Biochemical Properties and Phylogeny of Hydroxypyruvate Reductases from Methanotrophic Bacteria with Different C1-Assimilation Pathways
S. Y. But1*, S. V. Egorova2, V. N. Khmelenina1, and Y. A. Trotsenko1,2*
1Skryabin Institute of Biochemistry and Physiology of Microorganisms, Laboratory of Methylotrophy, Russian Academy of Sciences, 142290 Pushchino, Moscow Region, Russia; E-mail: flash20063@rambler.ru, trotsenko@ibpm.pushchino.ru2Pushchino State Institute of Natural Sciences, 142290 Pushchino, Moscow Region, Russia
* To whom correspondence should be addressed.
Received July 11, 2017; Revision received August 22, 2017
In the aerobic methanotrophic bacteria Methylomicrobium alcaliphilum 20Z, Methylococcus capsulatus Bath, and Methylosinus trichosporium OB3b, the biochemical properties of hydroxypyruvate reductase (Hpr), an indicator enzyme of the serine pathway for assimilation of reduced C1-compounds, were comparatively analyzed. The recombinant Hpr obtained by cloning and heterologous expression of the hpr gene in Escherichia coli catalyzed NAD(P)H-dependent reduction of hydroxypyruvate or glyoxylate, but did not catalyze the reverse reactions of D-glycerate or glycolate oxidation. The absence of the glycerate dehydrogenase activity in the methanotrophic Hpr confirmed a key role of the enzyme in utilization of C1-compounds via the serine cycle. The enzyme from Ms. trichosporium OB3b realizing the serine cycle as a sole assimilation pathway had much higher special activity and affinity in comparison to Hpr from Mm. alcaliphilum 20Z and Mc. capsulatus Bath assimilating carbon predominantly via the ribulose monophosphate (RuMP) cycle. The hpr gene was found as part of gene clusters coding the serine cycle enzymes in all sequenced methanotrophic genomes except the representatives of the Verrucomicrobia phylum. Phylogenetic analyses revealed two types of Hpr: (i) Hpr of methanotrophs belonging to the Gammaproteobacteria class, which use the serine cycle along with the RuMP cycle, as well as of non-methylotrophic bacteria belonging to the Alphaproteobacteria class; (ii) Hpr of methylotrophs from Alpha- and Betaproteobacteria classes that use only the serine cycle and of non-methylotrophic representatives of Betaproteobacteria. The putative role and origin of hydroxypyruvate reductase in methanotrophs are discussed.
KEY WORDS: hydroxypyruvate reductase, methanotrophs, serine cycle of carbon assimilationDOI: 10.1134/S0006297917110074
Abbreviations: Hpr, hydroxypyruvate reductase; RuBP, ribulose bisphosphate; RuMP, ribulose monophosphate.
Aerobic methane utilizing bacteria (methanotrophs) assimilate carbon
from methane via three biochemical pathways – the serine
pathway and the ribulose monophosphate (RuMP) and the ribulose
bisphosphate (RuBP) cycles. The serine pathway is initiated by
condensation of formaldehyde (as tetrahydrofolate derivative) and
glycine resulting in formation of serine from which hydroxypyruvate is
generated after transamination with glyoxylate. The reduction of
hydroxypyruvate into glycerate catalyzes hydroxypyruvate reductase
(Hpr), considered as an indicator enzyme of the serine pathway. The
serine pathway is characteristic for methanotrophs belonging to the
Alphaproteobacteria class (type II methanotrophs). In the RuMP pathway,
a C–C bond is created by condensation of formaldehyde with
ribulose-5-phosphate and forming hexulose-6-phosphate, which is
thereafter isomerized into fructose-6-phosphate. Hexosephosphate
cleavage leads to synthesis of trioses [1]. The
RuMP pathway functions in gammaproteobacterial methanotrophs (type I
methanotrophs). In the RuBP pathway realized by methanotrophs of the
Verrucomicrobia phylum, CO2 formed during oxidation of the
reduced C1-substrate is assimilated. After initial
deciphering of methylotrophic metabolism half century ago, these
pathways were considered as alternatives in each main group of
methanotrophic bacteria. Subsequent enzymatic studies provided evidence
that in methanotrophs of the Methylococcus and
Methylocaldum genera (type X methanotrophs) there are three
simultaneously functioning C1-assimilation pathways [2-5]. Furthermore, genes coding
for the serine pathway enzymes were revealed in methanotrophs
assimilating carbon via the RuMP pathway. However, distribution,
functionality, and physiological role of the enzymes in various
methanotrophs remain unclear. In this work, we present the results of
comparative characterization of Hpr in Methylomicrobium
alcaliphilum 20Z (representative of type I methanotrophs),
Methylosinus trichosporium OB3b (type II), and Methylococcus
capsulatus Bath (type X) and the distribution of hpr genes
in bacteria.
MATERIALS AND methods
Bacteria and growth conditions. Methylomicrobium alcaliphilum 20Z (VKM B-2133T = NCIMB 14124T), Ms. trichosporium OB3b (VKM B-2117 = NCIMB 11131), and Mc. capsulatus Bath (VKM B-2990 = NCIMB 11132) were grown under a methane–air atmosphere (1 : 1) in 2P mineral medium at 30°C (strains 20Z and OB3b) or 37°C (Bath). The medium for strain 20Z was supplemented with 0.1 M NaHCO3 and 0.3 M NaCl [6]. Escherichia coli Rosetta (DE3) obtained from Stratagene (USA) was grown in selective LB broth or agar (1.5%; Difco, USA) at 37°C [7]. Kanamycin (100 µg/ml) and chloramphenicol (25 µg/ml) were added if required.
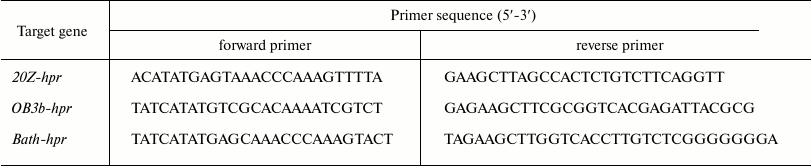
Cloning, expression, and purification of recombinant enzymes. DNA from methanotrophs was prepared by using a ZymoResearch Fungal/Bacterial DNA MiniPrep™ kit (Irvine, USA) according guidelines of the producer. Genes coding for putative Hpr in Mm. alcaliphilum 20Z (hpr20Z, MALCv4_3219), Ms. trichosporium OB3b (hprOB3b, ADVE02_v2_12566), and Mc. capsulatus Bath (hprBath, MCA1407) were amplified from the respective DNA using the primers listed in Table 1. The PCR-products were treated with endonucleases NdeI and HindIII and ligated in expression vector pET30(a)+ (hpr20Z and hprBath) or pET28b (hprOB3b) opened at the respective restriction sites. The resulting plasmids were transferred into E. coli Rosetta (DE3), and protein expression was induced by 0.5 mM isopropyl β-D-1-thiogalactopyranoside (Sigma-Aldrich, USA) added in the logarithmic phase of growth (OD600 = 0.6-0.8). After 18 h of growth at 18°C, cells were centrifuged (5000g for 30 min, 4°C). The cells were disrupted using an ultrasonic disintegrator (Misonix, USA) with 1 min cooling in ice 30 s after each 10-s sonication, then centrifuged for 30 min at 11,000g and 4°C. The recombinant proteins were purified on a column with Ni2+-NTA agarose as described earlier [8].
Table 1. Sequences of primers used in this
work
Hpr activity assay. Activity of Hpr was measured by registering velocity of NAD(P)H oxidation at λ = 340 nm using a Shimadzu UV1700 spectrophotometer (Shimadzu, Japan). The assay mixture (1 ml) contained 50 mM Tris-HCl (pH 7.0), 5 mM hydroxypyruvate, and 0.2 mM NADH. Activities of HprOB3b and Hpr20Z were measured at 30°C, and activity of HprBath at 37°C. Values of Km and Vmax were determined by measuring the activity with different concentrations of the substrate at saturated concentrations of the other. For calculation, the Enzyme Kinetics Module of SigmaPlot 12 was used, and the data approximated to the Michaelis–Menten equation:
or to the equation for substrate inhibition:
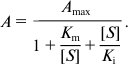
Influence of pH on Hpr activity was studied using following buffers: MES-KOH (pH 5.5-7.0), Tris-HCl (pH 7.0-9.0), and NaOH-glycine (pH 9.0-10.5).
Quaternary forms of the enzymes were analyzed by non-denaturing gel electrophoresis by using pore-limited gradient polyacrylamide (4-30%) [9]. The reference proteins thyroglobulin (667 kDa), ferritin (440 kDa), amylase (250 kDa), alcohol dehydrogenase (150 kDa), and bovine serum albumin (BSA, 66 kDa) were obtained from Sigma-Aldrich (USA).
RESULTS
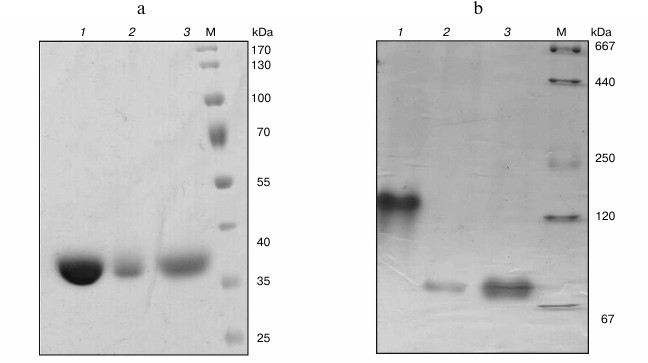
Cloning of hpr genes and purification of recombinant proteins. In the genomes of Mm. alcaliphilum 20Z, Ms. trichosporium OB3b, and Mc. capsulatus Bath, the open reading frames whose translated amino acid sequences showed homologies with earlier characterized Hpr from Hyphomicrobium methylovorum GM2 (37, 36, and 40% identity, respectively) [10] and Methylobacterium extorquens AM1 (43, 78, and 46%) [11] were found. These genes were cloned into vectors pET30(a)+ and pET28b appropriated for expression of proteins with six histidines on either N- or C-end under T7 promoter control. The His6-tagged proteins were purified from extracts of E. coli cells by single-step affinity chromatography on Ni2+-NTA agarose. Molecular masses of subunits of Hpr20Z-His6, HprOB3b-His6, and HprBath-His6 estimated by SDS-PAGE were in good agreement with the theoretically calculated subunit sizes (35-37 kDa) (Fig. 1a). Native gradient electrophoresis revealed single bands of 140 kDa for the M. trichosporium Hpr, whereas it was 70 kDa for the Mc. capsulatus Bath and Mm. alcaliphilum 20Z enzymes (Fig. 1b). Hence, the Hpr of strain OB3b exists as a homotetramer, whereas the enzymes from strains Bath and 20Z are homodimers.
Fig. 1. SDS-PAGE (a) and native electrophoresis in a polyacrylamide gradient (4-30%) (b) of recombinant Hpr from Ms. trichosporium OB3b (1), Mm. alcaliphilum 20Z (2), and Mc. capsulatus Bath (3). M, molecular mass marker.
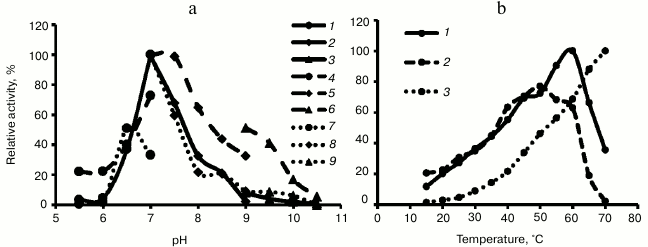
Properties of recombinant Hpr. The three recombinant enzymes reduced hydroxypyruvate into glycerate in the presence of either NADH or NADPH, showing maximal activity at pH 7.0 (Fig. 2). Hpr from mesophilic methanotrophs Mm. alcaliphilum 20Z and Ms. trichosporium OB3b demonstrated unexpectedly high temperature optima, and activity of Hpr from thermotolerant Mc. capsulatus Bath increased upon increasing temperature to at least 70°C (Fig. 2).
Fig. 2. a) pH dependence of activity of recombinant Hpr from Mm. alcaliphilum 20Z (1-3), Ms. trichosporium OB3b (4-6), and Mc. capsulatus Bath (7-9). Curves: 1, 4, 7) MES-KOH buffer; 2, 5, 8) Tris-HCl buffer; 3, 6, 9) Na-glycine. b) Temperature dependence of activity of recombinant Hpr from Mm. alcaliphilum 20Z (1), Ms. trichosporium OB3b (2), and Mc. capsulatus Bath (3).
Besides hydroxypyruvate, all the studied enzymes used glyoxylate as substrate but were not active with pyruvate, lactate, oxaloacetate, and α-ketoglutarate (Table 2). Such substrate specificity has been demonstrated for the Hpr from methylotrophic bacteria Hyphomicrobium methylovorum GM2 and Methylobacterium extorquens AM1 [10, 11]. However, in contrast to the above-mentioned paralogs, the Hpr of methanotrophs did not catalyze the reverse reaction of glycerate and glycolate oxidation.
Table 2. Substrate specificity of Hpr from
different methanotrophs
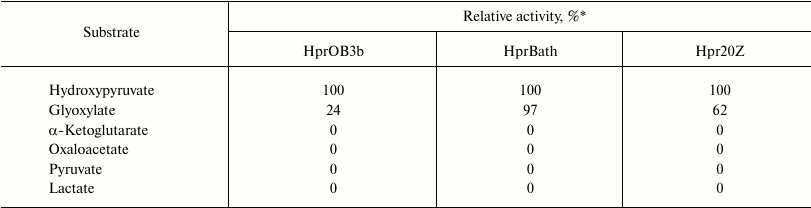
* Activity relatively to activity in the presence of 5 mM
hydroxypyruvate and 0.2 mM NADH.
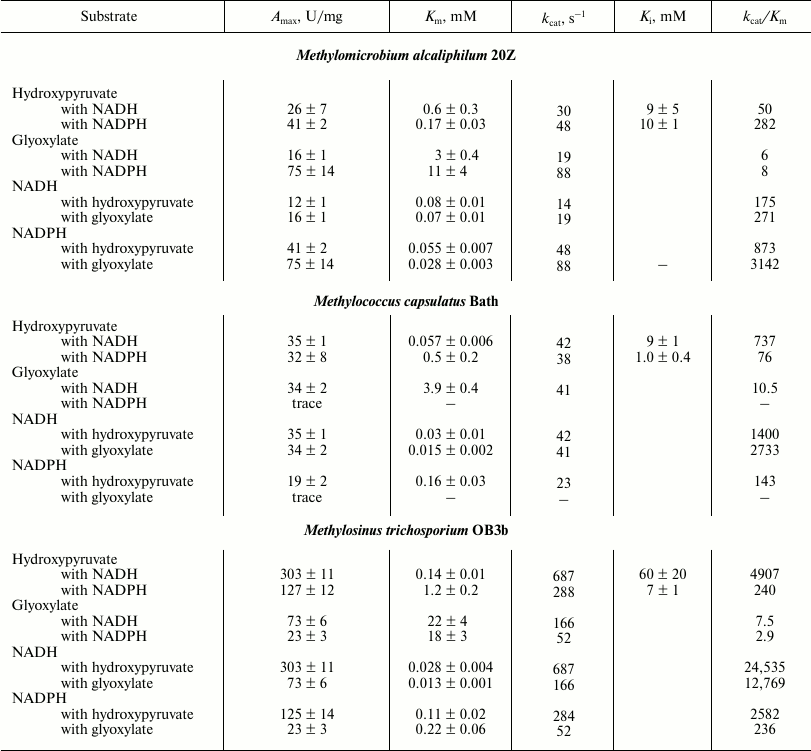
The substrate dependence of the activities of the three methanotrophic Hpr on glyoxylate and cofactors obeyed Michaelis–Menten kinetics. However, in the presence of hydroxypyruvate, substrate inhibition of enzyme activity occurred (Table 3). The activity of hydroxypyruvate reduction by the enzyme from Ms. trichosporium OB3b was 10-fold higher than those of the Hpr from Mm. alcaliphilum 20Z and Mc. capsulatus Bath. In the presence of NADH, the catalytic efficiency of HprOB3b with hydroxypyruvate was 7-fold higher than the catalytic efficiency of HprBath and almost 100-fold higher than that of Hpr20Z. However, in the presence of NADPH, the kcat/Km ratios for the enzymes from Ms. trichosporium OB3b and Mm. alcaliphilum 20Z were comparable and about 3-fold higher than the catalytic efficiency of HprBath. The efficiency of all three enzymes with glyoxylate as substrate was virtually the same independently of the cofactor used (Table 3). On the other hand, the enzymes had a differ preference depending on the cofactor. Thus, in the presence of NADH, Hpr of Ms. trichosporium OB3b used hydroxypyruvate three-orders of magnitude more efficiently than glyoxylate, but in the presence of NADPH only two-orders of magnitude more efficiently. The catalytic efficiency of the enzymes from Mc. capsulatus Bath and Mm. alcaliphilum 20Z was 10-fold higher with hydroxypyruvate in the presence of NADH, while in the presence of NADPH, HprBath was practically inactive with glyoxylate and Hpr20Z was 35 times more efficient with hydroxypyruvate (Table 3). We propose that the reduction of hydroxypyruvate is the main physiological role of Hpr in type II methanotrophs, while Hpr of type I and type X methanotrophs can also function as glyoxylate reductase. The ratio of these reactions can be regulated by concentrations of NADH and NADPH in cells.
Table 3. Kinetic parameters of Hpr from
different methanotrophs
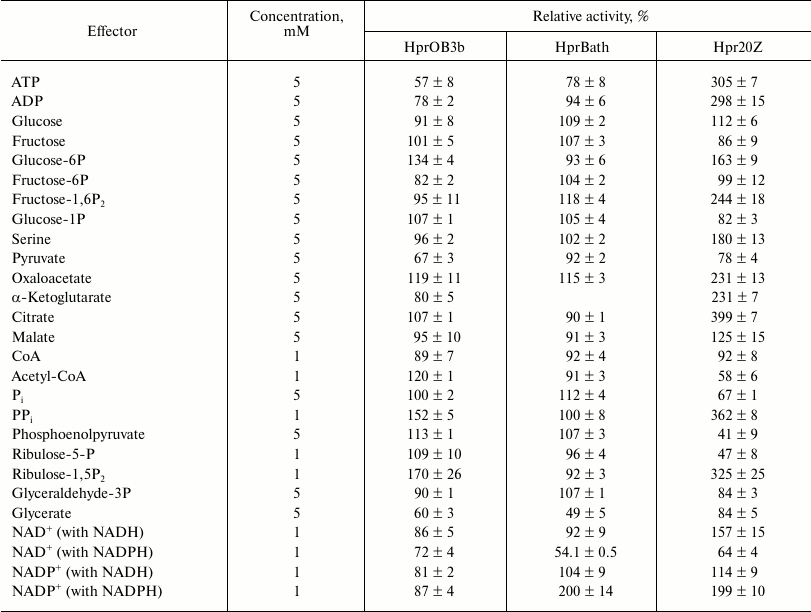
The Hpr from the three methanotrophs differed according to the influence of effectors on their activity. The activity of Hpr20Z was significantly enhanced in the presence of some intermediates of glycolysis and the TCA cycle, as well as serine, ADP, ATP, and inorganic pyrophosphate, but it was inhibited by Pi, pyruvate, acetyl-CoA, phosphoenolpyruvate, and ribulose-5-phosphate. At the same time, only minor stimulatory effect on the activity of Hpr from Ms. trichosporium OB3b was revealed in the presence of PPi, glucose-6-phosphate, ribulose-1,5-bisphosphate, oxaloacetate, and acetyl-CoA, whereas activity of the Mc. capsulatus Bath enzyme was dependent on the presence of fructose-1,6-bisphosphate and oxaloacetate only. Thus, HprBath and HprOB3b were inhibited by ADP and ATP (Table 4). The products of the reaction differently affected the activities of the Hpr. All three enzymes were inhibited by glycerate, Hpr20Z to a minor extent. NAD+ lowered the activity of the enzymes from Ms. trichosporium OB3b and Mc. capsulatus Bath, but Hpr20Z was inhibited by NAD+ only in the reaction with NADPH. In contrast, NAD+ stimulated NADH-dependent activity. NADP+ activated HprBath and Hpr20Z only in reaction with NADPH (Table 4).
Table 4. Influence of some metabolites on
activity of Hpr from different methanotrophs
Distribution and phylogenetic analysis. Genes coding for Hpr were found in all available genomes of methanotrophs with except for representatives of the phylum Verrucomicrobia. In Mm. alcaliphilum 20Z, the hpr gene is located in a single cluster with genes coding for other enzymes of the serine cycle: serine-glyoxylate aminotransferase (sga), glycerate-2-kinase (gck2), malate thiokinase (mtkAB), malyl-CoA lyase (mcl), malate dehydrogenase (mdh), serine hydroxymethyltransferase (glyA), as well as with genes responsible for formaldehyde oxidation: methylene-tetrahydromethanopterin/methylene-tetrahydrofolate dehydrogenase (mtdA) and formate-tetrahydrofolate ligase (ftfL). An analogous set of genes forming one or two identical gene clusters located in different chromosomal locus occurs in most gammaproteobacterial methanotrophs of genera Methylomicrobium, Methylobacter, Methylosarcina, and Methylomonas realizing the RuMP pathway of formaldehyde utilization. The genes of the serine cycle in alphaproteobacterial type II methanotrophs, including Ms. trichosporium OB3b, are organized analogously, but they additionally possess genes for phosphoenolpyruvate carboxylase (ppc) and methylene tetrahydrofolate cyclohydrolase (fch) and have separate location of gene mdh. At the same time, in Mc. capsulatus Bath the hpr gene is co-located with genes of serine-glyoxylate aminotransferase and a putative glycerate-3-kinase (gck3), and the gck2 gene is absent in the genome. Similar organization of genes for the serine pathway was also revealed in another type X methanotroph, Methylocaldum szegediense O12, as well as halophilic type I methanotroph Methylohalobius crimeensis 10Ki.
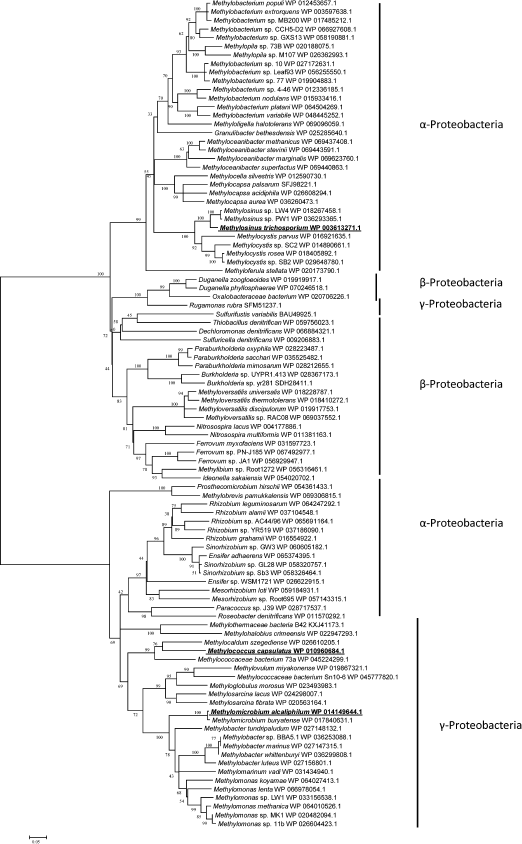
Fig. 3. Phylogenetic tree of translated amino acid sequences of Hpr.
The Hpr from Mm. alcaliphilum 20Z and Mc. capsulatus Bath shared 58% identity of amino acid sequences, and only 26 and 25.5% identities with the Ms. trichosporium OB3b enzyme. Phylogenetic analysis showed the occurrence of two different groups of Hpr presumably of different origin. One group included Hpr of methanotrophs of the Gammaproteobacteria class realizing three pathways of C1-assimilation (serine, RuMP, and RuBP cycles), as well as non-methylotrophic representatives of the Alphaproteobacteria class. Another phylogenetic group included Hpr of alphaproteobacterial methanotrophs, alphaproteobacterial methylotrophs which unable to grow on methane, as well as methylotrophic and non-methylotrophic representatives of the Betaproteobacteria class (Fig. 3).
DISCUSSION
In this report, we present the comparative characteristics of the Hpr from three methanotrophic bacteria differing in taxonomic position and C1-assimilation pathways. The recombinant enzymes from Mm. alcaliphilum 20Z, Ms. trichosporium OB3b, and Mc. capsulatus Bath catalyzed NAD(P)H-dependent reduction of hydroxypyruvate and glyoxylate. These enzymes had similar pH optima, while the strains are either neutrophilic (strains OB3b and Bath) or alkalophilic (strain 20Z). Their temperature optima were significantly higher than the optimal growth temperatures for these bacteria. Earlier, unusual high temperature optima were found for malate dehydrogenases from Mm. alcaliphilum 20Z and Ms. trichosporium OB3b, enzymes which are also involved in the serine cycle [12].
Phylogenetically, Hpr of methanotrophs can be divided on two groups where Hpr of gammaproteobacterial strains 20Z and Bath can be referred to one group, but the enzyme from alphaproteobacterium Ms. trichosporium OB3b belongs to another group. In these species, the gene encoding Hpr is a part of a rather conservative gene cluster coding for other serine cycle enzymes, implying “module” organization of the serine pathway in methanotrophs. Overall, topology of the phylogenetic tree corresponds to the taxonomic position of bacterial strains, suggesting an absence or few horizontal gene transfers and vertical evolution of the serine pathway enzymes. On the other hand, a significant divergence of Hpr in different representatives of methanotrophs indicate different functions of the enzyme. Tetrameric subunit structure of HprOB3b was not reported for any described Hpr. In contrast, all known Hprs are dimeric, similar to the enzymes from Mm. alcaliphilum 20Z and Mc. capsulatus Bath [10, 11, 13].
Activity and affinity to hydroxypyruvate of Hpr from Ms. trichosporium OB3b were much higher than of the other two enzymes. Also, its activity underwent less inhibition by hydroxypyruvate. This agrees with the serine cycle in the type II methanotrophs that is the single pathway of methane carbon assimilation. At the same time, the role of the Hpr in methanotrophs of types I and X remains unclear. Earlier, participation of Hpr in the synthesis of serine from phosphoglycerate in Mc. capsulatus Bath was proposed [4]. This hypothesis, however, contradicts the irreversibility of the reaction catalyzed by HprBath. Besides methylotrophs, Hpr is found in other bacteria (sometimes the name “glyceraldehyde dehydrogenases” is used). The translated amino acid sequences of methylotrophic enzymes share 37-43% identity with the enzymes from bacteria [14, 15], 32-37% with those from algae [16, 17], 35-65% from plants [18], and 26-46% from animals [13, 19]. It has been proposed that reductase activity of the enzyme is associated with serine degradation while glycerate dehydrogenase activity with biosynthesis of this amino acid [20].
The absence of glycerate dehydrogenase activity in the methanotrophic Hpr confirms involvement of the enzyme in assimilation of C1-substrate via serine and hydroxypyruvate forming in the reactions catalyzed by serine-hydroxymethyltransferase and serine-glyoxylate aminotransferase. In addition, Mc. capsulatus Bath has ribulose bisphosphate carboxylase/oxygenase (Rubisco) (MCA2583) that forms phosphoglycolate as a side product, and other enzymes for conversion of phosphoglycolate into glycine (MCA1499-1501) [4]. We suggest that the serine pathway in type X methanotrophs is involved in an analogous process of photorespiration in plants and cyanobacteria and serves for prevention of carbon deprivation as result of the Rubisco oxygenase activity. A stimulation of the activity of Hpr from Mm. alcaliphilum 20Z by the central metabolites can imply the mobilization of the serine cycle as an additional assimilation pathway under conditions of carbon source excess.Activity of glyoxylate reduction was found earlier for most described Hpr, including methylotrophic ones [10, 11, 13, 17, 18]. However, its physiological role is not always clear. It was suggested that the enzyme of Methylobacterium extorquens AM1 is needed for growth on either C1- or C2-compounds, since the Hpr mutant lost ability to grow on these substrates [11]. Importantly, in the genomes of the three studied methanotrophs there are genes coding for FAD-dependent glycolate oxidase (ADVE02_v2_14253-14256; MCA1499-1501; MALCv4_1676-1678). Perhaps a combination of glyoxylate reducing and glycolate oxidizing activities can form a futile cycle discharging reducing equivalent excess. Further studies using mutagenesis and metabolomic measurements can clarify role of Hpr and the serine pathway in methanotrophs.
Acknowledgments
This work was supported by the Russian Foundation for Basic Research (project No. 17-04-11113a).
REFERENCES
1.Trotsenko, Y. A., and Murrell, J. C. (2008)
Metabolic aspects of obligate aerobic methanotrophy, Adv. App.
Microbiol., 63, 183-229.
2.Whittenbury, R. (1980) The interrelationship of
autotrophy and methylotrophy as seen in Methylococcus capsulatus
(Bath), Proc. Int. Symp. “Microbial Growth on C1
compounds”, Heyden, London, pp. 181-190.
3.Baxter, N. J., Hirt, R. P., Bodrossy, L., Kovaks,
K. L., Embley, T. M., Prosser, J. I., and Murrell, J. C. (2002) The
ribulose-1,5-bisphosphate carboxylase/oxygenase gene cluster of
Methylococcus capsulatus (Bath), Arch. Microbiol.,
177, 279-289.
4.Ward, N., Larsen, Q., Sakwa, J., Bruseth, L.,
Khouri, H., Durkin, A. S., Dimitrov, G., Jiang, L., Scanlan, D., Kang,
K. H., Lewis, M., Nelson, K. E., Methe, B., Wu, M., Heidelberg, J. F.,
Paulsen, I. T., Fouts, D., Ravel, J., Tettelin, H., Ren, Q., Read, T.,
DeBoy, R. T., Seshadri, R., Salzberg, S. L., Jensen, H. B., Birkeland,
N. K., Nelson, W. C., Dodson, R. J., Grindhaug, S. H., Holt, I.,
Eidhammer, I., Jonasen, I., Vanaken, S., Utterback, T., Feldblyum, T.
V., Fraser, C. M., Lillehaug, J. R., and Eisen, J. A. (2004) Genomic
insights into methanotrophy: the complete genome sequence of
Methylococcus capsulatus (Bath), PLoS Biol., 2,
1616-1628.
5.Shishkina, V. N., Yurchenko, Y. Y., Romanovskaya,
V. A., Malashenko, Y. R., and Trotsenko, Y. A. (1976) About alternative
of methane assimilation pathways in obligate methylotrophs,
Mikrobiologiya, 45, 417-419.
6.Khmelenina, V. N., Kalyuzhnaya, M. G., Sakharovsky,
V. G., Suzina, N. E., Trotsenko, Y. A., and Gottschalk, G. (1999)
Osmoadaptation in halophilic and alkaliphilic methanotrophs, Arch.
Microbiol., 172, 321-329.
7.Sambrook, J., and Russell, D. W. (2001)
Molecular Cloning: A Laboratory Manual, 3rd Edn., Cold Spring
Harbor Laboratory, N.-Y.
8.Reshetnikov, A. S., Mustakhimov, I. I., Rozova, O.
N., Beschastny, A. P., Khmelenina, V. N., Murrell, J. C., and
Trotsenko, Y. A. (2008) Characterization of the pyrophosphate-dependent
6-phosphofructokinase from Methylococcus capsulatus Bath,
FEMS Microbiol. Lett., 288, 202-210.
9.Slater, G. G. (1969) Stable pattern formation and
determination of molecular size by pore-limit electrophoresis, Anal.
Chem., 41, 1039-1041.
10.Izumi, Y., Yoshida, T., Kanzaki, H., Toki, S.,
Miyazaki, S. S., and Yamada, H. (1990) Purification and
characterization of hydroxypyruvate reductase from a serine-producing
methylotroph Hyphomicrobium methylovorum GM2, Eur. J.
Biochem., 190, 279-284.
11.Chistoserdova, L., and Lidstrom, M. (1991)
Purification and characterization of hydroxypyruvate reductase from the
facultative methylotroph Methylobacterium extorquens AM1, J.
Bacteriol., 173, 7228-7232.
12.Rozova, O. N., Khmelenina, V. N., Bocharova, K.
A., Mustakhimov, I. I., and Trotsenko, Y. A. (2015) Role of
NAD+-dependent malate dehydrogenase in the metabolism of
Methylomicrobium alcaliphilum 20Z and Methylosinus
trichosporium OB3b, Microorganisms, 3, 47-59.
13.Ali, V., Shigeta, Y., and Nozaki, T. (2003)
Molecular and structural characterization of NADPH-dependent
D-glycerate dehydrogenase from the enteric parasitic protist
Entamoeba histolytica, Biochem. J., 375,
729-736.
14.Kohn, L. D., and Jakoby, W. B. (1968) Tartaric
acid metabolism. VII. Crystalline hydroxypyruvate reductase
(D-glycerate dehydrogenase), J. Biol. Chem., 243,
2494-2499.
15.Utting, J. M., and Kohn, L. D. (1975) Structural,
kinetic, and renaturation properties of an induced hydroxypyruvate
reductase from Pseudomonas acidovorans, J. Biol. Chem.,
250, 5233-5242.
16.Husic, D. W., and Tolbert, N. E. (1987)
NADH:hydroxypyruvate reductase and NADPH:glyoxylate reductase in algae:
partial purification and characterization from Chlamydomonas
reinhardtii, Arch. Biochem. Biophys., 252,
396-408.
17.Piotrowska, A., and Czerpak, R. (2009) Cellular
response of light/dark-grown green alga Chlorella vulgaris
Beijerinck (Chlorophyceae) to exogenous adenine- and phenylurea-type
cytokinins, Acta Physiol. Plant., 31, 573-585.
18.Kleczkowski, L. A., Randall, D. D., and Edwards,
G. E. (1991) Oxalate as a potent and selective inhibitor of spinach
(Spinacia oleracea) leaf NADPH-dependent hydroxypyruvate
reductase, Biochem. J., 276, 125-127.
19.Willis, J. E., and Saflach, H. J. (1962) Evidence
for mammalian D-glyceric dehydrogenase, J. Biol. Chem.,
237, 910-915.
20.Ho, C. L., Noji, M., Saito, M., and Saito, K.
(1999) Regulation of serine biosynthesis in Arabidopsis. Crucial
role of plastidic 3-phosphoglycerate dehydrogenase in nonphotosynthetic
tissues, J. Biol. Chem., 274, 397-402.