, therefore, are more likely to interact and generate rearrangements [74, 75]. Computer modeling has been instrumental in helping to understand the link between chromosome folding and chromosome rearrangements. Such models have demonstrated that specific loci are much closer to each other in the nuclear space than it could be expected based on their genomic locations, which explains their frequent involvement in chromosomal rearrangements [76, 77]. A study of the 3D organization of the yeast genome revealed that location of inter-chromosomal contacts correlates with chromosomal fragile sites where chromosomal breakpoints occur in evolution [78]. Another study detected rearrangement events in Saccharomyces species and observed increased frequency of the rearrangements between regions with higher frequency of contacts [79]. These results show how 3D chromosomal organization can influence evolutionary events. Chr–NE attachments might indirectly affect the frequency of evolutionary rearrangements by changing the probability of long-range Chr–Chr interactions.
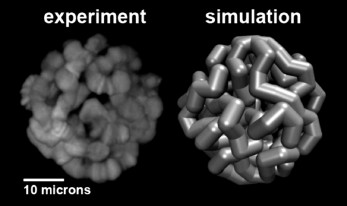
Computational modeling of the 3D organization of chromosomes: effects of Chr–NE attachments. 3D organization of eukaryotic genomes is extremely intricate and much more complex than organization of proteins or protein complexes. At the same time, all experimental methods available to study various aspects of 3D organization of eukaryotic genomes are inherently limited in their resolution, both spatial and temporal. Compared to structural biology, there is an additional complexity: both Chr–NE attachments and Chr–Chr contacts appear to be stochastic to some degree, as evidenced by DamID and Hi-C experiments [80-82]. All these factors combined make computational modeling an essential tool to complement the experimental studies. The use of computational modeling in close integration with experiments can generate verifiable hypotheses that otherwise would be very difficult or impossible to arrive at. Equally importantly, models can rationalize experimental observations and suggest explanations of the basic mechanisms of nuclear architecture. While computation is becoming commonplace in this field [24, 26, 83-85], Chr–NE attachments have received relatively little attention in nuclear architecture modeling studies compared to the Chr–Chr interactions [26, 86, 87] and other aspects of genomic architecture. Although a few published studies included Chr–NE attachments in their models [86, 88, 89], they provided no comprehensive analysis of the role of these attachments in the regulation of Chr–Chr contacts. Also, results of computational modeling of yeast genome architecture, which may lack chromosome territories and typical TADs [90], cannot be readily extrapolated to the 3D genome organization of higher eukaryotes. Recent simulation study [91] that generated thousands of models of Drosophila polytene nuclei (Fig. 2), allowed researchers to determine an objective threshold of statistical significance for Chr–NE attachments originally based on only 24 experimental nuclei [11]. This simulation work identified 33 additional positions of statistically significant Chr–NE attachments in the experimental nuclei [11]. Of the total 48 Chr–NE attachments, 34 were intercalary chromatin, 3 were euchromatin, and 11 Chr–NE attachments displayed some properties of intercalary chromatin by being late replicating regions. These results suggest that affinity for the NE can change gradually, with the highest affinity almost exclusively possessed by intercalary chromatin, and the lowest affinity for the NE being mostly, but not exclusively, a property of euchromatin. The newly identified Chr–NE attachments are real because they were confirmed in an independent DamID study [9]. The model of Drosophila polytene nucleus has been used to investigate if and how the presence and the number of Chr–NE attachments affect chromosome territories and Chr–Chr contacts. Model nuclei with more numerous Chr–NE attachments formed more distinct chromosome territories, and their chromosomes intertwined less frequently. They also displayed higher probabilities of intra-chromosome and intra-arm contacts. In contrast, inter-chromosome and inter-arm contacts become less common in nuclei with more numerous Chr–NE attachments [46]. Statistical models that generate snapshots of configurational ensembles have succeeded in reconstructing the folding of D. melanogaster chromosomes through the integration of experimental data on Chr–Chr and Chr–NE interactions [85]. However, dynamic models are needed to predict changes in Chr–NE and Chr–Chr interactions in the interphase nucleus.
Fig. 2. Representation of a simulated model nucleus (right) compared to experiment (left) for the D. melanogaster salivary glands [91].
Acknowledgments
The review of experimental and computational studies of Drosophila, yeast, and mammalian genomes was supported by the National Science Foundation grant MCB-1715207 to IVS and AVO. Our recent research on mosquitoes was supported by the grant 15-14-20011 to IVS from the Russian Science Foundation.
REFERENCES
1.Lemaitre, C., and Bickmore, W. A. (2015) Chromatin
at the nuclear periphery and the regulation of genome functions,
Histochem. Cell Biol., 144, 111-122.
2.Ciabrelli, F., and Cavalli, G. (2015)
Chromatin-driven behavior of topologically associating domains, J.
Mol. Biol., 427, 608-625.
3.Barton, L. J., Soshnev, A. A., and Geyer, P. K.
(2015) Networking in the nucleus: a spotlight on LEM-domain proteins,
Curr. Opin. Cell Biol., 34, 1-8.
4.Nguyen, H. Q., and Bosco, G. (2015) Gene
positioning effects on expression in eukaryotes, Annu. Rev.
Genet., 49, 627-646.
5.Grosberg, A. Y., Nechaev, S. K., and Shakhnovich,
E. I. (1988) The role of topological constraints in the kinetics of
collapse of macromolecules, J. Phys. (Paris), 49,
2095-2100.
6.Lebedev, D. V., Filatov, M. V., Kuklin, A. I.,
Islamov, A. K., Kentzinger, E., Pantina, R., Toperverg, B. P., and
Isaev-Ivanov, V. V. (2005) Fractal nature of chromatin organization in
interphase chicken erythrocyte nuclei: DNA structure exhibits biphasic
fractal properties, FEBS Lett., 579, 1465-1468.
7.Gursoy, G., Xu, Y., Kenter, A. L., and Liang, J.
(2014) Spatial confinement is a major determinant of the folding
landscape of human chromosomes, Nucleic Acids Res., 42,
8223-8230.
8.Mateos-Langerak, J., Bohn, M., de Leeuw, W.,
Giromus, O., Manders, E. M., Verschure, P. J., Indemans, M. H.,
Gierman, H. J., Heermann, D. W., van Driel, R., and Goetze, S. (2009)
Spatially confined folding of chromatin in the interphase nucleus,
Proc. Natl. Acad. Sci. USA, 106, 3812-3817.
9.Pickersgill, H., Kalverda, B., de Wit, E., Talhout,
W., Fornerod, M., and van Steensel, B. (2006) Characterization of the
Drosophila melanogaster genome at the nuclear lamina, Nat.
Genet., 38, 1005-1014.
10.Guelen, L., Pagie, L., Brasset, E., Meuleman, W.,
Faza, M. B., Talhout, W., Eussen, B. H., de Klein, A., Wessels, L., de
Laat, W., and van Steensel, B. (2008) Domain organization of human
chromosomes revealed by mapping of nuclear lamina interactions,
Nature, 453, 948-951.
11.Hochstrasser, M., Mathog, D., Gruenbaum, Y.,
Saumweber, H., and Sedat, J. W. (1986) Spatial organization of
chromosomes in the salivary gland nuclei of Drosophila
melanogaster, J. Cell. Biol., 102, 112-123.
12.Gonzalez-Sandoval, A., and Gasser, S. M. (2016)
On TADs and LADs: spatial control over gene expression, Trends
Genet., 32, 485-495.
13.Van Bemmel, J. G., Pagie, L., Braunschweig, U.,
Brugman, W., Meuleman, W., Kerkhoven, R. M., and van Steensel, B.
(2010) The insulator protein SU(HW) fine-tunes nuclear lamina
interactions of the Drosophila genome, PLoS One,
5, e15013.
14.Shevelyov, Y. Y., Lavrov, S. A., Mikhaylova, L.
M., Nurminsky, I. D., Kulathinal, R. J., Egorova, K. S., Rozovsky, Y.
M., and Nurminsky, D. I. (2009) The B-type lamin is required for
somatic repression of testis-specific gene clusters, Proc. Natl.
Acad. Sci. USA, 106, 3282-3287.
15.Kohwi, M., Lupton, J. R., Lai, S. L., Miller, M.
R., and Doe, C. Q. (2013) Developmentally regulated subnuclear genome
reorganization restricts neural progenitor competence in
Drosophila, Cell, 152, 97-108.
16.Amendola, M., and van Steensel, B. (2015) Nuclear
lamins are not required for lamina-associated domain organization in
mouse embryonic stem cells, EMBO Rep., 16, 610-617.
17.Hochstrasser, M., and Sedat, J. W. (1987)
Three-dimensional organization of Drosophila melanogaster
interphase nuclei. II. Chromosome spatial organization and gene
regulation, J. Cell Biol., 104, 1471-1483.
18.Hochstrasser, M., and Sedat, J. W. (1987)
Three-dimensional organization of Drosophila melanogaster
interphase nuclei. I. Tissue-specific aspects of polytene nuclear
architecture, J. Cell Biol., 104, 1455-1470.
19.Chen, H., Zheng, X., and Zheng, Y. (2014)
Age-associated loss of lamin-B leads to systemic inflammation and gut
hyperplasia, Cell, 159, 829-843.
20.Cremer, T., and Cremer, M. (2010) Chromosome
territories, Cold Spring Harb. Perspect. Biol., 2,
a003889.
21.Cremer, T., and Cremer, C. (2001) Chromosome
territories, nuclear architecture and gene regulation in mammalian
cells, Nat. Rev. Genet., 2, 292-301.
22.Bauer, C. R., Hartl, T. A., and Bosco, G. (2012)
Condensin II promotes the formation of chromosome territories by
inducing axial compaction of polyploid interphase chromosomes, PLoS
Genet., 8, e1002873.
23.Mathog, D., Hochstrasser, M., Gruenbaum, Y.,
Saumweber, H., and Sedat, J. (1984) Characteristic folding pattern of
polytene chromosomes in Drosophila salivary gland nuclei,
Nature, 308, 414-421.
24.Lieberman-Aiden, E., van Berkum, N. L., Williams,
L., Imakaev, M., Ragoczy, T., Telling, A., Amit, I., Lajoie, B. R.,
Sabo, P. J., Dorschner, M. O., Sandstrom, R., Bernstein, B., Bender, M.
A., Groudine, M., Gnirke, A., Stamatoyannopoulos, J., Mirny, L. A.,
Lander, E. S., and Dekker, J. (2009) Comprehensive mapping of
long-range interactions reveals folding principles of the human genome,
Science, 326, 289-293.
25.Sexton, T., Yaffe, E., Kenigsberg, E.,
Bantignies, F., Leblanc, B., Hoichman, M., Parrinello, H., Tanay, A.,
and Cavalli, G. (2012) Three-dimensional folding and functional
organization principles of the Drosophila genome, Cell,
148, 458-472.
26.Ulianov, S. V., Khrameeva, E. E., Gavrilov, A.
A., Flyamer, I. M., Kos, P., Mikhaleva, E. A., Penin, A. A., Logacheva,
M. D., Imakaev, M. V., Chertovich, A., Gelfand, M. S., Shevelyov, Y.
Y., and Razin, S. V. (2016) Active chromatin and transcription play a
key role in chromosome partitioning into topologically associating
domains, Genome Res., 26, 70-84.
27.Battulin, N., Fishman, V. S., Mazur, A. M.,
Pomaznoy, M., Khabarova, A. A., Afonnikov, D. A., Prokhortchouk, E. B.,
and Serov, O. L. (2015) Comparison of the three-dimensional
organization of sperm and fibroblast genomes using the Hi-C approach,
Genome Biol., 16, 77.
28.Dixon, J. R., Selvaraj, S., Yue, F., Kim, A., Li,
Y., Shen, Y., Hu, M., Liu, J. S., and Ren, B. (2012) Topological
domains in mammalian genomes identified by analysis of chromatin
interactions, Nature, 485, 376-380.
29.Ong, C. T., and Corces, V. G. (2014) CTCF: an
architectural protein bridging genome topology and function, Nat.
Rev. Genet., 15, 234-246.
30.Cmarko, D., Verschure, P. J., Martin, T. E.,
Dahmus, M. E., Krause, S., Fu, X. D., van Driel, R., and Fakan, S.
(1999) Ultrastructural analysis of transcription and splicing in the
cell nucleus after bromo-UTP microinjection, Mol. Biol. Cell,
10, 211-223.
31.Eagen, K. P., Hartl, T. A., and Kornberg, R. D.
(2015) Stable chromosome condensation revealed by chromosome
conformation capture, Cell, 163, 934-946.
32.Filion, G. J., van Bemmel, J. G., Braunschweig,
U., Talhout, W., Kind, J., Ward, L. D., Brugman, W., de Castro, I. J.,
Kerkhoven, R. M., Bussemaker, H. J., and van Steensel, B. (2010)
Systematic protein location mapping reveals five principal chromatin
types in Drosophila cells, Cell, 143, 212-224.
33.Kharchenko, P. V., Alekseyenko, A. A., Schwartz,
Y. B., Minoda, A., Riddle, N. C., Ernst, J., Sabo, P. J., Larschan, E.,
Gorchakov, A. A., Gu, T., Linder-Basso, D., Plachetka, A., Shanover,
G., Tolstorukov, M. Y., Luquette, L. J., Xi, R., Jung, Y. L., Park, R.
W., Bishop, E. P., Canfield, T. K., Sandstrom, R., Thurman, R. E.,
MacAlpine, D. M., Stamatoyannopoulos, J. A., Kellis, M., Elgin, S. C.,
Kuroda, M. I., Pirrotta, V., Karpen, G. H., and Park, P. J. (2011)
Comprehensive analysis of the chromatin landscape in Drosophila
melanogaster, Nature, 471, 480-485.
34.Osborne, C. S., Chakalova, L., Brown, K. E.,
Carter, D., Horton, A., Debrand, E., Goyenechea, B., Mitchell, J. A.,
Lopes, S., Reik, W., and Fraser, P. (2004) Active genes dynamically
colocalize to shared sites of ongoing transcription, Nat.
Genet., 36, 1065-1071.
35.Cheutin, T., and Cavalli, G. (2012) Progressive
polycomb assembly on H3K27me3 compartments generates polycomb bodies
with developmentally regulated motion, PLoS Genet., 8,
e1002465.
36.Li, H. B., Ohno, K., Gui, H., and Pirrotta, V.
(2013) Insulators target active genes to transcription factories and
polycomb-repressed genes to polycomb bodies, PLoS Genet.,
9, e1003436.
37.Finlan, L. E., Sproul, D., Thomson, I., Boyle,
S., Kerr, E., Perry, P., Ylstra, B., Chubb, J. R., and Bickmore, W. A.
(2008) Recruitment to the nuclear periphery can alter expression of
genes in human cells, PLoS Genet., 4, e1000039.
38.Reddy, K. L., Zullo, J. M., Bertolino, E., and
Singh, H. (2008) Transcriptional repression mediated by repositioning
of genes to the nuclear lamina, Nature, 452, 243-247.
39.Henikoff, S., and Dreesen, T. D. (1989)
Trans-inactivation of the Drosophila brown gene: evidence for
transcriptional repression and somatic pairing dependence, Proc.
Natl. Acad. Sci. USA, 86, 6704-6708.
40.Tolhuis, B., Blom, M., Kerkhoven, R. M., Pagie,
L., Teunissen, H., Nieuwland, M., Simonis, M., de Laat, W., van
Lohuizen, M., and van Steensel, B. (2011) Interactions among Polycomb
domains are guided by chromosome architecture, PLoS Genet.,
7, e1001343.
41.Bantignies, F., Roure, V., Comet, I., Leblanc,
B., Schuettengruber, B., Bonnet, J., Tixier, V., Mas, A., and Cavalli,
G. (2011) Polycomb-dependent regulatory contacts between distant Hox
loci in Drosophila, Cell, 144, 214-226.
42.Denholtz, M., Bonora, G., Chronis, C., Splinter,
E., de Laat, W., Ernst, J., Pellegrini, M., and Plath, K. (2013)
Long-range chromatin contacts in embryonic stem cells reveal a role for
pluripotency factors and polycomb proteins in genome organization,
Cell Stem Cell, 13, 602-616.
43.Papantonis, A., Kohro, T., Baboo, S., Larkin, J.
D., Deng, B., Short, P., Tsutsumi, S., Taylor, S., Kanki, Y.,
Kobayashi, M., Li, G., Poh, H. M., Ruan, X., Aburatani, H., Ruan, Y.,
Kodama, T., Wada, Y., and Cook, P. R. (2012) TNFalpha signals through
specialized factories where responsive coding and miRNA genes are
transcribed, EMBO J., 31, 4404-4414.
44.Schoenfelder, S., Sexton, T., Chakalova, L.,
Cope, N. F., Horton, A., Andrews, S., Kurukuti, S., Mitchell, J. A.,
Umlauf, D., Dimitrova, D. S., Eskiw, C. H., Luo, Y., Wei, C. L., Ruan,
Y., Bieker, J. J., and Fraser, P. (2010) Preferential associations
between co-regulated genes reveal a transcriptional interactome in
erythroid cells, Nat. Genet., 42, 53-61.
45.Kind, J., Pagie, L., Ortabozkoyun, H., Boyle, S.,
de Vries, S. S., Janssen, H., Amendola, M., Nolen, L. D., Bickmore, W.
A., and van Steensel, B. (2013) Single-cell dynamics of
genome–nuclear lamina interactions, Cell, 153,
178-192.
46.Kinney, N. A., Onufriev, A. V., and Sharakhov, I.
V. (2015) Quantified effects of chromosome–nuclear envelope
attachments on 3D organization of chromosomes, Nucleus,
6, 212-224.
47.Chubb, J. R., Boyle, S., Perry, P., and Bickmore,
W. A. (2002) Chromatin motion is constrained by association with
nuclear compartments in human cells, Curr. Biol., 12,
439-445.
48.Hubner, M., and Spector, D. (2010) Chromatin
dynamics, Annu. Rev. Biophys., 39, 471-489.
49.Agmon, N., Liefshitz, B., Zimmer, C., Fabre, E.,
and Kupiec, M. (2013) Effect of nuclear architecture on the efficiency
of double-strand break repair, Nat. Cell Biol., 15,
694-699.
50.Sexton, T., and Cavalli, G. (2015) The role of
chromosome domains in shaping the functional genome, Cell,
160, 1049-1059.
51.Fanucchi, S., Shibayama, Y., Burd, S., Weinberg,
M. S., and Mhlanga, M. M. (2013) Chromosomal contact permits
transcription between coregulated genes, Cell, 155,
606-620.
52.Hartl, T. A., Smith, H. F., and Bosco, G. (2008)
Chromosome alignment and transvection are antagonized by condensin II,
Science, 322, 1384-1387.
53.Zhimulev, I. F. (1996) Morphology and structure
of polytene chromosomes, in Advances in Genetics (Hall, J. C.,
ed.) Vol. 34, Academic Press, San Diego, pp. 1-490.
54.Demakov, S. A., Vatolina, T. Yu., Babenko, V. N.,
Semeshin, V. F., Belyaeva, E. S., and Zhimulev, I. F. (2011) Protein
composition of interband regions in polytene and cell line chromosomes
of Drosophila melanogaster, BMC Genom., 12,
566.
55.Belyaeva, E. S., Goncharov, F. P., Demakova, O.
V., Kolesnikova, T. D., Boldyreva, L. V., Semeshin, V. F., and
Zhimulev, I. F. (2012) Late replication domains in polytene and
non-polytene cells of Drosophila melanogaster, PLoS One,
7, e30035.
56.Vatolina, T. Y., Boldyreva, L. V., Demakova, O.
V., Demakov, S. A., Kokoza, E. B., Semeshin, V. F., Babenko, V. N.,
Goncharov, F. P., Belyaeva, E. S., and Zhimulev, I. F. (2011) Identical
functional organization of nonpolytene and polytene chromosomes in
Drosophila melanogaster, PLoS One, 6, e25960.
57.Zhimulev, I. F., Zykova, T. Y., Goncharov, F. P.,
Khoroshko, V. A., Demakova, O. V., Semeshin, V. F., Pokholkova, G. V.,
Boldyreva, L. V., Demidova, D. S., Babenko, V. N., Demakov, S. A., and
Belyaeva, E. S. (2014) Genetic organization of interphase chromosome
bands and interbands in Drosophila melanogaster, PLoS
One, 9, e101631.
58.Mathog, D., and Sedat, J. W. (1989) The
three-dimensional organization of polytene nuclei in male Drosophila
melanogaster with compound XY or ring X chromosomes,
Genetics, 121, 293-311.
59.Sharakhov, I. V., Sharakhova, M. V., Mbogo, C.
M., Koekemoer, L. L., and Yan, G. (2001) Linear and spatial
organization of polytene chromosomes of the African malaria mosquito
Anopheles funestus, Genetics, 159, 211-218.
60.Sharakhova, M. V., George, P., Brusentsova, I.
V., Leman, S. C., Bailey, J. A., Smith, C. D., and Sharakhov, I. V.
(2010 Genome mapping and characterization of the Anopheles
gambiae heterochromatin, BMC Genom., 11, 459.
61.Beliveau, B. J., Joyce, E. F., Apostolopoulos,
N., Yilmaz, F., Fonseka, C. Y., McCole, R. B., Chang, Y., Li, J. B.,
Senaratne, T. N., Williams, B. R., Rouillard, J. M., and Wu, C. T.
(2012) Versatile design and synthesis platform for visualizing genomes
with Oligopaint FISH probes, Proc. Natl. Acad. Sci. USA,
109, 21301-21306.
62.Cremer, M., Kupper, K., Wagler, B., Wizelman, L.,
von Hase, J., Weiland, Y., Kreja, L., Diebold, J., Speicher, M. R., and
Cremer, T. (2003) Inheritance of gene density-related higher order
chromatin arrangements in normal and tumor cell nuclei, J. Cell
Biol., 162, 809-820.
63.Artemov, G., Bondarenko, S., Sapunov, G., and
Stegniy, V. (2015) Tissue-specific differences in the spatial
interposition of X-chromosome and 3R chromosome regions in the malaria
mosquito Anopheles messeae Fall, PLoS One, 10,
e0115281.
64.Stegnii, V. N. (1987) Systemic reorganization of
the architectonics of polytene chromosomes in the onto- and
phylogenesis of malaria mosquitoes, Genetika, 23,
821-827.
65.Bondarenko, S. M., Artemov, G. N., Sharakhov, I.
V., and Stegniy, V. N. (2017) Tissue-specific features of the X
chromosome and nucleolus spatial dynamics in a malaria mosquito,
Anopheles atroparvus, PLoS One, 12, e0171290.
66.Stegnii, V. N. (1979) Reorganization of the
structure of interphase nuclei during the onto- and phylogeny of
malaria mosquitoes, Dokl. Akad. Nauk SSSR, 249,
1231-1234.
67.Stegnii, V. N., and Sharakhova, M. V. (1991)
Systemic reorganization of the architechtonics of polytene chromosomes
in onto- and phylogenesis of malaria mosquitoes. Structural features
regional of chromosomal adhesion to the nuclear membrane,
Genetika, 27, 828-835.
68.Ollion, J., Cochennec, J., Loll, F., Escude, C.,
and Boudier, T. (2013) TANGO: a generic tool for high-throughput 3D
image analysis for studying nuclear organization,
Bioinformatics, 29, 1840-1841.
69.Stegnii, V. N. (1987) Systemic reorganization of
the architectonics of polytene chromosomes in the onto- and
phylogenesis of malarial mosquitoes. II. Species specificity in the
pattern of chromosome relations with the nuclear envelope of nutrient
ovarian cells, Genetika, 23, 1194-1199.
70.Pombi, M., Caputo, B., Simard, F., Di Deco, M.
A., Coluzzi, M., della Torre, A., Costantini, C., Besansky, N. J., and
Petrarca, V. (2008) Chromosomal plasticity and evolutionary potential
in the malaria vector Anopheles gambiae sensu stricto: insights
from three decades of rare paracentric inversions, BMC Evol.
Biol., 8, 309.
71.Pevzner, P., and Tesler, G. (2003) Human and
mouse genomic sequences reveal extensive breakpoint reuse in mammalian
evolution, Proc. Natl. Acad. Sci. USA, 100,
7672-7677.
72.Alekseyev, M. A. (2008) Multi-break
rearrangements and breakpoint re-uses: from circular to linear genomes,
J. Comput. Biol., 15, 1117-1131.
73.Gandhi, M., Evdokimova, V., and Nikiforov, Y. E.
(2010) Mechanisms of chromosomal rearrangements in solid tumors: the
model of papillary thyroid carcinoma, Mol. Cell Endocrinol.,
321, 36-43.
74.Marshall, W. F. (2002) Order and disorder in the
nucleus, Curr. Biol., 12, R185-192.
75.Folle, G. A. (2008) Nuclear architecture,
chromosome domains and genetic damage, Mutat. Res., 658,
172-183.
76.Fudenberg, G., Getz, G., Meyerson, M., and Mirny,
L. A. (2011) High order chromatin architecture shapes the landscape of
chromosomal alterations in cancer, Nat. Biotechnol., 29,
1109-1113.
77.Gandhi, M., Medvedovic, M., Stringer, J. R., and
Nikiforov, Y. E. (2006) Interphase chromosome folding determines
spatial proximity of genes participating in carcinogenic RET/PTC
rearrangements, Oncogene, 25, 2360-2366.
78.Duan, Z., Andronescu, M., Schutz, K., McIlwain,
S., Kim, Y. J., Lee, C., Shendure, J., Fields, S., Blau, C. A., and
Noble, W. S. (2010) A three-dimensional model of the yeast genome,
Nature, 465, 363-367.
79.Khrameeva, E. E., Fudenberg, G., Gelfand, M. S.,
and Mirny, L. A. (2016) History of chromosome rearrangements reflects
the spatial organization of yeast chromosomes, J. Bioinform. Comput.
Biol., 14, 1641002.
80.Cheutin, T., Bantignies, F., Leblanc, B., and
Cavalli, G. (2010) Chromatin folding: from linear chromosomes to the 4D
nucleus, Cold Spring Harb. Symp. Quant. Biol., 75,
461-473.
81.Peric-Hupkes, D., and van Steensel, B. (2010)
Role of the nuclear lamina in genome organization and gene expression,
Cold Spring Harb. Symp. Quant. Biol., 75, 517-524.
82.Nagano, T., Lubling, Y., Stevens, T. J.,
Schoenfelder, S., Yaffe, E., Dean, W., Laue, E. D., Tanay, A., and
Fraser, P. (2013) Single-cell Hi-C reveals cell-to-cell variability in
chromosome structure, Nature, 502, 59-64.
83.Mirny, L. A. (2011) The fractal globule as a
model of chromatin architecture in the cell, Chromosome Res.,
19, 37-51.
84.Zhang, B., and Wolynes, P. G. (2015) Topology,
structures, and energy landscapes of human chromosomes, Proc. Natl.
Acad. Sci. USA, 112, 6062-6067.
85.Li, Q. J., Tjong, H., Li, X., Gong, K., Zhou, X.
J., Chiolo, I., and Alber, F. (2017) The three-dimensional genome
organization of Drosophila melanogaster through data
integration, Genome Biol., 18, 145.
86.Cook, P. R., and Marenduzzo, D. (2009) Entropic
organization of interphase chromosomes, J. Cell Biol.,
186, 825-834.
87.Mirny, L. (2011) The fractal globule as a model
of chromatin architecture in the cell, Chromosome Res.,
19, 37-51.
88.Wong, H., Marie-Nelly, H., Herbert, S.,
Carrivain, P., Blanc, H., Koszul, R., Fabre, E., and Zimmer, C. (2012)
A predictive computational model of the dynamic 3D interphase yeast
nucleus, Curr. Biol., 22, 1881-1890.
89.Wong, H., Arbona, J. M., and Zimmer, C. (2013)
How to build a yeast nucleus, Nucleus, 4, 361-366
90.Rowley, M. J., and Corces, V. G. (2016) The
three-dimensional genome: principles and roles of long-distance
interactions, Curr. Opin. Cell Biol., 40, 8-14.
91.Kinney, N., Sharakhov, I. V., and Onufriev, A. V.
(2014) Investigation of the chromosome regions with significant
affinity for the nuclear envelope in fruit fly – a model-based
approach, PLoS One, 9, e91943.