REVIEW: Adsorption of Bacteriophages on Bacterial Cells
A. V. Letarov1,2* and E. E. Kulikov1,3
1Winogradskii Institute of Microbiology, Biotechnology Federal Research Center, Russian Academy of Sciences, 117312 Moscow, Russia; E-mail: letarov@gmail.com, eumenius@gmail.com2Lomonosov Moscow State University, Faculty of Biology, 119991 Moscow, Russia
3Moscow Institute of Physics and Technology, Department of Molecular and Biological Physics, 141700 Moscow, Russia
* To whom correspondence should be addressed.
Received July 13, 2017; Revision received August 3, 2017
The biological functions of bacteriophage virions come down to the solution of three basic problems: to provide protection of viral nucleic acid from the factors of extracellular environment, to recognize a host suitable for phage replication, and to provide the delivery of nucleic acid through bacterial cell envelopes. This review considers the main regularities of phage–cell interaction at the initial stages of infection of tailed bacteriophages, from the reversible binding with receptors on the surface to the beginning of phage DNA entry. Data on the structure and functions of the phage adsorption apparatus, the main quantitative characteristics of the adsorption process, and the mechanisms of adaptation of phages and their hosts to each other effective at the stage of adsorption are presented.
KEY WORDS: bacteriophages, bacteriophage adsorption, bacteriophage adsorption kinetics, bacteriophage receptors, receptor-binding proteins, modulation of bacteriophage adsorptionDOI: 10.1134/S0006297917130053
Prokaryotes – bacteria and archaea – are the most important component of most natural ecosystems of the Earth throughout the entire history of the biosphere. It is prokaryotic life forms that prepared our planet for the life of eukaryotes, and they continue to support life on Earth by their involvement in various biogeochemical cycles [1]. In eukaryotic organisms, the cells of symbiotic bacteria are necessary for numerous processes, including nutrition and protection of a host organism [2-5]. All the currently known groups of prokaryotes have viruses – bacteriophages and archaeal viruses capable of specifically infecting their cells [6]. Viral infection leads to profound changes in different aspects of life of microorganisms, including their physiology, ecology, and the population dynamics of microbial communities. It is precisely bacteriophages that are the natural regulators of bacterial quantity and intra- and interspecies diversity, acting as selective bactericidal agents redistributing resources between various bacterial populations. This is especially important in aquatic ecosystems, where the availability of dissolved organic matter limits the growth of most heterotrophic bacteria [7-9]. Phage lysis has an effect also on spatial cell distribution, e.g. by influencing the properties and sedimentation of bacterial aggregates [10]. Viral infection is also a factor of natural selection, which maintains the high diversity of surface structures and intracellular antiviral systems of bacteria and archaea [11-13]. The bacteriophages and viruses of archaea also directly influence the evolution of genomes of their hosts (temperate phages capable of integration into the genome of infected cells and direct transfer of genetic information, as well as transducing virulent phages and transposable phages) [14]. Undoubtedly, the quantitatively dominating group of prokaryotic viruses is the so-called tailed phages (order Caudovirales). This group includes up to 95% of the currently characterized isolates of prokaryotic viruses [15].
One of the key aspects of bacteriophage function is the following: phage particles are capable of highly specific affinity interaction with certain conservative surface structures of bacterial cells possessing biochemical and physiological potential for replication or integration of the genome of this bacteriophage [16, 17].
The adsorption of phage particles to a bacterium leads to the activation of molecular mechanisms of infection (tail contraction, DNA ejection, etc.) in the virion, and it is exactly the interaction between proteins of the adsorption apparatus of bacteriophages and receptor structures of the bacterial cell surface that is necessary for conformational triggering of phage infection [18-20]. Thus, the specificity of phage virion adsorption, as well as adsorption kinetics, primarily determines whether this host strain will be sensitive to the respective bacteriophage and how efficiently the population of this strain can be controlled by the phage population. The latter circumstance is especially important not only for modeling ecological dynamics in natural microbial communities but also for practical applications, among which we should mention in the first place phage infection control in food and microbiological industries [21] and phage therapy for bacterial infections, which is currently considered an important alternative to antibacterial chemotherapy. In some cases, phage therapy makes it possible to solve the problems of treating infections caused by the pathogens with high resistance to the available antibacterial drugs [22]. This review considers the peculiar features of bacteriophage adsorption to the cells of bacterial hosts and the accompanying biological effects.
NATURE OF PHAGE RECEPTORS OF BACTERIAL CELLS
The diversity of bacteriophage receptors is determined by the surface layer structure of host cells. Examples of receptors for different bacteriophages can be found in the table. In eubacteria, the two predominant types of cell wall structure are Gram-positive and Gram-negative (Gr+, Gr–) [23].
Examples of some bacteriophage receptors
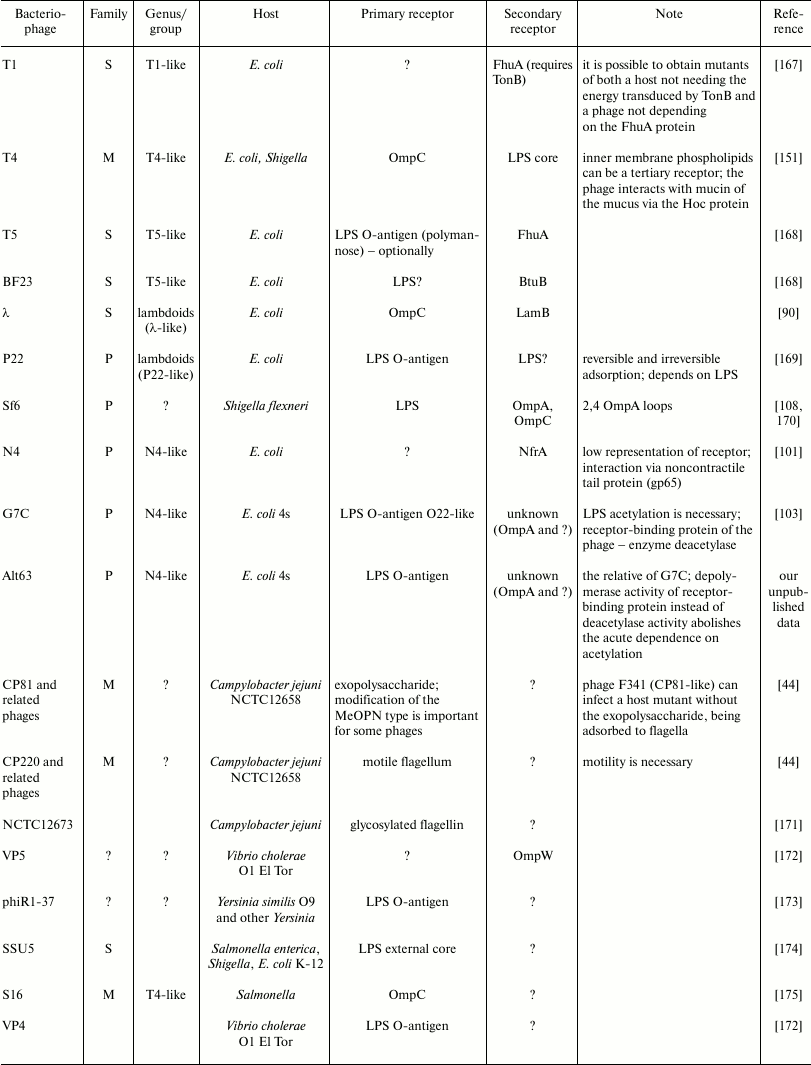
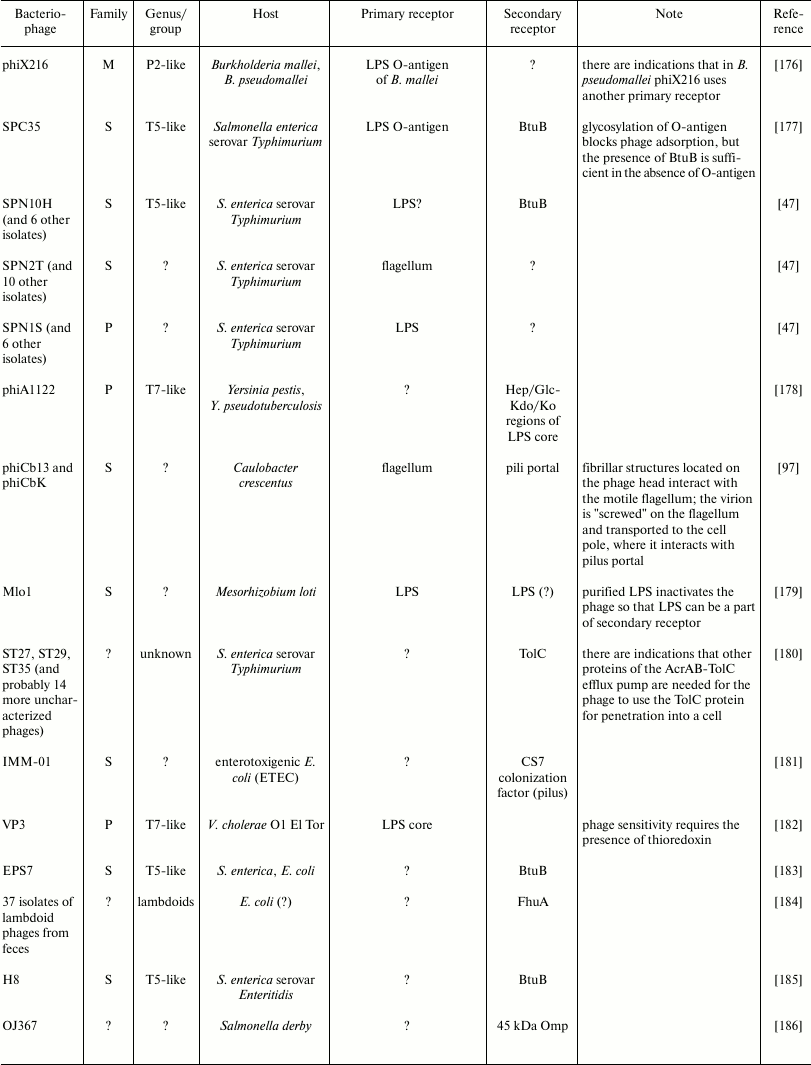
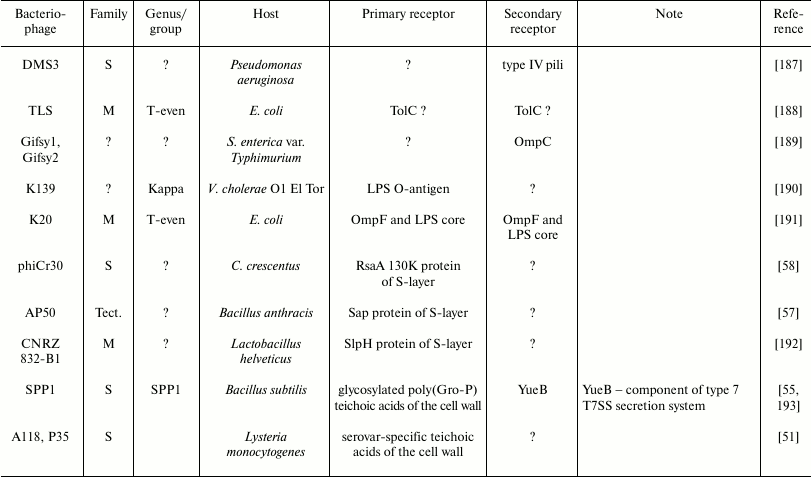
Note: Symbol “?” means that corresponding receptor or
characteristic of bacteriophage is unknown or absent in
publications.
Mycobacteria, e.g. the tuberculosis pathogen Mycobacterium tuberculosis, have a peculiar cell wall structure. These microorganisms belong to the branch of Gr+ bacteria but have additional cell wall layers with some properties similar to those of the outer membrane [24]. In mycoplasmas, the cell wall is absent, and their phages apparently recognize as receptors the molecules localized directly on the cytoplasmic membrane surface.
Phage receptors in Gram-negative bacteria. Lipopolysaccharides. Phage receptors have been best studied in Gram-negative bacteria. The surface of Gram-negative cells is formed by the outer membrane (OM) of the cell envelope.
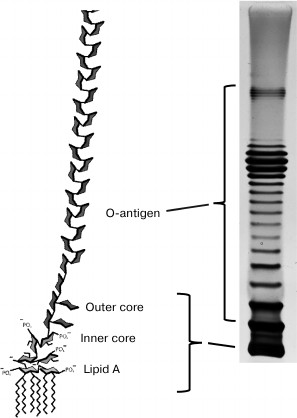
The inner leaflet of the OM is composed mainly of phospholipids that are analogous to the cytoplasmic membrane (CPM) phospholipids, while the outer leaflet contains mainly lipopolysaccharides (LPSs), i.e. lipid–polysaccharide conjugates (Fig. 1).
Fig. 1. Structure of lipopolysaccharide molecules (© CC BY-SA 3.0 image).
The LPS molecule consists of the highly polar phospholipid A (the most conservative part), the core oligosaccharide dividing it into the inner and outer cores, and the O-antigen [25].
Lipid A is a vitally important component of the OM [26]. Mutants of most species of Gram-negative bacteria that are incapable of its synthesis are unviable (except for some mutants of Haemophilus influenzae). The presence of at least some part of the inner core-PS (one or two Kdo (keto-deoxyoctulosonate) residues in Escherichia coli) is also necessary for vitality. O-polysaccharide (OPS, O-antigen), on the contrary, is not necessary for cell viable under laboratory conditions, notwithstanding that polysaccharide residues substantially increase the hydrophilicity of the outer surface of bacteria. Mutants incapable of OPS synthesis are called R (rough) mutants because they often have less regularly shaped colonies and rather dry biomass consistency, in contrast to S (smooth) strains synthesizing normal OPS. The structures of OPS units in many organisms are extremely varied. Therefore, about 200 O-serotypes are distinguished in Escherichia coli species pool, probably due to the selection of bacteria by the immune system (for pathogenic strains) and bacteriophages. However, only five variants of the core-OS structure are known [27]. The O-antigens of Gram-negative bacteria are often recognized by phages as primary receptors [28]. The OPS often effectively masks the underlying structures so that phages first must bind to the O-antigen and, in some cases, enzymatically degrade this structure to provide access to secondary receptors. The LPS core can also be a bacteriophage receptor; in addition, it can be used as both primary and secondary receptor. Some phages, e.g. the Salmonella enterica P22 phage, can use LPS as both primary and secondary receptors [29, 30].
Capsular polysaccharides, exopolysaccharides, and other polysaccharide molecules. Many Gram-negative as well as Gram-positive bacteria produce a polysaccharide material that forms a morphologically structured layer above the cell wall surface – a capsule, or a layer less tightly bound to the cells – exopolysaccharides (that can act as the principal component of the biofilm matrix). Capsular and exopolysaccharides usually have longer carbohydrate chains than OPS. They can be bound to outer membrane lipids but often lose direct covalent bonding to the cell. The organization of the fine structure of bacterial capsules is still unknown. The structures of the repeating elements of capsular PS (K-antigen) are also highly variable, though usually less so than OPS structures. Some E. coli strains produce polysialic acid (K-antigen) and chondroitin-like polysaccharides mimicking the polysaccharides of mammalian cells; in addition, colanic acid production frequently occurs in bacteria [31, 32]. In Klebsiella pneumoniae, there are more than 80 types of K-antigen, which is one of the key factors of pathogenicity of this dangerous infectious agent in hospitals. Capsular polysaccharides very efficiently screen the cell surface from bacteriophages, but they can be used as a primary receptor [33].
In addition to OPS, K-antigen, and exo-PS, some bacteria produce additional types of polysaccharides as, e.g. enterobacterial common antigen (ECA). This polysaccharide is conservative in different representatives of Enterobacteriaceae but confined to this family [34, 35]. The ECA chain length is usually comparable to that of OPS or is slightly less. This polysaccharide can be anchored in the outer membrane due to covalent bonding with a phospholipid, but sometimes it can be attached to the core-PS of PLS instead of O-antigen. Some strains also have a cyclic form of ECA. There are still no data on the possibility of using ECA as a phage receptor.
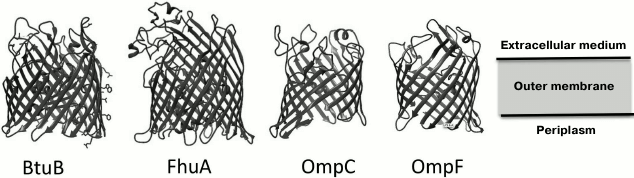
Porins and other proteins of the outer membrane. The outer membrane of Gram-negative bacteria is highly permeable to water, ions, and most low molecular weight compounds due to the presence of a great number of integral membrane proteins – porins. In contrast to the CPM proteins enriched in transmembrane α-helical structures, the OM porins are constructed mainly of β-structural elements forming a barrel, with the central cavity permeable to certain low molecular weight substances [36] (Fig. 2).
Fig. 2. Examples of porin structures in the Escherichia coli outer membrane that are used as receptors by bacteriophages (© CC BY-SA 3.0 image).
The loop-shaped regions of the secondary structure of porins, which connect transmembrane segments on the outer side of the membrane, can be phage receptors [36, 37].
Porins can be both primary and secondary receptors.
In some cases, the proteins of specific transport systems can be bound to CPM through periplasmic proteins – energy carriers. For some phages, the energy delivered in this way is necessary for successful internalization of viral DNA; in other cases, one outer membrane protein acting as a receptor is enough. It is interesting that not only the well-represented porins such as OmpA synthesized in E. coli in amounts up to 105 copies per cell [38] are used as receptors but also rare proteins such as NfrA (phage N4 receptor), the amount of which is about 5 copies per cell [39]. Representation of the proteins on the cell surface makes a tangible contribution to the adsorption kinetics of bacteriophage particles but, theoretically, only one receptor molecule is sufficient for successful infection.
Lipoproteins occur infrequently on the surface of most Gram-negative bacteria due to their natural hydrophobicity, being localized mainly on the periplasmic side of OM and CPM; however, they are present in considerable amounts in some spirochetes. Nevertheless, some amphipathic lipoproteins are exposed on the surface even in enterobacteria; thus, 4 out of 92 known outer membrane lipoproteins can be exposed to extracellular space in E. coli [40]. There are also numerous decoration proteins of the outer membrane surface, sometimes referred to as surface lipoproteins [41], which can be involved in the binding of various extracellular ligands and are probably significant for the pathogenesis of infections caused by Gr– bacteria. Theoretically, lipoproteins can also be used by bacteriophages as receptors, but there are no convincing data on such systems.
It is reported that some surface adhesins of bacteria, in particular Ag43 in E. coli, stimulate phage adsorption in the presence of bile salts (natural amphiphilic surfactants) inhibiting this process [42], but there are still no data on specific recognition of such structures by phages.
Pili, flagella, and other protuberances on OM surface. Pili and flagella are quite often used as bacteriophage receptors. However, flagellar filaments per se and tubular structures of the pili in most cases are used as primary receptors. Many phages use the rotation of flagella for transporting phage particles to the cell surface [43-48]. The portal structures of the pili can be secondary receptors.
Receptors of phages of Gram-positive bacteria. The cell wall of Gr+ bacteria consists of multilayered peptidoglycan enveloping teichoic acid molecules [49]. Teichoic acids of the cell wall make up to 50% of the total wall mass. Teichoic acids can covalently bind to CPM lipids (lipoteichoic acids), as well as form covalent and noncovalent bonds with peptidoglycan [50]. Teichoic acids are very often used as bacteriophage receptors [16, 51-56]. Like Gr– bacteria, the function of phage receptors can be performed by capsular and other surface polysaccharides, as well as the surface proteins of Gram-positive cells. The S-layer – a paracrystalline layer of protein subunits covering the cell surface – is a structure typical of many strains of both Gr+ and Gr– bacteria. Though the representativeness of S-layer subunits on the surface is very high (the order of 105 subunits for a large cell such as B. anthracis), there are very few data on the use of this structure by bacteriophages (see table). It has been shown that the S-layer is a receptor for at least B. anthracis phage AP50 (tectivirus) [57], Caulobacter crescentus siphovirus phiCr30 [58], and Lactobacillus helveticus myovirus CNRZ 832-B1 [59] (see table). It seems that S-layer-specific phages often escape the attention of researchers, because many strains lose this structure after being cultivated for a long time under laboratory conditions and, therefore, the occurrence of such viruses in nature may be higher.
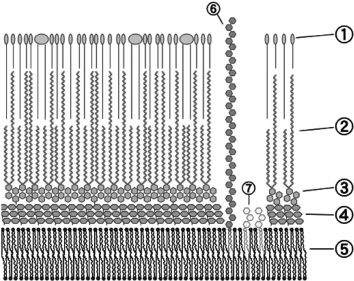
Mycobacterial phage receptors. The phage receptors of mycobacteria have been studied very poorly. Though mycobacteria belong to the phylogenetic branch of Gr+ organisms, the structure of their cell wall is substantially different from other Gr+ microbes (Fig. 3).
Fig. 3. Envelope structure of M. tuberculosis: 1) external lipids; 2) mycolic acids; 3) polysaccharide (arabinogalactan); 4) peptidoglycan; 5) plasma membrane; 6) lipoarabinomannan (LAM); 7) phosphatidylinositol mannoside (© CC BY-SA 3.0 image).
The bulk of the M. tuberculosis cell wall consists of covalently bound peptidoglycan, arabinogalactan, and mycolic acid, which form the so-called mycolyl–arabinogalactan–peptidoglycan complex (mAGP). In addition to mAGP, the cell wall comprises several variants of different free, not covalently bound lipids, including glycolipids, peptidoglycolipids, sulfolipids, phenol glucolipids, etc. Mycolic acid molecules covalently bound to arabinogalactan and free lipid molecules form a lipid bilayer on the cell surface, being virtually a cell outer membrane (mycobacterial outer membrane). This membrane also includes proteins with structural organization similar to that of porins of Gram-negative bacteria [60].
Mycobacterial cells are often surrounded by capsules over the outer membrane.
The entire cell wall is transpierced by molecules of lipoarabinomannan – a lipopolysaccharide anchored in the outer leaflet of the plasma membrane.
According to the available data, the phage receptors of mycobacteria are more often the lipid compounds of the cell wall. The Mycobacterium smegmatis phage I3 uses as receptors molecules of glycopeptolipid – a tetrapeptide covalently bound to the lipid moieties and a methyl-rhamnose residue [61]. However, receptors have not yet been identified for the overwhelming majority of mycobacteriophages.
RECEPTOR-BINDING PROTEINS OF BACTERIOPHAGES
The adsorption apparatus of phages includes proteins called adhesins or receptor-binding proteins (RBPs), as well as other virion components receiving signals from RBP binding and responding to them by certain conformational rearrangements. It also includes additional components involved in modulation of the function of the adsorption apparatus in some viruses. In tailed phages, the entire phage tail, together with some capsid proteins involved in DNA penetration through cell surface structures and probably able to interact with certain receptor structures on bacterial surface (decoration proteins), can be considered as adsorption apparatus in the broad sense of this term.
Phage adhesins and their locations. In most cases, both primary and secondary receptors are recognized by phage tail adhesins. In the long-tailed phages of Gram-negative bacteria, the adhesins are mostly fibrillar. The basis of phage fibers is always one or several trimeric proteins with polypeptide chains arranged parallel to each other and forming a helical structure.
Such proteins usually have one or several β-helical elements [62-66], see also [67] and the references given in that report.
The N-terminal domains of the fibers are used for attachment to the virion or, in the case of composite fibers, to more proximal elements. The C-terminal domains are oriented away from the virion and can form receptor-binding centers or attach a separate receptor-binding protein that may have different degree of oligomerization (most often monomer).
The siphoviruses of Gram-negative hosts usually have an axial (central) tail fiber used for the recognition of secondary receptor [19]. This fiber is usually formed by a single protein. In the λ phage, it is a product of the J gene. At the C-terminus of this fiber there is a domain recognizing the maltose transporter protein LamB [68]. In the T5 phage, the central fiber formed by the pb4 protein at the C-terminus carries a monomer of the receptor-binding protein pb5 [69].
The lateral tail fibers of siphoviruses are usually needed for recognition of primary receptors, and their binding is not necessary for interaction with secondary receptor. Therefore, after long-term cultivation of bacteriophage in laboratory strains, lateral fibers are often lost due to the selection of phage lines with inactivated fiber genes, because cells can be infected directly through the interaction between the axial fiber and the respective receptor. Thus, in many laboratory strains of the λ phage, lateral fibers are not formed because of mutation in the stf gene [70]. In the siphoviruses of E. coli, the targets of lateral fibers are usually LPS O-polysaccharides. The adsorption constant on the strains, which produce OPS recognized by the fibers, is much higher compared to R-mutants [71]. Moreover, on many strains OPSs fully screen the surface receptors of OM, making infection impossible without the involvement of lateral fibers.
In myoviruses, e.g. in T-even phages, long tail fibers (LTFs) attached to the baseplate also perform the function of primary receptor recognition.
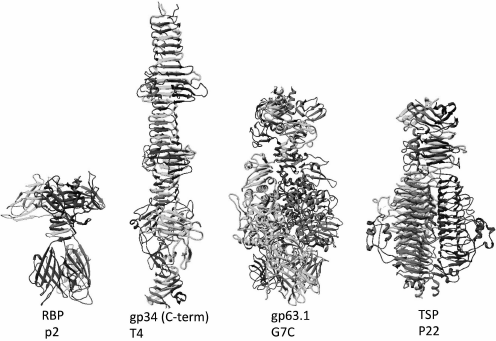
Podoviruses can also have fibrillar adhesins like, e.g. the T7 phage; however, tailspike proteins are more widespread. Structurally, the spikes of bacteriophages are usually parallel homotrimers but, in contrast to fibers, are constructed mainly of globular domains (Fig. 4). Quite often, tail spikes have an enzymatic activity allowing them to penetrate through the polysaccharide layers of cell envelope.
Fig. 4. Structures of the receptor-binding protein (RBP) of lactophage p2, the C-terminal fragment of the proximal half of LTF of the T4 phage containing β-helix elements, the RBP of the G7C phage gp63.1 possessing an acetyl esterase activity, the modifying O-antigen of E. coli, and the tailspike protein of the Salmonella P22 phage having enzymatic activity of O-antigen depolymerase (the enzymatic domain also has β-helical structure).
Until recently, adhesins shaped as tail spikes were considered an exceptional attribute of podoviruses, but now bacteriophages of other families carrying such proteins have been found. Thus, the enterobacterial myovirus Vi-I and the related phages (Det7, CBA120, etc.) have branched-chain adhesins as fibers carrying several different types of spikes. On electron microphotographs, such adhesins look like a palm with fingers spread apart.
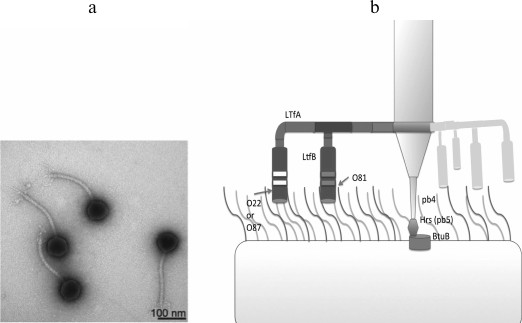
In the T5-like coliphages DT57C and DT571/2, the LTFs have a branched structure: the shorter protein LtfB is attached to the long fibrillar protein LtfA. Each of these proteins carries a receptor-binding center specifically interacting with a certain type of O-antigen. As a result, the phage initiates the hosts with two types of O-antigen. In the DT57C and DT571/2 phages, LtfA proteins recognize E. coli O22 and E. coli O87, respectively, while LtfB proteins are identical and recognize the O81 serotype [72].
Most other phages also seem to use branched adhesins to extend the range of phage hosts recognizing different receptors. However, it is likely that in some cases such structures are necessary to increase the number of binding centers for enhancing the affinity of a phage to a cell surface due to the effect of avidity.
Branched-chain adhesins have also been found in podoviruses, e.g. in K1-5, phiEco32, and G7C coliphages ([73, 74], P. Leiman, personal communication). However, in G7C both proteins forming a branched adhesin are necessary for cell infection because they recognize the primary and secondary receptors.
At the terminals of six-tail fibers of the BPP1 phage of the whooping cough pathogen Bordetella pertussis, there are two interconnected trimers of receptor-binding protein Mtd, which make the distal part of the fiber look like a flattened blade [75]. This phage has an original system for enhancing the variability of adhesin by the retrohoming mechanism.
The variants described here do not exhaust the structural diversity of adhesions even of the phages of Gr– bacteria.
Many various morphologies of protuberances of the baseplates in different phages that also seem capable of performing the functions of adhesins have been described using data of electron and cryoelectron microscopy.
Classical fibrillar adhesins are less typical for the phages of Gr+ hosts (except for myoviruses). In most cases, they carry receptor-binding proteins similar to tail spikes in organization. Some siphoviruses of Gr+ bacteria have something like a baseplate with 6 to 18 trimers of receptor-binding proteins [76, 77] rather similar to tail spikes.
In Lactobacillus J-1 phage and many other lactophages, the classical receptor-binding proteins are absent, and their functions are performed by “additional” polysaccharide-binding domains present in the Dit protein forming the lower ring of the basal structure of the tail [78].
Nevertheless, some phages of Clostridium difficile siphoviruses were shown to carry the genes of fibrillar adhesins involved in cell surface recognition (M. R. J. Clokie, Leicester University, personal communication).
RECEPTION OF BACTERIOPHAGES ON HOST CELLS – GENERAL
ISSUES
Specific adsorption of bacteriophages is a major function of a virion, i.e. recognition of a host cell suitable for virus replication and delivery of viral nucleic acid into this cell. From a chemical standpoint, bacteriophage adsorption is a specific, usually noncovalent binding of phage receptor-binding proteins (viral adhesins) with certain molecules on the bacterial cell surface, which are called phage receptors.
The molecular recognition of a receptor by adhesin occurs because of sufficiently exact steric correspondence between the receptor-binding center of the phage adhesin and a certain part of the receptor molecule [79, 80]. Thus, not the whole receptor molecule is directly involved in the recognition by phage adhesin, but a comparatively small region sterically corresponding to the receptor-binding center of the viral protein (by analogy to the recognition by antibodies of separate epitopes on the surface of large antigen molecules). Primary recognition seems to be followed by induced correspondence of adhesin and receptor structures, which leads to a conformational signal activating phage infection and the subsequent molecular rearrangements in the virion finally resulting in cell infection.
For example, the Oad (pb5) adhesin protein of T5 phage is bound to a short region of about seven bp in one of the loops of the FhuA protein, a siderophore transporter of the E. coli outer membrane [81]. It approximately corresponds to the size of a typical epitope recognized by antibodies, makes it recognizes with a high degree of specificity (theoretically, 1/207). The bacteriophages recognizing the polysaccharides of bacterial cell surface usually recognize the sequences of 2-3 monosaccharide residues due to the presence of lectin domains in their adhesins [82, 83]. The high degree of molecular match between adhesin and receptor achieves rather tight binding due to electrostatic, hydrogen bonding, and dispersion interactions. Transient covalent binding is possible in the case of processive adhesins having their own enzymatic activities and chemically modifying the receptor (glycosidases, peptidases, etc.) [84-86]. The specificity of molecular recognition of receptors substantially limits the range of hosts infected by a given bacteriophage, simultaneously being a guarantee that the phage DNA will be delivered to cells capable of supporting the propagation of the virus. Because direct recognition involves only a small region of the surface receptor molecule, the adsorption specificity of phages may be rather narrow – within a group of bacterial strains with similar surface structures. The more conservative is the structure recognized by a certain bacteriophage, the broader is its adsorption specificity, which however can be limited by masking the respective receptor by other structures or the presence in the cells of resistance mechanisms blocking the stages of infection after the adsorption.
The interaction between the phage and the cell surface can usually be divided into two phases: reversible and irreversible adsorption [17, 87, 88]. Reversible adsorption provides primary cell recognition, which sometimes is less specific compared to irreversible adsorption. At this stage, phage particle dissociation from the cell surface is possible with preservation of the viability of the viral particle. The transition to irreversible adsorption usually requires additional binding of a phage protein to some secondary receptor, either using the same RBP or another versus that employed for reversible binding. Secondary receptor recognition is followed by rearrangements in the virion structure, which are necessary to create a channel for phage genome delivery through the cell envelope and for nucleic acid ejection. The mechanisms of interaction between tailed phages and cell receptors will be considered below.
ADSORPTION KINETICS
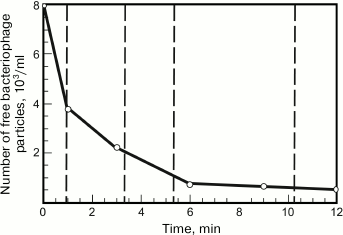
Bacteriophage adsorption can be assessed by measuring a decrease in the biological titer of the bacteriophage in solution due to its adsorption on introduced cells. To determine such dependence, the phage is mixed with the cells at a great excess of the latter (low multiplicity of infection). Then the infected cells are removed by centrifugation at different intervals or killed with chloroform, and the remaining free bacteriophage particles are titrated by inoculation on plates [89]. A typical bacteriophage adsorption curve is presented in Fig. 5.
Fig. 5. Time dependence of the concentration of free G7C bacteriophage particles. The concentration of host cells is about 107 PFU/ml.
In the first approximation, phage adsorption shows second-order kinetics [16, 87], i.e. the reaction rate depends on the first-degree concentrations of adsorbent and adsorbate (in this case, phages and bacteria):
–dP/dt = K[B][P],
where K (ml/min) is the adsorption constant depending only on temperature (as well as on the physiological state of the cells and the medium composition), [B] and [P] are the current bacterial and cell concentrations at time t. From this differential equation, it follows that under conditions of great excess of bacterial cells, when their concentration does not vary during the experiment,
K = B × t × ln(P0/P),
where B is the concentration of bacterial cells, t is the time (min) from the start of the experiment, P0 is the initial phage concentration at time t = 0, and P is the free phage concentration at time t. The characteristic K values for phages are usually of the order of 10–9 ml/min. Higher constants, up to 10–8 ml/min or slightly more, are typical of the viruses using molecules with very high representativeness on the cell surface as receptors, e.g. polysaccharides, so that almost any random collision of a phage particle and a cell leads to successful infection [17]. The low values of adsorption constant (about 10–11 ml/min) are typical of filamentous phages adsorbed at the ends of sex pili of bacterial cells, as well as some other phages using lowly represented receptors or having a low affinity to phage receptors (e.g. due to a mutation-induced change in the latter) [90].
Many significant consequences derive from the adsorption isotherm equation presented above. At sufficiently low concentrations of phages and host bacteria, the time spent on the infection of a certain portion of cells necessary for the predetermined increase in phage population can be considerably greater than the latent period per se. Consequently, increasing the phage population in the experiment will be observed only upon reaching a certain critical density of a host population. For example, if we record phage growth at a 2-fold increase in the PFU number between two consecutive measurements made, e.g. once per hour, for successful registration of phage growth it is necessary that no less than 1/n of all phage particles would be adsorbed during the time (1 – t), where t is the length of the latent period (h) and n is the phage yield per infected cell. It is not difficult to calculate that, with a typical value of adsorption constant of 10–9 ml/min, the latent period of 20 min and the phage yield of 100 particles per cell, the required density of bacterial population will be about 3·105 cells/ml. It accounts for the apparent threshold concentrations of bacteria observed in experiments that are necessary for the beginning of phage growth [91, 92]. Analogously, if we set a goal to stop the growth of a bacterial population, no less than 50% of them must be infected during a bacterial generation. Consequently, the threshold phage concentration for a generation time of 20 min will be about 1.8·107 particles per ml. The results of these calculations are very constructive, as it is well known that even a single phage particle in a test tube with a growing culture containing millions of cells can cause massive bacterial lysis. However, this contradiction is merely apparent: under such experimental conditions, phages are neither destroyed nor carried beyond the system; therefore, their concentration rapidly increases. In practice, upon addition of a small number of phage to a culture, bacterial growth continues for some time, until the viral concentration exceeds the threshold value. The terms “multiplication threshold” and “inundation threshold” were proposed to denote the threshold cell concentration required for the apparent growth and threshold concentration of a phage, respectively [93].
From these calculations, it also follows that the frequently used value of multiplicity of infection (the number of phages per cell) makes sense only under conditions of high phage and cell concentrations. Indeed, if the absolute concentrations are low, even at a formally high ratio of the number of viral particles to the cell number in the system, the effective multiplicity of infection within a given time can be extremely low.
It should also be noted that it is extremely important to consider the threshold concentrations of phages and bacteria when using phage infection as a tool of biological control of microbial populations, including phage therapy for bacterial infections [93, 94], the application of phage preparations for decontamination or preservation of foods [95, 96], and other similar applications.
CHAIN OF EVENTS DURING PHAGE INFECTION OF A CELL
The early stages of infection are known in detail only for few phages of Gram-negative bacteria.
As mentioned above, at rather low concentrations of bacterial hosts observed in many natural ecosystems, the frequency of occurrence of phage particles and sensitive cells can be very low. Therefore, some phages use an adsorption strategy, when initially there is an interaction aimed at merely retaining the virion close to the host cell. In contrast to the classical interaction with primary receptors, which is described below, these interactions seem to have no direct functional relation to the final host recognition and initiation of the infection.
Though most receptor-binding proteins are associated with phage tails (in podoviruses, they usually form a ring around the tail, which is attached directly to the phage neck), in some cases adsorption can also involve capsid proteins. Thus, the Caulobacter crescentus siphovirus phiCb13 has filamentous protuberances of the head decoration protein localized at the vertices of its icosahedral capsid. These filaments interact with flagellin from the flagella of the motile cells of C. crescentus and wind around the flagellar filament in such a way that, because of rotation of the flagellum, the phage particle descends to the cell surface like a nut moving around the thread of a bolt [97].
Near the cell pole, the phage recognizes a secondary receptor (the portal structure of the pilus) due to the “common” adhesins located at the basal structure of its tail. It should be noted that in many other phages using flagellum rotation the respective adhesins are located on the basal structure of the tail.
The decoration protein Hoc occupying the centers of hexamers of the T4 phage capsid protein was also shown capable of binding polysaccharide ligands. However, it turned out that they were not the receptors on the bacterial surface but the polysaccharide mucin – a component of crypt cell production in the intestines [98]. This supposedly leads to the concentration of phage particles in the parietal layer of the intestines, where the content of metabolically active hosts (E. coli) is much higher. It is interesting that the Hoc protein is one of the phage proteins containing immunoglobulin-like domains [99, 100]. Such domains have been found in many phages and are always located exposed to the external medium. For example, such domains have been found on many head decoration proteins like in the T4 and T5 phages, on tail tube proteins in the λ and T5 phages, on the ends of collar strands of “whiskers” in the RB43 phage, and on other proteins of virions. The Ig-like domains are often highly variable with respect to their sequences even in closely related phages, suggesting them to be instruments of adaptation to specific habitat conditions. Before discovery of the binding of the Hoc protein to mucin, the function of these domains was unknown. Now there are grounds to suggest that at least some of them can recognize biomolecules, increasing the concentration of phages near potential hosts.
In a more typical case, however, reversible and irreversible adsorption is performed by adhesins associated with the phage tail, being stages of the cell infection process. As consistent with the two-stage pattern of adsorption, the recognitions of primary and secondary receptors in most bacteriophages are differentiated. In some bacteriophages, the recognition of primary and secondary receptors is functionally dissociated: the primary receptor binding seems to be necessary only for phage fixation on the cell surface, which increases the probability of successful cell infection at each collision. This binding is not functionally necessary for the secondary receptor recognition and infection, so that, in the absence of cell surface structures masking the secondary receptor, the infection can occur directly, without the involvement of adhesins recognizing the primary receptor.
For example, the N4 phage can directly bind to its protein receptor NfrA via protein pg65, forming a kind of sheath around the short tail of this podovirus [101]. This noncontractile sheath acts as a receptor recognizing the NfrA porin [39]. Interestingly, in most other N4-like phages this function is performed by spike proteins [74, 102]. However, many such phages use another strategy (see below).
The T5-like viruses can also infect bacterial strains lacking surface O-polysaccharide due to direct recognition of the outer membrane porin proteins by the phage adhesin Oad (pb5) [81]. However, even with such strategy, the primary receptor recognition by the classical adhesins bound to the phage tail often play a key role in providing a contact between the respective RBP and the secondary receptor. Thus, during infection by T5-like coliphages DT57C and DT571/2 of E. coli strains lacking O-antigen, the lateral tail fibers (LTFs) are not involved, and phage mutants lacking this structure infect the cells at the same rate as the wild-type viruses. However, O-antigens of many serotypes provide 100% screening of the terminal receptor (BtuB), which results in complete resistance of cells carrying these antigens to the given phages. However, specific recognition of O-antigen by the receptor-binding domains of LTFs provides effective infection of the respective strains [72] (Fig. 6).
Fig. 6. a) Morphology of DT571/2 phage particles (DT57C phage morphology is identical). b) Model of LTF organization and their interaction with O-antigens. A branched fibril consists of the trimers of LtfA and LtfB proteins. The receptor-recognizing regions of LtfA of the DT571/2 and DT57C phages bind to O-antigens of type O87 and O22, respectively. LtfB proteins with similar specificity allow both phages to infect a host with O81-type O-antigen. With modifications from [72].
In other cases, the primary receptor binding is necessary for infection. Mutation-induced inactivation of the primary receptor imparts resistance to the bacterial strain. In this case, it is indicated that the phage is dependent on the primary receptor [18, 103].
This phenomenon is observed mainly in myoviruses, where the binding to the primary receptor causes a conformational signal for rearrangement of the baseplate and for tail contraction [64, 104, 105]. In addition, such relationships have been described for podoviruses, including those possessing enzymatically active adhesins [18, 73, 104, 106, 107].
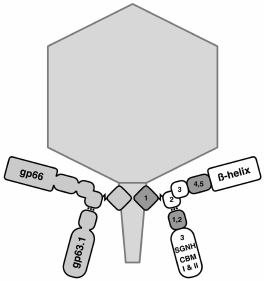
It is interesting that adsorption strategies can strongly vary within a group of related viruses. In the N4-like phage G7C and many other related phages (in contrast to the N4 phage described above), secondary receptor binding is impossible without preliminary recognition of the primary receptor, i.e. the success of phage infection depends on the presence of both types of receptors in a cell [74, 103]. The available data indirectly show that the binding of RBP of such podoviruses to the primary receptor generates a conformational signal that puts the tail apparatus into a certain “triggered” state, when it can interact with the secondary receptor. In the G7C phage, binding to the primary receptor (O-antigen) is a necessary stage of infection [103]. However, during infection the enzymatically active RBP of the phage deacetylates O-polysaccharide (OPS) by removing only one acetyl group per six residues of the repeating element of OPS. For reasons not quite understood, this reaction allows phage penetration to unidentified secondary receptors and successful infection of the cells. At the same time, in a strain where O-acetylation of the O-antigen is inhibited by mutation in the acetylase gene wclK, it is resistant to the G7C phage [103] and does not interact with the recombinant RBP (pg63.1) of the virus [74]. The strains that have completely lost the O-antigen are also completely resistant to the phage. According to indirect data, Prokhorov et al. [74] supposed that deacetylation is a processive reaction providing transport of the virion to the membrane surface and its activation. The latter circumstance manifests itself as follows: the phage can be inactivated by LPS preparation but only if the preparation has been obtained by harsh phenol extraction. The LPS obtained by proteinase K treatment does not activate the phage. However, preparations of outer membrane vesicles (without any aggressive procedures used for its isolation) inactivate the phage very efficiently. Based on these results, the following model was proposed: the interaction of G7C phage pg63.1 provides penetration of the phage tail to an unidentified secondary receptor (supposedly one of the outer membrane proteins); simultaneously, there is a conformational change in the phage tail apparatus, which is necessary for the final receptor recognition. Long-term contact with concentrated LPS preparations can initiate DNA release without contact with the secondary receptor. These data are consistent with the results of cryoelectron reconstruction of the G7C phage (S. Nazarov, EPFL, Switzerland, personal communication). The pg63.1-lacking bacteriophage particles were obtained in [74] by infecting a nonpermissive host with the respective amber mutant. In these particles, in contrast to the wild-type phage, there is a morphological change (opening) of the distal end of the tubular tail. The conformational signal from the binding of gp63.1 to the O-antigen is transmitted via the second RBP protein of the gp66 phage, through which gp63.1 is attached to the virion (Fig. 7).
Fig. 7. Scheme of organization of branched-chain adhesins of G7C phage that are formed by gp66 and gp63.1 (according to [64]).
It is very interesting that the pg66 domain where gp63.1 is attached according to data of bioinformatics analysis is similar to the gp10 domain of the T4 bacteriophage [64], which performs an analogous role in this phage, providing the attachment of short- and (via the adaptor protein gp9) long-tail fibers (LTFs). This protein passes the signal of LTF binding to the receptors on to the internal proteins of the baseplate, which leads to its rearrangement and tail contraction.
Like G7C phage, the Sf6 phage infection of Salmonella cells is accompanied by particle activation with the primary receptor. It has been shown that its receptors are LPSs, as well as OmpA and OmpC proteins [108]. However, inactivation of the phage by protein receptor preparations is effective only in the presence of LPS.
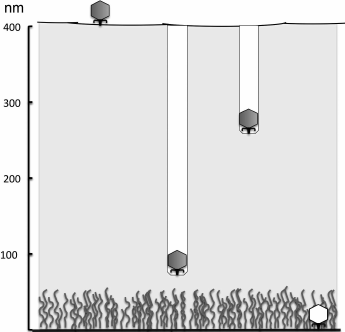
In cases when the bacterial cell surface is protected with a sufficiently thick exopolysaccharide layer, phage adsorption to the surface is preceded by EPS recognition by phage proteins and enzymatic degradation of the exopolysaccharide [86]. It seems that bacteriophages have no alternative solutions for overcoming very thick capsules or intercellular matrix of biofilms. For penetration through capsules, they apparently use processive hydrolysis, when the breaking of additional polymer bonds or molecules occurs without dissociation of the virus. This results in directed movement of a particle into the polysaccharide layer. The K1 phage infecting the K1-capsular strains of E. coli due to consecutive work of its six enzymatically active tail spikes drills a “well” in the capsule, which is easily discernible on electron microphotographs (Fig. 8). After reaching the cell surface, the phage binds to the next receptor.
Fig. 8. Penetration of K-specific coliphages to the cell surface due to processive hydrolysis of K-polysaccharide by enzymatically active tail spikes (according to [73]).
It should be noted that capsules in many cases are the primary receptor necessary for infection. For the above-mentioned K1 phage [109, 110], as well as many phages of the hypermucoid strains of Klebsiella, successful infection requires the presence of a certain type of capsular polysaccharide [111, 112].
In the absence of bacterial strains devoid of massive capsules due to the respective phages, infection begins directly with interaction with the primary receptor located close to the cell surface. However, in this case the surface polysaccharides (e.g. O-antigen) are also often exposed to enzymatic hydrolysis by phage proteins. This strategy is more typical of podoviruses, though enzymatically active adhesins have been found in some representatives of the Myoviridae family [86, 113].
The primary adsorption of many phages occurs mainly near the poles of rod-like cells, probably due to nonuniform arrangement of protein receptors on the bacterial surface [114]. After adsorption, there is a lateral shift of some phages along the cell surface due to dissociation and reassociation of adhesins with neighboring receptor molecules. This behavior is typical of phages that recognize the receptors highly represented on the surface. Because of the shift, phages often migrate to the circumpolar regions of cells. The lateral shifts of T7 phage were reconstructed according to the data of cryoelectron tomography of phage particles infecting E. coli minicells [115]. This process resembles a dance, when the phage alternately binds to the receptor (core-OS) via one and then another of its six fibers until the final recognition of unknown secondary receptor and DNA ejection take place. The λ phage directly recognizing the secondary receptor – the maltose transporter LamB – during the infection of laboratory E. coli strains that also moves along the cell surface, apparently migrating from one LamB molecule to another until irreversible binding occurs [68].
The scenario of events occurring after the binding of secondary receptor is sufficiently clear only for myoviruses. The binding of baseplate adhesins (as a rule, fibrillar) with receptors on cell surface is accompanied by rearrangement of the baseplate [20, 104] and contraction of the tail sheath. As a result, the internal tube of the tail is submerged into the cell wall and reaches the surface of the plasma membrane. The phages of Gram-negative microorganisms have a protein or a protein complex at the tube end that looks like a rigid needle, where there may be peptidoglycan-hydrolase domains at its bottom. For example, in the T4 phage this device (cell puncturing device [64]) consists of a pg5 trimer having a lysozyme domain and forming a rigid trimeric β-helix (needle) at the C-end and a pg5.4 monomer forming a needlepoint.
The phages of Gram-positive bacteria also have peptidoglycan hydrolases associated with the knob of the baseplate, which forms a tip of the tube during tail contraction. Detailed description of the mechanism of baseplate rearrangement and the molecular events underlying tail contraction can be found in other works [20, 64].
There are at least two strategies of interaction between myoviruses and cell receptors. The T4 phage and numerous related viruses [116, 117] have two sets of fibrillar adhesins: six long-tail fibers (LTFs) attached to the corners of the baseplate and six short-tail fibers (STF) stacked under the baseplate. LTF binding is a signal for the beginning of baseplate rearrangement, which is accompanied by the unfolding of short-tail fibers tightly fixing the baseplate on cell surface [20, 64, 104] and preventing repulsion of the phage during tail contraction. Thus, the T4-like phage recognizes the two types of receptors. LTFs of the T4 phage, e.g. they bind to the OmpC protein, while STFs interact with LPSs [118]. In the P1 phage, short-tail fibers are absent and, therefore, the virus is retained near the cell surface due to the binding of several LTFs with the core polysaccharide of LPS of the host, which is a signal for the beginning of tail contraction. Interestingly, in this phage the baseplate is repulsed from the cell surface to approximately 53 nm at the beginning of contraction to 90 nm in the phages with completely contracted tail. This 37-nm distance seems to appear due to elastic straightening of LTFs composed of the angled halves (like in T4). As a result, though the long (220 nm) sheath of the P1 phage tail is contracted by 125 nm, penetration of the tail tube into the cell wall is limited to about 30 nm, which approximately corresponds to the distance to the CPM surface [105].
It seems that myoviruses infecting hosts with a massive capsule use a more complex strategy. Recognition of the capsular polysaccharide and its degradation by enzymatically active adhesins may be a necessary stage of infection [112] but, at the same time, rearrangement of the baseplate and tail contraction probably occur as a result of interaction with the receptor located directly on the cell surface. The details of this strategy have not yet been determined.
In the podoviruses of Gram-negative hosts, DNA ejection is preceded by the ejection of internal capsid proteins forming a channel through the periplasmic space. Usually, among these proteins there are molecules with peptidoglycan-hydrolase domains that form a hole in the murein layer of the cell envelope. The existence of such channel has been shown experimentally for the T7 phage [115]. It is formed by the pg14, pg15, and pg16 proteins, which localize in the capsid interior before the infection and form an inner body [119]. Other podoviruses also seem to have internal proteins providing the transperiplasmic transport of genomic DNA. In the P22 phage, ejection of internal proteins also precedes DNA ejection, suggesting that they also function to create a transperiplasmic channel [120]. One or several such proteins must have peptidoglycan-hydrolase activity [121] to overcome the rigid layer of the cell envelope. In N4-like phages, the ejected protein is an RNA polymerase providing the transcription of immediate early genes [39]. About six copies of this protein are delivered to a cell. A single-subunit virion RNA polymerase (vRNAP) is a very large protein comprising more than 3000 a.a. At present, it is the largest polypeptide known to be transported via transmembrane systems in prokaryotes. It is notable that a protein fragment of about 1000 a.a. size is sufficient for the polymerase activity of vRNAP [122]. It would be reasonable to assume that vRNAP, simultaneously with the enzymatic function, can act as a structure protein of the transperiplasmic channel.
The fine structure of the channel of podoviruses is still unknown, though it is supposed to be similar to the channel formed by a pilot protein of phiX174 phage belonging to the Microviridae family. This protein forms a ten-stranded α-helical structure (coiled-coil) within which there is a channel of sufficient size to ensure the passage of a DNA molecule [123]. At both ends, the polypeptide chains have transmembrane domains that anchor the protein in the outer and inner membranes of a host cell.
The mechanisms of penetration of podoviruses of Gram-positive hosts into cells have been poorly studied. In the Bacillus subtilis φ29 phage, the gp9 protein forming the lower thickened part of the short tubular tail has hydrophobic loops located inside the tail channel, close to its distal end. At the instant of infection, the loops turn and come out, submerging with their hydrophobic α-helices into the cytoplasmic membrane of the cell [124]. The phage probably needs for infection the presence of pores in the massive cell envelope, through which its tail can reach the membrane surface. It has been suggested that analogous membrane-associated loops can be involved in the interaction between the ends of tail tubes of many myoviruses, e.g. T4, and the cytoplasmic membranes of cells [124].
Siphoviruses are also supposed to have a channel formed by the ejected proteins. The tape-measure protein (TMP) located in the tail channel plays a key role in this process [18]. Many TMPs have peptidoglycan-hydrolase activity and can cause membrane fusion. In the T5 phage, the interaction between pb5 adhesin located at the end of the axial fiber of the tail and the receptor (porin FhuA) is followed by deformation of the axial fiber (pb4) and its submergence into the membrane near the receptor molecule. Then the TMP (pb2) protein is released and submerged into the membrane. The channel that appears crosses the periplasmic space of the cell. The murein layer is overcome by the enzymatic activity of the pb2 protein [69].
The available data show that TMPs of some phages can interact with periplasmic proteins of the host for successful infection (i.e. there is in fact an additional periplasmic receptor) [125].
After channel formation, the genomic DNA of the phage is transported into the cell.
The mechanisms providing DNA ejection from the head of bacteriophage into the cell are not quite clear. The beginning of ejection seems to be a merely physicochemical process. The release of DNA from T4 phage particles can be induced in vitro. The process is very rapid: 168 kb of genomic DNA are released through the tail channel in several tens of seconds. In vivo, internalization of the T4 phage DNA takes several minutes and depends on the cell membrane potential [126]. One of the energy sources that provide DNA transport from the capsid to the cytoplasm is DNA pressure caused by the very high packing density of the charged polymeric molecule. Probably, it is precisely this force that is responsible for the beginning of DNA release from the capsid. However, it could be expected that DNA transport would slow as the head is emptied, which in fact does not happen. An additional energy source could be an osmotic gradient appearing due to consumption of a certain amount of water for rehydration of the decompacted DNA, as well as due to the movement of counter-ions compensating for DNA charge. It is supposed that the penetration of some part of a DNA molecule into a bacterial cell results in appearance of an additional osmotic gradient, along which a certain amount of water penetrates the cell after DNA. Water molecules can diffuse both through the membrane and through the phage capsid wall. The water penetrating the capsid displaces DNA and is possibly transported together with DNA along the tail channel, thereby completing the entry of the whole DNA molecule into the cytoplasm (Fig. 9).
Fig. 9. Model of phage DNA penetration into a cell (according to [126]).
The physicochemical mechanisms have a sufficient potential for phage DNA transport and seem to ensure this process in most phages (e.g. phages T4 and λ). However, some viruses still use a two-stage strategy of genome internalization. After the entry of the first several portions of DNA, the process is suspended. Then there is transcription of the early or pre-early genes located on the genome segment that has penetrated the cell. The products of these genes ensure the “hijacking” of cellular metabolism by the phage. For example, the T7 phage has genes of inhibitors of the restriction-modification systems of host cells at the left end of its genome, and the T5 phage causes cellular DNA degradation even before the internalization of the phage genome is completed. The full penetration of DNA requires the activity of phage proteins expressed from the “leader” segment of the genome and depends on cellular metabolic energy. In the T7 phage, the engine that pushes DNA into the cell is T7-RNA polymerase. Therefore, the genes of this virus have the same orientation (against the direction of DNA entry). In the N4 phage, transcription of the early genes by their own RNA polymerase penetrating the cell together with DNA can also contribute to the transport of the genome, but the involvement of other mechanisms is needed to complete this process. The details and potential diversity of the mechanisms of active DNA transport by early phage proteins are yet unknown. However, it should be noted that in many phages with such mechanism of genome internalization it is possible to obtain mutants introducing the whole DNA at once (which often leads to a decrease in the survival rate of phages during the infection) [126], so that active DNA transport in these viruses is an adaptive attainment rather than a way of compensating for energy deficiency of the physicochemical mechanisms of genome penetration.
ADAPTATION OF BACTERIAL CELLS TO EFFECTS OF BACTERIOPHAGES
One of the strategies used by bacteria to modulate bacteriophage adsorption to their cells is the blocking of receptors via structural or conformational changes in the receptor molecules. For example, Staphylococcus aureus produces protein A anchored in the outer membrane that can specifically bind to the Fc domain of class G immunoglobulins [127]. It has been shown that phage adsorption is improved under a reduced level of protein A expression by the cells, indicating that this protein masks phage receptors [128]. The T5 bacteriophage infecting E. coli produces lipoprotein Llp capable of blocking the phage receptor – the ferrichrome iron transport protein FhuA. The Llp protein is expressed in the very beginning of the bacteriophage life cycle, providing the possibility to prevent superinfection of the cells and impeding the inactivation of bacteriophage progeny through the binding to cellular debris after phage release [129]. Some bacteria can carry plasmids containing the genes of proteins that can mask or alter the conformation of phage receptors, e.g. the F plasmid-encoded transporter protein TraT incorporated into the outer membrane can mask the OmpT receptor necessary for the infection by a wide range of T-even bacteriophages of E. coli [130].
Phase variation is another mechanism altering cell surface structures in some bacteria, e.g. Bordetella spp. It is necessary for successful colonization of different niches by these bacteria [131]. The production of various toxins and adhesion and virulence factors by these cells is controlled by the two-component BvgAS system. The cells in the Bvg+ state express colonization and virulence factors including secretion systems and proteins acting as phage receptors (e.g. transporter protein Prn) [132]. The cells in the Bvg– state do not express this protein, and the efficiency of phage inoculation onto such cells decreases by approximately a million times [133]. Despite the absence of the primary phage receptor, phage infection occurs though at very low efficiency – indicating that the Bordetella phages (e.g. BPP-1) have a mechanism for overcoming the absence of a primary receptor on the cell surface.
The synthesis of structured extracellular biopolymers in some bacteria can enhance the survival rate of bacteria in different ecological niches, protecting them from unfavorable physicochemical factors and simultaneously creating a physical barrier between the phage and the bacterial surface receptor [134-138]. In addition to capsular polysaccharides, we also mention polyalginates produced by some species of the genus Pseudomonas [136] and hyaluronates produced by some pathogenic streptococci [139]. It is interesting that the genomes of many species of streptococci contain prophages encoding hyaluronidase, the enzyme capable of degrading not only the surface hyaluronate of bacterial cells but also the hyaluronate of connective tissues in an organism affected by the pathogen, which contributes to its deeper colonization by pathogenic microbes [140]. The hyaluronic layer on the bacterial surface prevents the infection of the cells by most virulent phages but allows the temperate phages of streptococci, which express amounts of hyaluronidase higher by several orders of magnitude, to overcome the physical barrier [141, 142].
Free biomolecules and their complexes normally present in environments with high density of microbial life can interact with receptors, masking or attenuating them. For example, microcin J25 produced by some E. coli cells under starvation can specifically bind to the already known iron transporter protein FhuA, which is simultaneously used as a receptor by many coliphages, and to prevent phage infection. This microcin is used by cells as a starvation signal and inhibits the growth of phylogenetically related bacterial strains, simultaneously preventing phage-induced cell death due to competitive inhibition of phage adsorption [143].
All these systems of bacterial adaptation to the presence of phages in the environment are notable for the fact that the maintenance of their active state requires substantial amounts of energy and metabolites. Also, the physical barrier around the cells impedes mass exchange between the bacterium and the environment. In this context, phase variations seem to be the maximally effective mechanism of transient resistance of bacteria to bacteriophages, allowing simultaneous solution of other problems of bacterial biology.
ADAPTATIONS OF ADSORPTION APPARATUSES OF BACTERIOPHAGES
Phage adsorption specificity is one of the most significant factors determining the range of hosts of these viruses. Therefore, most ecological adaptations of phages in natural microbial communities occur due to microevolutionary changes in the adsorption apparatus. However, in some cases it is necessary to provide preferential infection of cells in a physiological state or phase of the life cycle that would be most suitable for viral replication. Some phages may have for this purpose simple molecular sensors of environmental conditions for changing the activity of the adsorption apparatus. These mechanisms are referred to as phenotypic adaptations. It should be noted that the presence of specialized mechanisms of phenotypic adaptation of the adsorption apparatus is a rather exotic property inherent in only a few phages, Therefore, the microevolutionary processes based on random and/or programmed variability of the genes of receptor-binding proteins play a key role in adaptation of the adsorption apparatus of phages to the current ecological situation.
Variability of phage adhesins. Like other bacteriophage genes, the genes of receptor-binding proteins are exposed to natural mutations. Point amino-acid substitutions sometimes can drastically change the affinity for receptors. In some cases, the formation of new specificity with respective changes in phage host range can be observed even in evolving microcosms under laboratory conditions. For example, the joint cultivation of a λ virulent mutant and E. coli resulted in the selection of resistant bacteria, and then phage variants recognizing the new receptor. Instead of the LamB protein responsible for maltose transport, some genetic lines of the phage became capable of recognizing the porin OmpF [144, 145]. To this end, it is necessary to accumulate several point mutations. Analogously, the T4-like phage PP01 infecting E. coli O157:H7, which recognizes the porin OmpC with its LTF, was adapted to recognize an unidentified alternative primary receptor, providing the phage the ability to grow on strains lacking the OmpC protein [146]. For such alteration of the specificity, 2-3 (in different clones) amino acid substitutions in the receptor-binding protein pg38 were sufficient. Such examples have been described for a great number of other phages.
In addition to point substitutions, recombination events (a modular evolution mechanism) make a considerable contribution to variability of receptor-binding proteins [147]. Due to the common structural design of the main types of phage adhesins, the genes of these proteins can recombine with the formation of functional chimeric molecules. The fibrillar proteins of phages usually contain considerable amounts of β-helices. In the case of typical trimeric fibers, the β-helical structure is a triangular prism, each face being formed by a β-layer of one of the three polypeptide chains. This prism is twisted relative to its longitudinal axis [148]. The α-helical coiled-coil structures are rare in phage fibers (except for collar filaments of T-even bacteriophages) [149]. Thus, since fibrillar structures are composed of repeating protein motifs arranged similarly in different proteins, many recombination events take place among them. The general structural design as a parallel homotrimer implies that the tertiary and quaternary contacts of amino acid residues in such proteins are limited by the closely spaced amino acids in the sequences of the same protein chain or by amino acids in the analogous positions of the neighboring chains. Therefore, the formed chimeric structures retain nearly all the tertiary and quaternary interactions during recombinations and are often quite functional. Analogous reasoning is applicable to spike proteins, where recombinations can rather freely occur at the sites separating globular domains. As a result, most sequences of phage adhesins have a mosaic origin (are natural chimeric proteins), as is easily revealed by comparing them with the data bases, e.g. using the blastP algorithm (Fig. 10).
Fig. 10. Results of protein sequence analysis of the lateral fiber of LtfA of the DT57C phage (AJA41628.1) using BlastP software at ncbi.nlm.nih.gov. The sequence length is 1076 a.a. Black and gray lines show the similarity between certain parts of this sequence and other proteins (mainly bacteriophage proteins). Different shades of gray correspond to different degrees of similarity between the sequences (score). For details, see the above-mentioned site.
The described recombinations often lead to changes in the host range of the phage. For example, the structure of the distal part of the distal half of LTF in a classical object of molecular biology, phage T4, is untypical of most T-even phages. In the T2 and T6 (and many other) phages, the C-terminal part of pg37 forming the last two thirds of the distal half of the LTF is meant for the interaction with pg38, which really carries the receptor-recognizing regions, while the C-terminal fragment of the 37 gene and the 38 gene in T4 are replaced by the analogous fiber region of the λ-like phages and the chaperon necessary for successful folding of this protein, respectively [150, 151]. As a result, pg38 (the chaperon acquired by modular transfer) in T4 is not a component of the viral particle, and the receptor-recognizing region is located directly at the C-end of pg37. The high level of similarity between the nucleotide sequences of the T4 phage and the T2 and T6 phages demonstrates that replacement of this module occurred in the very recent evolutionary past. By analogy, a variety of recombination events within pg37 between different T4-like phages have been described, which also resulted in the transfer of alternative allele variants of the 38 gene, obviously accompanied by a change in adsorption specificity [151-153]. It has not yet been properly investigated how significant the contribution of such recombination events to the current mutual adaptation of phage and bacterial populations in real natural communities in time intervals comparable with the natural cycles of these communities is (in most cases, from days to several years). According to some data, it can true for the coliphages in intestinal ecosystems. For example, in a small group of N4-like phage isolates from the feces of horses held in the same stable for seven years, we found numerous substitutions of receptor-recognizing regions of tailspike proteins, including substitution of the enzymatically active domain (O-antigen deacetylase and depolymerase) ([154] and our unpublished data). It seems that the contribution of divergence due to accumulation of mutations and modular mechanisms can vary significantly depending on availability of the pool of the respective genes for bacteriophages, which in turn depends on the biological diversity of the bacteria and phages in a community, as well as on the peculiarities of ecological interactions within it.
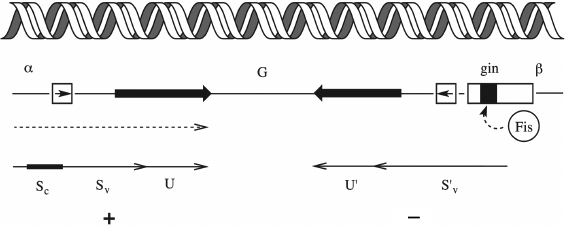
The variability of phage adhesins can be determined not only by the stochastic processes of mutation or random recombinations. Some viruses have specially programmed mechanisms for enhancing the variability of receptor-recognizing proteins. The Mu phage, which is a temperate transposable phage of E. coli, has an inverted genomic segment, the so-called G-segment (Fig. 11). The G-segment encodes two alternative variants of the C-terminal domain of the gene of the receptor-recognizing fiber: Sv and S´v (an ortholog of the stf gene of lateral fibers of the λ phage), as well as two variants of chaperone U (an ortholog of the tfa gene of the λ phage): U and U´, respectively.
Fig. 11. Mechanism of switching of host specificity in the Mu phage due to genomic segment inversion. See text for explanation.
The inverted segment is flanked by recognition sites of the site-specific recombinase Gin. The mutual arrangement of the transcribed gene S and the G-segment is such that its inversion is accompanied by substitution of a certain version of the C-terminal segment as well as the corresponding variant of the U chaperone into the active gene copy. Since site-specific recombination is an accurate process, there is no reading frame failure. As a result, the Mu phage population always contains a small portion of particles carrying adhesin for recognizing the alternative host range. At position G+, with the Sv+U variant in the active locus, the phage can infect E. coli K12 and Salmonella arizonae, while in the G– orientation the phage can grow on the strains Citrobacter freundi, Shigella sonnei, Erwinia, and Enterobacter [155, 156]. Such modules, the so-called shufflons (from “shuffle”), occur in other phages too, though not always associated with phage variations of the host range. Shufflons are also found in bacteria, where they control phase variations of different characteristics, e.g. the type of antigen of the flagella (flagellin) [157].
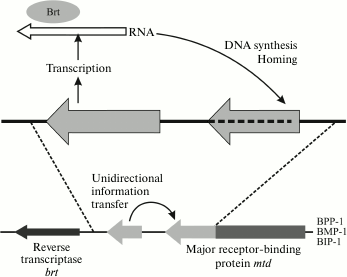
The Bordetella pertussis BPP1, BMP1, and BIP1 phages increase the variability of adhesin by using the mechanism of reverse retrohoming, where high frequency of errors during reverse transcription is used as a generator of variability (Fig. 12).
Fig. 12. Improvement of variability of the Mtd adhesin gene of the BPP1 phage by retrohoming.
The genome of this virus contains the active mtd locus (from “major tropism determinant”) and, additionally, the TR (template region) locus encoding the C-terminal segment of Mtd protein. This additional copy of the gene region is transcribed, and then the phage-encoded reverse transcriptase Brt converts it into cDNA [133] by the method of recombinant retrohoming (this mechanism is described in a review [158]).
The cDNA copy substitutes with a certain frequency for the sequence of the C-terminal region of the mtd gene. This process requires bRT and bAvd proteins [158]. As a result, because of the high frequency of errors during reverse transcription in certain nucleotide positions (some adenine residues), new mtd gene variants are formed, which determine a different variant of host specificity. For the most part, these phase variations are necessary for the phage to adapt to infection of a host in a human organism or in the environment, because the main phage receptor – pertactin (Prn) – is expressed only in an organism under the control of the two-component BvgAS system, and in the external medium the phage must switch over to other receptors.
Such systems have been denoted as DGR (diversity generating retron) elements. They are slightly less widespread than shufflons [158]. Similar DGRs have been found in some Mu-like prophages of Syntrophobotulus glycolicus [159]. However, it is interesting that, according to the data of metagenomics, DRGEs are very widespread in the phages associated with the human intestinal microbiome, where they determine the variability of some proteins, many of them with immunoglobulin-like domains [160, 161]. As mentioned above, the phage Ig-like domains are always exposed on the surface and, at least in T4, they are used for the binding to different molecules, increasing the probability of meeting with the host cells (in T4, the hoc protein recognizes mucin of the intestinal mucus [98], but it is quite possible that some Ig-like domains of the phages recognize additional receptors on bacterial cells).
Phenotypic adaptations of the adsorption apparatuses of bacteriophages. The changes in the host range or adsorption rate as a result of structural rearrangements in the protein complex of the adsorption apparatus not associated with mutations in the respective genes are phenotypic adaptations. Very few such mechanisms are known, but it is probable that more detailed investigation of adsorption modulation events in different phages will change our ideas about the prevalence rate of this phenomena.
The so-called molecular sensor of the T4 bacteriophage and many related viruses is a classical example of phenotypic adaptation of the virus. T4 is almost not adsorbed at low pH values and low ionic strength. The transition can be artificially blocked by adding polyethylene glycol (6% PEG 6000). In the T2 phage, adsorption activation requires the presence of the free amino acid tryptophan.
Though this phenomenon has been studied since the 1970s, the exact mechanism of sensor operation is still unclear. Until recently, it has been thought that LTFs of a free phage particle can be in the two alternative states: “down” and “up”, pressed into the contractile tail sheath [149]. It was supposed that LTFs are competent for interaction with a receptor in the “down” position but inactive in the “up” position. In the “up” position, the proximal and distal halves of the fiber contact the proteins of pg18 sheath and phage head, respectively [162]. In addition, the distal half makes two contacts with fibritin (pg wac) – the collar and collar fiber protein. The T4 phage particle includes 12 trimeric fibritin molecules (but not 6 as it was supposed until recently), composed mainly of α-helical coiled-coil structure. Six fibritin molecules bend down and can be seen as collar filaments. They contact LTF in the region of its bend, binding via its C-terminal regions to the pg36 protein; other six molecules are bent in the plane perpendicular to the tail axis and form a collar interacting with a more distal region of LTF formed by pg37 [162]. It is still unclear which of these numerous contacts shows sensitivity to environmental factors. In some works, the function of sensor element was attributed to fibritin; however, recent data suggest that it facilitates the down→up transition rather than controlling LTF release. New data of cryoelectron microscopy show that LTFs are in the “up” state in most particles (at least under the specific conditions of the experiment). This probably indicates that the sensor model needs to be modified. For example, what is meant here may be not the physical up→down transition in free particles, but the higher or lower stabilization of the “up” position, which prohibits or permits the turn of the LTF at the moment of infection. It is interesting that the transition of fibers into the “down” position only at the time of contact with the cell surface has been also described for the T7 phage. Until the moment of adsorption, its fibers are pressed to the capsid [104]. It may be supposed that the molecular sensor of T-even phages controls not the change in LTF position in free particles, but the possibility of their unfolding after the primary reversible contact with the cell surface.
The mechanism of the Lactococcus lactis siphovirus p2 is somewhat similar. Its six receptor-binding proteins (trimers similar to tail spikes) can rotate on the baseplate by almost 200° in response to increase in the concentration of Ca2+, passing from the “up” orientation to the “down” orientation suitable for interaction with the receptor.
This mechanism probably allows the adsorption apparatus to be activated near the cell surface, where the concentration of Ca2+ is higher due to the presence of negatively charged teichoic acids of the cell wall. It is also possible that such mechanism provides preferential infection of cells in milk, whereas in the external environment, where host bacteria endure unfavorable conditions, the phage “prefers” to remain free [66, 77].
Two other lactophages, Tp901-1 and Tuc2009, also have a rather peculiar adsorption apparatus that can adapt phenotypically to the infection of cells in the logarithmic or stationary phase. Their round baseplates carry six receptor-recognizing complexes called tripods: each of them included three trimers of proper RBP (BppL) joined together by proteins BppU and BppA. Thus, the baseplate carries altogether 18 RBP trimers, which leads to intensification of adsorption due to the effect of avidity. In addition to RBP, the interaction with the cell involves the Tal (tail associated lysine) protein forming a short and a thick central fiber. This protein was named as such due to the presence of a domain with peptidoglycan-hydrolase (peptidase) activity at its C-end, which makes its way through the thick cell envelope of Lactococcus lactis. However, this domain turned out to be essential only for the infection of cells in the stationary phase, with their densely packed peptidoglycan being maximally cross-linked by peptide bridges. In some particles of the population, the Tal protein is autoprocessed due to the activity of the available peptidase domain. At the same time, the C-terminal part with the peptidoglycan-hydrolyzing domain is removed. Surprisingly, the phages with the shortened Tal protein are adsorbed noticeably faster but are much less capable of infecting the cells close to the stationary phase [163].
Probably, along with these highly specific molecular mechanisms, there are some “hidden” mechanisms of phenotypic adaptation inherent in a much broader range of phages. The adsorption curves of most bacteriophages (see, e.g. Fig. 3 from [154]) demonstrate the following two phases: a relatively fast one, for which the adsorption constants are usually calculated, and a slow one, which in some cases passes into a plateau [16]. In fact, these phases overlap, i.e. in the initial phage population there is a rapidly adsorbed predominant fraction and a minor fraction with very slow adsorption kinetics. Nevertheless, the phages from the “slow” or “residual” fraction are viable and form normal plaques during sample inoculation. For some phage–host systems, e.g. for the λ and E. coli K-12 phages, it has been shown that the progeny of the “slow” fraction has the same properties as the initial phage, i.e. the difference in adsorption rates is not associated with any gene mutations [164]. It seems to be true for most phages, though it has been shown that in the T4 phage the above-described phenomenon is accounted for (at least partially) by the emergence of specific, frequently occurring mutations in the 37 gene (the distal half of LTF) [165].
In some phages, the plateau can be much higher than in the λ phage or the G7C phage and reach tens of percent of unadsorbed particles. In the Alt63 phage closely related to the G7C phage but different in the variant of the protein recognizing the primary receptor (O-antigen), the height of the plateau reaches 40-80% but, which is especially interesting, depends on cell culture density used in the experiment (the lesser the cell number, the higher the plateau). The mechanism of such dependence has not yet been determined. The mechanisms of formation of the “slow” fraction in other phages, except for T4, are not clear either. In some cases, it may be accounted for by the presence of the incomplete set of RBPs because of aberrations of assembly or damage of particles. In the case of the λ phage adsorbed to E. coli K-12 through a single axial fiber, this explanation is obviously unacceptable. It has been supposed that the J protein of the axial fiber of the λ phage may have two different structural conformations: the native one attained in the main pathway of folding and the aberrant one reflecting the local energy minimum where some molecules are trapped during folding [164]. Finally, there may be a relatively tight but reversible binding of adhesins to the blocking molecules, e.g. receptor fragments.
The adaptive significance of the “residual” fraction seems to consist in reservation of some part of the phage population in the event of emergence of conditions extremely unfavorable for viral replication in the cells. Therefore, the λ phage can be adsorbed to cells in the stationary phase, but in this case viral production does not occur and the phage dies. The “residual” fraction of particles allows the virus to survive transition of the culture to the stationary phase under experimental conditions. In a sense, the phenomenon of “residual fraction” is like the phenomenon of persister cells, i.e. cells with suppressed metabolism that occur with a low frequency in many bacterial populations [166]. These temporarily “dormant” cells do not multiply but are tolerant to the effects of most antibiotics, because the biosynthetic processes impaired by antibiotics are switched off in them. In contrast to resistant mutants, the progeny of persister cells has the same antibiotic sensitivity as the initial strain.
The event of adsorption is the climax of existence of a free viral particle, when the free energy reserve inherent in the structure of the virion during morphogenesis is realized. In bacteriophages, adsorption specificity seems to be the determining factor that limits the available host range. At first glance, it might seem that the reduction of this range due to recognition of rather variable structures is a wrong solution developed under the pressure of some additional selecting factors. However, if we take into account that the sizes of receptor-recognizing sites of phage adhesins comprise only a few carbohydrate or amino acid residues of the receptor and, at the same time, the virus is at a sufficiently close distance to the cell for only 5 ms during the collision between the phage particle and the cell, with its adsorption apparatus being turned towards the cell for only 1.6 ms (see review [17]), the efficiency of cell surface recognition, which keeps the adsorption constants in some phages close to the maximally possible values (about 10–8 ml/min), causes no doubts. Despite the great diversity of adsorption apparatuses of phages and component proteins, their similar structural design offers great possibilities for combinatorial variability, inter alia, due to recombinations between rather distant viral genomes. This allows us to rely on the development of effective artificial systems for controlling host specificity in bacteriophages, in the first place relevant for the development of next-generation agents for phage therapy and other applications.
Acknowledgments
This work was supported by the Russian Science Foundation (project No. 15-15-00134).
REFERENCES
1.Knoll, A. H. (2015) Paleobiological perspectives on
early microbial evolution, Cold Spring Harb. Perspect. Biol.,
7, a018093.
2.Postler, T. S., and Ghosh, S. (2017) Understanding
the holobiont: how microbial metabolites affect human health and shape
the immune system, Cell Metab., 26, 110-130.
3.Putnam, H. M., Barott, K. L., Ainsworth, T. D., and
Gates, R. D. (2017) The vulnerability and resilience of reef-building
corals, Curr. Biol., 27, R528-R540.
4.Martin, F. M., Uroz, S., and Barker, D. G. (2017)
Ancestral alliances: plant mutualistic symbioses with fungi and
bacteria, Science, 356.
5.Takeshita, K., and Kikuchi, Y. (2017) Riptortus
pedestris and Burkholderia symbiont: an ideal model system
for insect-microbe symbiotic associations, Res. Microbiol.,
168, 175-187.
6.Clokie, M. R., Millard, A. D., Letarov, A. V., and
Heaphy, S. (2011) Phages in nature, Bacteriophage, 1,
31-45.
7.Sime-Ngando, T. (2014) Environmental
bacteriophages: viruses of microbes in aquatic ecosystems, Front.
Microbiol., 5, 355.
8.Weinbauer, M. G. (2004) Ecology of prokaryotic
viruses, FEMS Microbiol. Rev., 28, 127-181.
9.Weinbauer, M. G., and Rassoulzadegan, F. (2004) Are
viruses driving microbial diversification and diversity? Environ.
Microbiol., 6, 1-11.
10.Weinbauer, M. G., Bettarel, Y., Cattaneo, R.,
Luef, B., Maier, C., Motegi, C., Peduzzi, P., and Mari, X. (2009) Viral
ecology of organic and inorganic particles in aquatic systems: avenues
for further research, Aquat. Microb. Ecol., 57,
321-341.
11.Shabbir, M. A., Hao, H., Shabbir, M. Z., Wu, Q.,
Sattar, A., and Yuan, Z. (2016) Bacteria vs. bacteriophages: parallel
evolution of immune arsenals, Front. Microbiol., 7,
1292.
12.Dy, R. L., Richter, C., Salmond, G. P., and
Fineran, P. C. (2014) Remarkable mechanisms in microbes to resist phage
infections, Annu. Rev. Virol., 1, 307-331.
13.Samson, J. E., Magadan, A. H., Sabri, M., and
Moineau, S. (2013) Revenge of the phages: defeating bacterial defences,
Nat. Rev. Microbiol., 11, 675-687.
14.Touchon, M., Moura de Sousa, J. A., and Rocha, E.
P. (2017) Embracing the enemy: the diversification of microbial gene
repertoires by phage-mediated horizontal gene transfer, Curr. Opin.
Microbiol., 38, 66-73.
15.King, A. M. Q., Adams, M. J., Carstens, E. B.,
and Lefkowitz, E. J. (2012) Virus Taxonomy: Classification and
Nomenclature of Viruses. Ninth Report of the International Committee on
Taxonomy of Viruses, Academic Press, London-Waltham-San Diego.
16.Bertozzi Silva, J., Storms, Z., and Sauvageau, D.
(2016) Host receptors for bacteriophage adsorption, FEMS Microbiol.
Lett., 363, fnw002.
17.Storms, Z. J., and Sauvageau, D. (2015) Modeling
tailed bacteriophage adsorption: insight into mechanisms,
Virology, 485, 355-362.
18.Casjens, S. R., and Molineux, I. J. (2012) Short
noncontractile tail machines: adsorption and DNA delivery by
podoviruses, Adv. Exp. Med. Biol., 726, 143-179.
19.Davidson, A. R., Cardarelli, L., Pell, L. G.,
Radford, D. R., and Maxwell, K. L. (2012) Long noncontractile tail
machines of bacteriophages, Adv. Exp. Med. Biol., 726,
115-142.
20.Leiman, P. G., and Shneider, M. M. (2012)
Contractile tail machines of bacteriophages, Adv. Exp. Med.
Biol., 726, 93-114.
21.Mahony, J., and Van Sinderen, D. (2015) Novel
strategies to prevent or exploit phages in fermentations, insights from
phage–host interactions, Curr. Opin. Biotechnol.,
32, 8-13.
22.Letarov, A. V., Golomidova, A. K., and Tarasyan,
K. K. (2010) Ecological basis for rational phage therapy, Acta
Naturae, 2, 60-72.
23.Auer, G. K., and Weibel, D. B. (2017) Bacterial
cell mechanics, Biochemistry, 56, 3710-3724.
24.Jankute, M., Cox, J. A., Harrison, J., and Besra,
G. S. (2015) Assembly of the mycobacterial cell wall, Annu. Rev.
Microbiol., 69, 405-423.
25.Knirel, Y. A., and Valvano, M. A. (2011)
Bacterial Lipopolysaccharides: Structure, Chemical Synthesis,
Biogenesis, and Interaction with Host Cells, Springer, Wien.
26.Steimle, A., Autenrieth, I. B., and Frick, J. S.
(2016) Structure and function: lipid A modifications in commensals and
pathogens, Int. J. Med. Microbiol., 306, 290-301.
27.Amor, K., Heinrichs, D. E., Frirdich, E.,
Ziebell, K., Johnson, R. P., and Whitfield, C. (2000) Distribution of
core oligosaccharide types in lipopolysaccharides from
Escherichia coli, Infect. Immun., 68,
1116-1124.
28.Nikaido, H., and Vaara, M. (1985) Molecular basis
of bacterial outer membrane permeability, Microbiol. Rev.,
49, 1-32.
29.Andres, D., Baxa, U., Hanke, C., Seckler, R., and
Barbirz, S. (2010) Carbohydrate binding of Salmonella phage P22
tailspike protein and its role during host cell infection, Biochem.
Soc. Trans., 38, 1386-1389.
30.Andres, D., Hanke, C., Baxa, U., Seul, A.,
Barbirz, S., and Seckler, R. (2010) Tailspike interactions with
lipopolysaccharide effect DNA ejection from phage P22 particles in
vitro, J. Biol. Chem., 285, 36768-36775.
31.Ranjit, D. K., and Young, K. D. (2016) Colanic
acid intermediates prevent de novo shape recovery of
Escherichia coli spheroplasts, calling into question biological
roles previously attributed to colanic acid, J. Bacteriol.,
198, 1230-1240.
32.Ren, G., Wang, Z., Li, Y., Hu, X., and Wang, X.
(2016) Effects of lipopolysaccharide core sugar deficiency on colanic
acid biosynthesis in Escherichia coli, J. Bacteriol.,
198, 1576-1584.
33.Porter, N. T., and Martens, E. C. (2017) The
critical roles of polysaccharides in gut microbial ecology and
physiology, Annu. Rev. Microbiol., doi:
10.1146/annurev-micro-102215-095316.
34.Kuhn, H. M., Meier-Dieter, U., and Mayer, H.
(1988) ECA, the enterobacterial common antigen, FEMS Microbiol.
Rev., 4, 195-222.
35.Gozdziewicz, T. K., Lugowski, C., and
Lukasiewicz, J. (2014) First evidence for a covalent linkage between
enterobacterial common antigen and lipopolysaccharide in Shigella
sonnei phase II ECALPS, J. Biol. Chem., 289,
2745-2754.
36.Slusky, J. S. (2016) Outer membrane protein
design, Curr. Opin. Struct. Biol., 45, 45-52.
37.Power, M. L., Ferrari, B. C., Littlefield-Wyer,
J., Gordon, D. M., Slade, M. B., and Veal, D. A. (2006) A naturally
occurring novel allele of Escherichia coli outer membrane
protein A reduces sensitivity to bacteriophage, Appl. Environ.
Microbiol., 72, 7930-7932.
38.Smith, S. G., Mahon, V., Lambert, M. A., and
Fagan, R. P. (2007) A molecular Swiss army knife: OmpA structure,
function and expression, FEMS Microbiol. Lett., 273,
1-11.
39.Choi, K. H., McPartland, J., Kaganman, I.,
Bowman, V. D., Rothman-Denes, L. B., and Rossmann, M. G. (2008) Insight
into DNA and protein transport in double-stranded DNA viruses: the
structure of bacteriophage N4, J. Mol. Biol., 378,
726-736.
40.Szewczyk, J., and Collet, J. F. (2016) The
journey of lipoproteins through the cell: one birthplace, multiple
destinations, Adv. Microb. Physiol., 69, 1-50.
41.Hooda, Y., Lai, C. C. L., and Moraes, T. F.
(2017) Identification of a large family of slam-dependent surface
lipoproteins in gram-negative bacteria, Front. Cell Infect.
Microbiol., 7, 207.
42.Wegrzyn, G., and Thomas, M. S. (2002) Modulation
of the susceptibility of intestinal bacteria to bacteriophages in
response to Ag43 phase variation – a hypothesis, Med. Sci.
Monit., 8, HY15-18.
43.Baldvinsson, S. B., Sorensen, M. C., Vegge, C.
S., Clokie, M. R., and Brondsted, L. (2014) Campylobacter
jejuni motility is required for infection of the flagellotropic
bacteriophage F341, Appl. Environ. Microbiol., 80,
7096-7106.
44.Sorensen, M. C., Gencay, Y. E., Birk, T.,
Baldvinsson, S. B., Jackel, C., Hammerl, J. A., Vegge, C. S., Neve, H.,
and Brondsted, L. (2015) Primary isolation strain determines both phage
type and receptors recognised by Campylobacter jejuni
bacteriophages, PLoS One, 10, e0116287.
45.Choi, Y., Shin, H., Lee, J. H., and Ryu, S.
(2013) Identification and characterization of a novel
flagellum-dependent Salmonella-infecting bacteriophage, iEPS5,
Appl. Environ. Microbiol., 79, 4829-4837.
46.Lee, T. J., Schwartz, C., and Guo, P. (2009)
Construction of bacteriophage phi29 DNA packaging motor and its
applications in nanotechnology and therapy, Ann. Biomed. Eng.,
37, 2064-2081.
47.Shin, H., Lee, J. H., Kim, H., Choi, Y., Heu, S.,
and Ryu, S. (2012) Receptor diversity and host interaction of
bacteriophages infecting Salmonella enterica serovar
typhimurium, PLoS One, 7, e43392.
48.Yen, J. Y., Broadway, K. M., and Scharf, B. E.
(2012) Minimum requirements of flagellation and motility for infection
of Agrobacterium sp. strain H13-3 by flagellotropic
bacteriophage 7-7-1, Appl. Environ. Microbiol., 78,
7216-7222.
49.Siegel, S. D., Liu, J., and Ton-That, H. (2016)
Biogenesis of the Gram-positive bacterial cell envelope, Curr. Opin.
Microbiol., 34, 31-37.
50.Van der Es, D., Hogendorf, W. F., Overkleeft, H.
S., Van der Marel, G. A., and Codee, J. D. (2017) Teichoic acids:
synthesis and applications, Chem. Soc. Rev., 46,
1464-1482.
51.Bielmann, R., Habann, M., Eugster, M. R., Lurz,
R., Calendar, R., Klumpp, J., and Loessner, M. J. (2015) Receptor
binding proteins of Listeria monocytogenes bacteriophages A118
and P35 recognize serovar-specific teichoic acids, Virology,
477, 110-118.
52.Chang, Y., Shin, H., Lee, J. H., Park, C. J.,
Paik, S. Y., and Ryu, S. (2015) Isolation and genome characterization
of the virulent Staphylococcus aureus bacteriophage SA97,
Viruses, 7, 5225-5242.
53.Uchiyama, J., Takemura-Uchiyama, I., Kato, S.,
Sato, M., Ujihara, T., Matsui, H., Hanaki, H., Daibata, M., and
Matsuzaki, S. (2014) In silico analysis of AHJD-like viruses,
Staphylococcus aureus phages S24-1 and S13′, and study of
phage S24-1 adsorption, Microbiology Open, 3,
257-270.
54.Xia, G., Corrigan, R. M., Winstel, V., Goerke,
C., Grundling, A., and Peschel, A. (2011) Wall teichoic acid-dependent
adsorption of staphylococcal siphovirus and myovirus, J.
Bacteriol., 193, 4006-4009.
55.Baptista, C., Santos, M. A., and Sao-Jose, C.
(2008) Phage SPP1 reversible adsorption to Bacillus
subtilis cell wall teichoic acids accelerates virus recognition
of membrane receptor YueB, J. Bacteriol., 190,
4989-4996.
56.Ainsworth, S., Sadovskaya, I., Vinogradov, E.,
Courtin, P., Guerardel, Y., Mahony, J., Grard, T., Cambillau, C.,
Chapot-Chartier, M. P., and Van Sinderen, D. (2014) Differences in
lactococcal cell wall polysaccharide structure are major determining
factors in bacteriophage sensitivity, MBio, 5,
e00880-00814.
57.Plaut, R. D., Beaber, J. W., Zemansky, J., Kaur,
A. P., George, M., Biswas, B., Henry, M., Bishop-Lilly, K. A., Mokashi,
V., Hannah, R. M., Pope, R. K., Read, T. D., Stibitz, S., Calendar, R.,
and Sozhamannan, S. (2014) Genetic evidence for the involvement of the
S-layer protein gene sap and the sporulation genes spo0A, spo0B, and
spo0F in phage AP50c infection of Bacillus anthracis, J.
Bacteriol., 196, 1143-1154.
58.Edwards, P., and Smit, J. (1991) A transducing
bacteriophage for Caulobacter crescentus uses the
paracrystalline surface layer protein as a receptor, J.
Bacteriol., 173, 5568-5572.
59.Callegari, M. L., Riboli, B., Sanders, J. W.,
Cocconcelli, P. S., Kok, J., Venema, G., and Morelli, L. (1998) The
S-layer gene of Lactobacillus helveticus CNRZ 892:
cloning, sequence and heterologous expression, Microbiology,
144, 719-726.
60.Abrahams, K. A., and Besra, G. S. (2016)
Mycobacterial cell wall biosynthesis: a multifaceted antibiotic target,
Parasitology, doi: 10.1017/S0031182016002377.1-18.
61.Chen, J., Kriakov, J., Singh, A., Jacobs, W. R.,
Jr., Besra, G. S., and Bhatt, A. (2009) Defects in glycopeptidolipid
biosynthesis confer phage I3 resistance in Mycobacterium
smegmatis, Microbiology, 155, 4050-4057.
62.Garcia-Doval, C., Caston, J. R., Luque, D.,
Granell, M., Otero, J. M., Llamas-Saiz, A. L., Renouard, M., Boulanger,
P., and Van Raaij, M. J. (2015) Structure of the receptor-binding
carboxy-terminal domain of the bacteriophage T5 L-shaped tail fibre
with and without its intra-molecular chaperone, Viruses,
7, 6424-6440.
63.Granell, M., Namura, M., Alvira, S., Kanamaru,
S., and Van Raaij, M. J. (2017) Crystal structure of the
carboxy-terminal region of the bacteriophage T4 proximal long tail
fiber protein Gp34, Viruses, 9, 168.
64.Taylor, N. M., Prokhorov, N. S.,
Guerrero-Ferreira, R. C., Shneider, M. M., Browning, C., Goldie, K. N.,
Stahlberg, H., and Leiman, P. G. (2016) Structure of the T4 baseplate
and its function in triggering sheath contraction, Nature,
533, 346-352.
65.Koc, C., Xia, G., Kuhner, P., Spinelli, S.,
Roussel, A., Cambillau, C., and Stehle, T. (2016) Structure of the
host-recognition device of Staphylococcus aureus phage varphi11,
Sci. Rep., 6, 27581.
66.Sciara, G., Bebeacua, C., Bron, P., Tremblay, D.,
Ortiz-Lombardia, M., Lichiere, J., Van Heel, M., Campanacci, V.,
Moineau, S., and Cambillau, C. (2010) Structure of lactococcal phage p2
baseplate and its mechanism of activation, Proc. Natl. Acad. Sci.
USA, 107, 6852-6857.
67.Garcia-Doval, C., and Van Raaij, M. J. (2012)
Structure of the receptor-binding carboxy-terminal domain of
bacteriophage T7 tail fibers, Proc. Natl. Acad. Sci. USA,
109, 9390-9395.
68.Chatterjee, S., and Rothenberg, E. (2012)
Interaction of bacteriophage l with its E. coli receptor, LamB,
Viruses, 4, 3162-3178.
69.Zivanovic, Y., Confalonieri, F., Ponchon, L.,
Lurz, R., Chami, M., Flayhan, A., Renouard, M., Huet, A., Decottignies,
P., Davidson, A. R., Breyton, C., and Boulanger, P. (2014) Insights
into bacteriophage T5 structure from analysis of its morphogenesis
genes and protein components, J. Virol., 88,
1162-1174.
70.Gallet, R., Shao, Y., and Wang, I. N. (2009) High
adsorption rate is detrimental to bacteriophage fitness in a
biofilm-like environment, BMC Evol. Biol., 9, 241.
71.Heller, K., and Braun, V. (1979) Accelerated
adsorption of bacteriophage T5 to Escherichia coli F, resulting
from reversible tail fiber-lipopolysaccharide binding, J.
Bacteriol., 139, 32-38.
72.Golomidova, A. K., Kulikov, E. E., Prokhorov, N.
S., Guerrero-Ferreira, R., Knirel, Y. A., Kostryukova, E. S., Tarasyan,
K. K., and Letarov, A. V. (2016) Branched lateral tail fiber
organization in T5-like bacteriophages DT57C and DT571/2 is revealed by
genetic and functional analysis, Viruses, 8, 26.
73.Leiman, P. G., Battisti, A. J., Bowman, V. D.,
Stummeyer, K., Muhlenhoff, M., Gerardy-Schahn, R., Scholl, D., and
Molineux, I. J. (2007) The structures of bacteriophages K1E and K1-5
explain processive degradation of polysaccharide capsules and evolution
of new host specificities, J. Mol. Biol., 371,
836-849.
74.Prokhorov, N. S., Riccio, C., Zdorovenko, E. L.,
Shneider, M. M., Browning, C., Knirel, Y. A., Leiman, P. G., and
Letarov, A. V. (2017) Function of bacteriophage G7C esterase tailspike
in host cell adsorption, Mol. Microbiol., doi:
10.1111/mmi.13710.
75.Dai, W., Hodes, A., Hui, W. H., Gingery, M.,
Miller, J. F., and Zhou, Z. H. (2010) Three-dimensional structure of
tropism-switching Bordetella bacteriophage, Proc. Natl. Acad.
Sci. USA, 107, 4347-4352.
76.Veesler, D., Spinelli, S., Mahony, J., Lichiere,
J., Blangy, S., Bricogne, G., Legrand, P., Ortiz-Lombardia, M.,
Campanacci, V., Van Sinderen, D., and Cambillau, C. (2012) Structure of
the phage TP901-1 1.8 MDa baseplate suggests an alternative host
adhesion mechanism, Proc. Natl. Acad. Sci. USA, 109,
8954-8958.
77.Bebeacua, C., Tremblay, D., Farenc, C.,
Chapot-Chartier, M. P., Sadovskaya, I., Van Heel, M., Veesler, D.,
Moineau, S., and Cambillau, C. (2013) Structure, adsorption to host,
and infection mechanism of virulent lactococcal phage p2, J.
Virol., 87, 12302-12312.
78.Dieterle, M. E., Spinelli, S., Sadovskaya, I.,
Piuri, M., and Cambillau, C. (2017) Evolved distal tail carbohydrate
binding modules of Lactobacillus phage J-1: a novel type of
anti-receptor widespread among lactic acid bacteria phages, Mol.
Microbiol., 104, 608-620.
79.Steinbacher, S., Baxa, U., Miller, S., Weintraub,
A., Seckler, R., and Huber, R. (1996) Crystal structure of phage P22
tailspike protein complexed with Salmonella sp. O-antigen
receptors, Proc. Natl. Acad. Sci. USA, 93,
10584-10588.
80.Lubkowski, J., Hennecke, F., Pluckthun, A., and
Wlodawer, A. (1999) Filamentous phage infection: crystal structure of
g3p in complex with its coreceptor, the C-terminal domain of TolA,
Structure, 7, 711-722.
81.Mondigler, M., Holz, T., and Heller, K. J. (1996)
Identification of the receptor-binding regions of pb5 proteins of
bacteriophages T5 and BF23, Virology, 219, 19-28.
82.Broeker, N. K., and Barbirz, S. (2017) Not a
barrier but a key: how bacteriophages exploit host’s O-antigen as
an essential receptor to initiate infection, Mol. Microbiol.,
doi: 10.1111/mmi.13729.
83.Zaccheus, M. V., Broeker, N. K., Lundborg, M.,
Uetrecht, C., Barbirz, S., and Widmalm, G. (2012) Structural studies of
the O-antigen polysaccharide from Escherichia coli TD2158 having
O18 serogroup specificity and aspects of its interaction with the
tailspike endoglycosidase of the infecting bacteriophage HK620,
Carbohydr. Res., 357, 118-125.
84.Fischetti, V. A. (2010) Bacteriophage endolysins:
a novel anti-infective to control Gram-positive pathogens, Int. J.
Med. Microbiol., 300, 357-362.
85.Fischetti, V. A. (2005) Bacteriophage lytic
enzymes: novel anti-infectives, Trends Microbiol., 13,
491-496.
86.Pires, D. P., Oliveira, H., Melo, L. D.,
Sillankorva, S., and Azeredo, J. (2016) Bacteriophage-encoded
depolymerases: their diversity and biotechnological applications,
Appl. Microbiol. Biotechnol., 100, 2141-2151.
87.Delbruck, M. (1940) The growth of bacteriophage
and lysis of the host, J. Gen. Physiol., 23, 643-660.
88.Krueger, A. P. (1931) The sorption of
bacteriophage by living and dead susceptible bacteria. I. Equilibrium
conditions, J. Gen. Physiol., 14, 493-516.
89.Clokie, M. R. J., and Kropinski, A. M. (2009)
Bacteriophages: Methods and Protocols, Humana Press, New
York.
90.Gallet, R., Kannoly, S., and Wang, I. N. (2011)
Effects of bacteriophage traits on plaque formation, BMC
Microbiol., 11, 181.
91.Wiggins, B. A., and Alexander, M. (1985) Minimum
bacterial density for bacteriophage replication: implications for
significance of bacteriophages in natural ecosystems, Appl. Environ.
Microbiol., 49, 19-23.
92.Kasman, L. M., Kasman, A., Westwater, C., Dolan,
J., Schmidt, M. G., and Norris, J. S. (2002) Overcoming the phage
replication threshold: a mathematical model with implications for phage
therapy, J. Virol., 76, 5557-5564.
93.Abedon, S. (2011) Phage therapy pharmacology:
calculating phage dosing, Adv. Appl. Microbiol., 77,
1-40.
94.Payne, R. J., and Jansen, V. A. (2001)
Understanding bacteriophage therapy as a density-dependent kinetic
process, J. Theor. Biol., 208, 37-48.
95.Kazi, M., and Annapure, U. S. (2016)
Bacteriophage biocontrol of foodborne pathogens, J. Food Sci.
Technol., 53, 1355-1362.
96.Sulakvelidze, A. (2013) Using lytic
bacteriophages to eliminate or significantly reduce contamination of
food by foodborne bacterial pathogens, J. Sci. Food Agric.,
93, 3137-3146.
97.Guerrero-Ferreira, R. C., Viollier, P. H., Ely,
B., Poindexter, J. S., Georgieva, M., Jensen, G. J., and Wright, E. R.
(2011) Alternative mechanism for bacteriophage adsorption to the motile
bacterium Caulobacter crescentus, Proc. Natl. Acad. Sci.
USA, 108, 9963-9968.
98.Barr, J. J., Auro, R., Furlan, M., Whiteson, K.
L., Erb, M. L., Pogliano, J., Stotland, A., Wolkowicz, R., Cutting, A.
S., Doran, K. S., Salamon, P., Youle, M., and Rohwer, F. (2013)
Bacteriophage adhering to mucus provide a non-host-derived immunity,
Proc. Natl. Acad. Sci. USA, 110, 10771-10776.
99.Fraser, J. S., Maxwell, K. L., and Davidson, A.
R. (2007) Immunoglobulin-like domains on bacteriophage: weapons of
modest damage? Curr. Opin. Microbiol., 10, 382-387.
100.Fraser, J. S., Yu, Z., Maxwell, K. L., and
Davidson, A. R. (2006) Ig-like domains on bacteriophages: a tale of
promiscuity and deceit, J. Mol. Biol., 359, 496-507.
101.McPartland, J., and Rothman-Denes, L. B. (2009)
The tail sheath of bacteriophage N4 interacts with the
Escherichia coli receptor, J. Bacteriol.,
191, 525-532.
102.Chan, J. Z., Millard, A. D., Mann, N. H., and
Schafer, H. (2014) Comparative genomics defines the core genome of the
growing N4-like phage genus and identifies N4-like roseophage specific
genes, Front. Microbiol., 5, 506.
103.Knirel, Y. A., Prokhorov, N. S., Shashkov, A.
S., Ovchinnikova, O. G., Zdorovenko, E. L., Liu, B., Kostryukova, E.
S., Larin, A. K., Golomidova, A. K., and Letarov, A. V. (2015)
Variations in O-antigen biosynthesis and O-acetylation associated with
altered phage sensitivity in Escherichia coli 4s, J.
Bacteriol., 197, 905-912.
104.Hu, B., Margolin, W., Molineux, I. J., and Liu,
J. (2015) Structural remodeling of bacteriophage T4 and host membranes
during infection initiation, Proc. Natl. Acad. Sci. USA,
112, E4919-4928.
105.Liu, J., Chen, C. Y., Shiomi, D., Niki, H., and
Margolin, W. (2011) Visualization of bacteriophage P1 infection by
cryo-electron tomography of tiny Escherichia coli,
Virology, 417, 304-311.
106.Andres, D., Hanke, C., Baxa, U., Seul, A.,
Barbirz, S., and Seckler, R. (2010) Tailspike interactions with
lipopolysaccharide effect DNA ejection from phage P22 particles in
vitro, J. Biol. Chem., 285, 36768-36775.
107.Molineux, I. J. (2006) The T7 group, in The
Bacteriophages (Calendar, R., ed.) 2nd Edn., Oxford University
Press, Oxford-New York, pp. 277-301.
108.Parent, K. N., Erb, M. L., Cardone, G., Nguyen,
K., Gilcrease, E. B., Porcek, N. B., Pogliano, J., Baker, T. S., and
Casjens, S. R. (2014) OmpA and OmpC are critical host factors for
bacteriophage Sf6 entry in Shigella, Mol. Microbiol.,
92, 47-60.
109.Scholl, D., Adhya, S., and Merril, C. (2005)
Escherichia coli K1′s capsule is a barrier to
bacteriophage T7, Appl. Environ. Microbiol., 71,
4872-4874.
110.Scholl, D., and Merril, C. (2005) The genome of
bacteriophage K1F, a T7-like phage that has acquired the ability to
replicate on K1 strains of Escherichia coli, J.
Bacteriol., 187, 8499-8503.
111.Zhan, L., Wang, S., Guo, Y., Jin, Y., Duan, J.,
Hao, Z., Lv, J., Qi, X., Hu, L., Chen, L., Kreiswirth, B. N., Zhang,
R., Pan, J., Wang, L., and Yu, F. (2017) Outbreak by hypermucoviscous
Klebsiella pneumoniae ST11 isolates with carbapenem resistance
in a tertiary hospital in China, Front. Cell Infect. Microbiol.,
7, 182.
112.Pan, Y. J., Lin, T. L., Chen, C. C., Tsai, Y.
T., Cheng, Y. H., Chen, Y. Y., Hsieh, P. F., Lin, Y. T., and Wang, J.
T. (2017) Klebsiella phage PhiK64-1 encodes multiple
depolymerases for multiple host capsular types, J. Virol.,
91, e02457-16.
113.Drulis-Kawa, Z., Majkowska-Skrobek, G., and
Maciejewska, B. (2015) Bacteriophages and phage-derived proteins
– application approaches, Curr. Med. Chem., 22,
1757-1773.
114.Edgar, R., Rokney, A., Feeney, M., Semsey, S.,
Kessel, M., Goldberg, M. B., Adhya, S., and Oppenheim, A. B. (2008)
Bacteriophage infection is targeted to cellular poles, Mol.
Microbiol., 68, 1107-1116.
115.Hu, B., Margolin, W., Molineux, I. J., and Liu,
J. (2013) The bacteriophage t7 virion undergoes extensive structural
remodeling during infection, Science, 339, 576-579.
116.Comeau, A. M., Bertrand, C., Letarov, A.,
Tetart, F., and Krisch, H. M. (2007) Modular architecture of the T4
phage superfamily: a conserved core genome and a plastic periphery,
Virology, 362, 384-396.
117.Desplats, C., and Krisch, H. M. (2003) The
diversity and evolution of the T4-type bacteriophages, Res.
Microbiol., 154, 259-267.
118.Mosig, G., and Eiserling, F. (2006) T4 and
related phages: structure and development, in The Bacteriophages
(Calendar, R., ed.) 2nd Edn., Oxford University Press, Oxford-New York,
pp. 225-267.
119.Leptihn, S., Gottschalk, J., and Kuhn, A.
(2016) T7 ejectosome assembly: a story unfolds, Bacteriophage,
6, e1128513.
120.Jin, Y., Sdao, S. M., Dover, J. A., Porcek, N.
B., Knobler, C. M., Gelbart, W. M., and Parent, K. N. (2015)
Bacteriophage P22 ejects all of its internal proteins before its
genome, Virology, 485, 128-134.
121.Moak, M., and Molineux, I. J. (2004)
Peptidoglycan hydrolytic activities associated with bacteriophage
virions, Mol. Microbiol., 51, 1169-1183.
122.Davydova, E. K., Kazmierczak, K. M., and
Rothman-Denes, L. B. (2003) Bacteriophage N4-coded, virion-encapsulated
DNA-dependent RNA polymerase, Methods Enzymol., 370,
83-94.
123.Sun, L., Rossmann, M. G., and Fane, B. A.
(2014) High-resolution structure of a virally encoded DNA-translocating
conduit and the mechanism of DNA penetration, J. Virol.,
88, 10276-10279.
124.Xu, J., Gui, M., Wang, D., and Xiang, Y. (2016)
The bacteriophage varphi29 tail possesses a pore-forming loop for cell
membrane penetration, Nature, 534, 544-547.
125.Cumby, N., Reimer, K., Mengin-Lecreulx, D.,
Davidson, A. R., and Maxwell, K. L. (2015) The phage tail tape measure
protein, an inner membrane protein and a periplasmic chaperone play
connected roles in the genome injection process of E. coli phage
HK97, Mol. Microbiol., 96, 437-447.
126.Molineux, I. J., and Panja, D. (2013) Popping
the cork: mechanisms of phage genome ejection, Nat. Rev.
Microbiol., 11, 194-204.
127.Foster, T. J. (2005) Immune evasion by
staphylococci, Nat. Rev. Microbiol., 3, 948-958.
128.Nordstrom, K., and Forsgren, A. (1974) Effect
of protein A on adsorption of bacteriophages to Staphylococcus
aureus, J. Virol., 14, 198-202.
129.Pedruzzi, I., Rosenbusch, J. P., and Locher, K.
P. (1998) Inactivation in vitro of the Escherichia
coli outer membrane protein FhuA by a phage T5-encoded
lipoprotein, FEMS Microbiol. Lett., 168, 119-125.
130.Riede, I., and Eschbach, M. L. (1986) Evidence
that TraT interacts with OmpA of Escherichia coli,
FEBS Lett., 205, 241-245.
131.Uhl, M. A., and Miller, J. F. (1996)
Integration of multiple domains in a two-component sensor protein: the
Bordetella pertussis BvgAS phosphorelay, EMBO J.,
15, 1028-1036.
132.Beier, D., Fuchs, T. M., Graeff-Wohlleben, H.,
and Gross, R. (1996) Signal transduction and virulence regulation in
Bordetella pertussis, Microbiologia, 12,
185-196.
133.Liu, M., Deora, R., Doulatov, S. R., Gingery,
M., Eiserling, F. A., Preston, A., Maskell, D. J., Simons, R. W.,
Cotter, P. A., Parkhill, J., and Miller, J. F. (2002) Reverse
transcriptase-mediated tropism switching in Bordetella
bacteriophage, Science, 295, 2091-2094.
134.Stummeyer, K., Schwarzer, D., Claus, H., Vogel,
U., Gerardy-Schahn, R., and Muhlenhoff, M. (2006) Evolution of
bacteriophages infecting encapsulated bacteria: lessons from
Escherichia coli K1-specific phages, Mol. Microbiol.,
60, 1123-1135.
135.Kennedy, L., and Sutherland, I. W. (1994)
Gellan lyases – novel polysaccharide lyases, Microbiology,
140, 3007-3013.
136.Sutherland, I. W. (1995) Polysaccharide lyases,
FEMS Microbiol. Rev., 16, 323-347.
137.Sutherland, I. W. (1987) Xanthan lyases –
novel enzymes found in various bacterial species, J. Gen.
Microbiol., 133, 3129-3134.
138.Sutherland, I. W., Hughes, K. A., Skillman, L.
C., and Tait, K. (2004) The interaction of phage and biofilms, FEMS
Microbiol. Lett., 232, 1-6.
139.Hynes, W. L., Hancock, L., and Ferretti, J. J.
(1995) Analysis of a second bacteriophage hyaluronidase gene from
Streptococcus pyogenes: evidence for a third hyaluronidase
involved in extracellular enzymatic activity, Infect. Immun.,
63, 3015-3020.
140.McClean, D. (1941) Studies on diffusing
factors: the hyaluronidase activity of testicular extracts, bacterial
culture filtrates and other agents that increase tissue permeability,
Biochem. J., 35, 159-183.
141.Kjems, E. (1955) Studies on streptococcal
bacteriophages. I. Technique of isolating phage-producing strains,
Acta Pathol. Microbiol. Scand., 36, 433-440.
142.Benchetrit, L. C., Gray, E. D., and Wannamaker,
L. W. (1977) Hyaluronidase activity of bacteriophages of group A
streptococci, Infect. Immun., 15, 527-532.
143.Destoumieux-Garzon, D., Duquesne, S., Peduzzi,
J., Goulard, C., Desmadril, M., Letellier, L., Rebuffat, S., and
Boulanger, P. (2005) The iron-siderophore transporter FhuA is the
receptor for the antimicrobial peptide microcin J25: role of the
microcin Val11-Pro16 beta-hairpin region in the recognition mechanism,
Biochem. J., 389, 869-876.
144.Meyer, J. R., Dobias, D. T., Weitz, J. S.,
Barrick, J. E., Quick, R. T., and Lenski, R. E. (2012) Repeatability
and contingency in the evolution of a key innovation in phage lambda,
Science, 335, 428-432.
145.Burmeister, A. R., Lenski, R. E., and Meyer, J.
R. (2016) Host coevolution alters the adaptive landscape of a virus,
Proc. R. Soc. Lond. Ser. B Biol. Sci., 283, 20161528.
146.Morita, M., Fischer, C. R., Mizoguchi, K.,
Yoichi, M., Oda, M., Tanji, Y., and Unno, H. (2002) Amino acid
alterations in Gp38 of host range mutants of PP01 and evidence for
their infection of an ompC null mutant of Escherichia
coli O157:H7, FEMS Microbiol. Lett., 216,
243-248.
147.Sandmeier, H. (1994) Acquisition and
rearrangement of sequence motifs in the evolution of bacteriophage tail
fibres, Mol. Microbiol., 12, 343-350.
148.Buth, S. A., Menin, L., Shneider, M. M., Engel,
J., Boudko, S. P., and Leiman, P. G. (2015) Structure and biophysical
properties of a triple-stranded beta-helix comprising the central spike
of bacteriophage T4, Viruses, 7, 4676-4706.
149.Letarov, A., Manival, X., Desplats, C., and
Krisch, H. M. (2005) gpwac of the T4-type bacteriophages: structure,
function, and evolution of a segmented coiled-coil protein that
controls viral infectivity, J. Bacteriol., 187,
1055-1066.
150.George, D. G., Yeh, L. S., and Barker, W. C.
(1983) Unexpected relationships between bacteriophage lambda
hypothetical proteins and bacteriophage T4 tail-fiber proteins,
Biochem. Biophys. Res. Commun., 115, 1061-1068.
151.Trojet, S. N., Caumont-Sarcos, A., Perrody, E.,
Comeau, A. M., and Krisch, H. M. (2011) The gp38 adhesins of the T4
superfamily: a complex modular determinant of the phage’s host
specificity, Genome Biol. Evol., 3, 674-686.
152.Tetart, F., Desplats, C., Kutateladze, M.,
Monod, C., Ackermann, H. W., and Krisch, H. M. (2001) Phylogeny of the
major head and tail genes of the wide-ranging T4-type bacteriophages,
J. Bacteriol., 183, 358-366.
153.Yoichi, M., Abe, M., Miyanaga, K., Unno, H.,
and Tanji, Y. (2005) Alteration of tail fiber protein gp38 enables T2
phage to infect Escherichia coli O157:H7, J.
Biotechnol., 115, 101-107.
154.Kulikov, E., Kropinski, A. M., Golomidova, A.,
Lingohr, E., Govorun, V., Serebryakova, M., Prokhorov, N., Letarova,
M., Manykin, A., Strotskaya, A., and Letarov, A. (2012) Isolation and
characterization of a novel indigenous intestinal N4-related coliphage
vB_EcoP_G7C, Virology, 426, 93-99.
155.Plasterk, R. H., Brinkman, A., and Van de
Putte, P. (1983) DNA inversions in the chromosome of Escherichia
coli and in bacteriophage Mu: relationship to other site-specific
recombination systems, Proc. Natl. Acad. Sci. USA, 80,
5355-5358.
156.Grundy, F. J., and Howe, M. M. (1984)
Involvement of the invertible G segment in bacteriophage mu tail fiber
biosynthesis, Virology, 134, 296-317.
157.Johnson, R. C. (1991) Mechanism of
site-specific DNA inversion in bacteria, Curr. Opin. Genet.
Dev., 1, 404-411.
158.Guo, H., Arambula, D., Ghosh, P., and Miller,
J. F. (2014) Diversity-generating retroelements in phage and bacterial
genomes, Microbiol. Spectr., 2, doi:
10.1128/microbiolspec.MDNA3-0029-2014.
159.Toussaint, A. (2013) Transposable Mu-like
phages in Firmicutes: new instances of divergence generating
retroelements, Res. Microbiol., 164, 281-287.
160.Ye, Y. (2014) Identification of
diversity-generating retroelements in human microbiomes, Int. J.
Mol. Sci., 15, 14234-14246.
161.Minot, S., Grunberg, S., Wu, G. D., Lewis, J.
D., and Bushman, F. D. (2012) Hypervariable loci in the human gut
virome, Proc. Natl. Acad. Sci. USA, 109, 3962-3966.
162.Fokine, A., Zhang, Z., Kanamaru, S., Bowman, V.
D., Aksyuk, A. A., Arisaka, F., Rao, V. B., and Rossmann, M. G. (2013)
The molecular architecture of the bacteriophage T4 neck, J. Mol.
Biol., 425, 1731-1744.
163.Stockdale, S. R., Mahony, J., Courtin, P.,
Chapot-Chartier, M. P., Van Pijkeren, J. P., Britton, R. A., Neve, H.,
Heller, K. J., Aideh, B., Vogensen, F. K., and Van Sinderen, D. (2013)
The lactococcal phages Tuc2009 and TP901-1 incorporate two alternate
forms of their tail fiber into their virions for infection
specialization, J. Biol. Chem., 288, 5581-5590.
164.Gallet, R., Lenormand, T., and Wang, I. N.
(2012) Phenotypic stochasticity protects lytic bacteriophage
populations from extinction during the bacterial stationary phase,
Evolution, 66, 3485-3494.
165.Storms, Z. J., and Sauvageau, D. (2014)
Evidence that the heterogeneity of a T4 population is the result of
heritable traits, PLoS One, 9, e116235.
166.Fisher, R. A., Gollan, B., and Helaine, S.
(2017) Persistent bacterial infections and persister cells, Nat.
Rev. Microbiol., 5, 453-464.
167.Langenscheid, J., Killmann, H., and Braun, V.
(2004) A FhuA mutant of Escherichia coli is infected by phage
T1-independent of TonB, FEMS Microbiol. Lett., 234,
133-137.
168.Sayers, J. (2006) Bacteriophage T5, in The
Bacteriophages (Calendar, R., ed.) 2nd Edn., Oxford University
Press, Oxford-New York, pp. 268-276.
169.Andres, D., Roske, Y., Doering, C., Heinemann,
U., Seckler, R., and Barbirz, S. (2012) Tail morphology controls DNA
release in two Salmonella phages with one lipopolysaccharide
receptor recognition system, Mol. Microbiol., 83,
1244-1253.
170.Porcek, N. B., and Parent, K. N. (2015) Key
residues of S. flexneri OmpA mediate infection by bacteriophage
Sf6, J. Mol. Biol., 427, 1964-1976.
171.Javed, M. A., van Alphen, L. B., Sacher, J.,
Ding, W., Kelly, J., Nargang, C., Smith, D. F., Cummings, R. D., and
Szymanski, C. M. (2015) A receptor-binding protein of
Campylobacter jejuni bacteriophage NCTC 12673 recognizes
flagellin glycosylated with acetamidino-modified pseudaminic acid,
Mol. Microbiol., 95, 101-115.
172.Xu, D., Zhang, J., Liu, J., Xu, J., Zhou, H.,
Zhang, L., Zhu, J., and Kan, B. (2014) Outer membrane protein OmpW is
the receptor for typing phage VP5 in the Vibrio cholerae O1 El
Tor biotype, J. Virol., 88, 7109-7111.
173.Beczala, A., Ovchinnikova, O. G., Datta, N.,
Mattinen, L., Knapska, K., Radziejewska-Lebrecht, J., Holst, O., and
Skurnik, M. (2015) Structure and genetic basis of Yersinia
similis serotype O:9 O-specific polysaccharide, Innate
Immun., 21, 3-16.
174.Kim, M., Kim, S., Park, B., and Ryu, S. (2014)
Core lipopolysaccharide-specific phage SSU5 as an auxiliary component
of a phage cocktail for Salmonella biocontrol, Appl. Environ.
Microbiol., 80, 1026-1034.
175.Marti, R., Zurfluh, K., Hagens, S., Pianezzi,
J., Klumpp, J., and Loessner, M. J. (2013) Long tail fibres of the
novel broad-host-range T-even bacteriophage S16 specifically recognize
Salmonella OmpC, Mol. Microbiol., 87, 818-834.
176.Kvitko, B. H., Cox, C. R., DeShazer, D.,
Johnson, S. L., Voorhees, K. J., and Schweizer, H. P. (2012) phiX216, a
P2-like bacteriophage with broad Burkholderia pseudomallei and
B. mallei strain infectivity, BMC Microbiol., 12,
289.
177.Kim, M., and Ryu, S. (2012) Spontaneous and
transient defence against bacteriophage by phase-variable glucosylation
of O-antigen in Salmonella enterica serovar typhimurium,
Mol. Microbiol., 86, 411-425.
178.Kiljunen, S., Datta, N., Dentovskaya, S. V.,
Anisimov, A. P., Knirel, Y. A., Bengoechea, J. A., Holst, O., and
Skurnik, M. (2011) Identification of the lipopolysaccharide core of
Yersinia pestis and Yersinia pseudotuberculosis as
the receptor for bacteriophage phiA1122, J. Bacteriol.,
193, 4963-4972.
179.Turska-Szewczuk, A., Pietras, H., Pawelec, J.,
Mazur, A., and Russa, R. (2010) Morphology and general characteristics
of bacteriophages infectious to Robinia pseudoacacia
mesorhizobia, Curr. Microbiol., 61, 315-321.
180.Ricci, V., and Piddock, L. J. (2010) Exploiting
the role of TolC in pathogenicity: identification of a bacteriophage
for eradication of Salmonella serovars from poultry, Appl.
Environ. Microbiol., 76, 1704-1706.
181.Begum, Y. A., Chakraborty, S., Chowdhury, A.,
Ghosh, A. N., Nair, G. B., Sack, R. B., Svennerholm, A. M., and Qadri,
F. (2010) Isolation of a bacteriophage specific for CS7-expressing
strains of enterotoxigenic Escherichia coli, J. Med.
Microbiol., 59, 266-272.
182.Zhang, J., Li, W., Zhang, Q., Wang, H., Xu, X.,
Diao, B., Zhang, L., and Kan, B. (2009) The core oligosaccharide and
thioredoxin of Vibrio cholerae are necessary for binding and
propagation of its typing phage VP3, J. Bacteriol., 191,
2622-2629.
183.Hong, J., Kim, K. P., Heu, S., Lee, S. J.,
Adhya, S., and Ryu, S. (2008) Identification of host receptor and
receptor-binding module of a newly sequenced T5-like phage EPS7,
FEMS Microbiol. Lett., 289, 202-209.
184.Hernandez-Sanchez, J., Bautista-Santos, A.,
Fernandez, L., Bermudez-Cruz, R. M., Uc-Mass, A., Martinez-Penafiel,
E., Martinez, M. A., Garcia-Mena, J., Guarneros, G., and Kameyama, L.
(2008) Analysis of some phenotypic traits of feces-borne temperate
lambdoid bacteriophages from different immunity groups: a high
incidence of cor+, FhuA-dependent phages, Arch.
Virol., 153, 1271-1280.
185.Rabsch, W., Ma, L., Wiley, G., Najar, F. Z.,
Kaserer, W., Schuerch, D. W., Klebba, J. E., Roe, B. A., Laverde Gomez,
J. A., Schallmey, M., Newton, S. M., and Klebba, P. E. (2007) FepA- and
TonB-dependent bacteriophage H8: receptor binding and genomic sequence,
J. Bacteriol., 189, 5658-5674.
186.Ko, Y. T. (2005) The receptor of an oyster
juice-borne coliphage OJ367 in the outer membrane of Salmonella
derby, J. Microbiol. Immunol. Infect., 38,
399-408.
187.Budzik, J. M., Rosche, W. A., Rietsch, A., and
O’Toole, G. A. (2004) Isolation and characterization of a
generalized transducing phage for Pseudomonas aeruginosa strains
PAO1 and PA14, J. Bacteriol., 186, 3270-3273.
188.German, G. J., and Misra, R. (2001) The TolC
protein of Escherichia coli serves as a cell-surface
receptor for the newly characterized TLS bacteriophage, J. Mol.
Biol., 308, 579-585.
189.Ho, T. D., and Slauch, J. M. (2001) OmpC is the
receptor for Gifsy-1 and Gifsy-2 bacteriophages of Salmonella,
J. Bacteriol., 183, 1495-1498.
190.Nesper, J., Kapfhammer, D., Klose, K. E.,
Merkert, H., and Reidl, J. (2000) Characterization of Vibrio
cholerae O1 antigen as the bacteriophage K139 receptor and
identification of IS1004 insertions aborting O1 antigen biosynthesis,
J. Bacteriol., 182, 5097-5104.
191.Traurig, M., and Misra, R. (1999)
Identification of bacteriophage K20 binding regions of OmpF and
lipopolysaccharide in Escherichia coli K-12, FEMS
Microbiol. Lett., 181, 101-108.
192.Sechaud, L., Rousseau, M., Fayard, B.,
Callegari, M. L., Quenee, P., and Accolas, J. P. (1992) Comparative
study of 35 bacteriophages of Lactobacillus helveticus:
morphology and host range, Appl. Environ. Microbiol., 58,
1011-1018.
193.Sao-Jose, C., Baptista, C., and Santos, M. A.
(2004) Bacillus subtilis operon encoding a membrane receptor for
bacteriophage SPP1, J. Bacteriol., 186, 8337-8346.