Intracellular Localization of Apoptotic Endonuclease EndoG and Splice-Variants of Telomerase Catalytic Subunit hTERT
D. D. Zhdanov1,2*, V. S. Pokrovsky1,3, E. V. Orlova4, V. S. Orlova2, M. V. Pokrovskaya1, S. S. Aleksandrova1, and N. N. Sokolov1
1Orekhovich Institute of Biomedical Chemistry, 119121 Moscow, Russia; E-mail: zhdanovdd@mail.ru2Russian University of People’s Friendship, Ecological Faculty, 117198 Moscow, Russia
3Blokhin Russian Cancer Research Center, 115478 Moscow, Russia; E-mail: pokrovskiy@ronc.ru
4Institute of Theoretical and Experimental Biophysics, 142290 Pushchino, Moscow Region, Russia
* To whom correspondence should be addressed.
Received February 20, 2017; Revision received March 29, 2017
The activity of telomerase catalytic subunit hTERT (human telomerase reverse transcriptase) can be regulated by alternative splicing of its mRNA. The mechanism of hTERT splicing is not understood in detail. Apoptotic endonuclease EndoG is known to participate in this process. In the present work, the intracellular colocalization and mRNA levels of EndoG and hTERT splice-variants in normal and apoptotic cancer cells were studied. We found that the development of apoptosis increased the expression of EndoG and changed the ratio of hTERT splice-variants, which decreased the telomerase activity in the cells. The development of apoptosis was accompanied by changes in the amount of mRNA and in the localization of EndoG and hTERT splice-variants in the cytoplasm, nuclei, and mitochondria of the cells. The suppression of EndoG expression using RNA interference prevented induction of the α+β– splice-variant of hTERT and inhibition of the telomerase activity. A high degree of the intracellular colocalization of EndoG and hTERT was shown. The changes in the expression and localization of EndoG corresponded with changes in the amount and localization of hTERT splice-variants. These data confirm the participation of EndoG in the alternative splicing of mRNA of the telomerase catalytic subunit and in regulation of the telomerase activity.
KEY WORDS: EndoG, telomerase, hTERT, alternative splicing, apoptosisDOI: 10.1134/S0006297917080041
Abbreviations: AS, alternative splicing; BSA, bovine serum albumin; hTERT, human telomerase reverse transcriptase; siRNA, small interfering RNA; TRAP, telomeric repeat amplification protocol.
Telomeres are DNA–protein complexes located on the ends of
chromosomes. Telomeres prevent the recognition of chromosome ends by
the DNA repair system as damaged regions [1]. Each
human chromosome ends by 15-20 kb of telomere repeats TTAGGG. The
length of telomere ends is shortened at each replication cycle. The
shortening of telomeres to critical values leads to activation of the
DNA repair system, inhibition of the cell division, transition of the
cells to the replicative aging state, and to activation of apoptotic
processes [2, 3]. In the
majority of tumor type cells, as well as in embryonic cells, germ
cells, stem cells, and activated lymphocytes, a multienzyme complex of
telomerase is functioning [4]. Telomerase
synthesizes telomere repeats on the chromosome ends, which allows the
cells to maintain the telomere length at a level that is sufficient for
unlimited proliferation [5]. The main components of
human telomerase are hTR, which includes a template for synthesis of
telomeric DNA, and hTERT that has reverse transcriptase activity and
can synthesize telomere repeats on the hTR template [6]. It is known that the telomerase activity is
regulated by the level of synthesis of the telomerase complex main
subunits [7] and by the alternative splicing (AS)
of its hTERT mRNA [8, 9].
Regulation of hTERT gene expression has been studied rather well
[10], whereas the regulation of telomerase during
the AS of its mRNA is studied insufficiently. At present, 22
splice-variants of hTERT are known, although only the full-size variant
of hTERT has catalytic activity [11, 12]. Two splice-variants constitute the major part of
the common hTERT mRNA. Deletion of 36 nucleotides in exon 6 (the
variant α–) leads to removal of a part of reverse
transcriptase domain A of the protein molecule and to loss of catalytic
activity. Deletion of 182 nucleotides in exons 7 and 8 (the variant
β–) leads to frameshift and appearance of a stop-codon in
exon 10, which results in synthesis of a shortened form of hTERT [13, 14]. This splice-variant of
hTERT functions as dominant negative [8]. We showed
earlier that the overexpression of apoptotic endonuclease EndoG is
involved in the AS of hTERT pre-mRNA [15]: it
causes a decrease in the amount of mRNA of the full-size
α+β+ variant and an increase in the amount of mRNA of the
α+β– splice-variant. Changes in the ratio of
splice-variants result in inhibition of the telomerase activity,
shortening of telomeres, transition of tumor cells to the replicative
aging state, and to the cell death through apoptosis [16, 17]. It is known that
normally EndoG is localized in mitochondria and is internalized into
the nucleus only during the development of apoptosis [18], whereas in most cases AS occurs in the cell
nuclei [19]. Therefore, the question of EndoG and
hTERT colocalization in the cell and of localization of AS of hTERT is
very interesting. The purpose of this work was to study the
intracellular localization, expression, and amount of mRNA of EndoG and
of hTERT splice-variants in the cell compartments in norm and during
apoptosis of tumor cells.
MATERIALS AND METHODS
Tissue culture, induction, and detection of apoptotic cells. Cells of human intestine tumor CaCo-2 (ATCC, USA) were cultured in medium RPMI-1640 (Gibco, USA) containing 10% embryonic calf serum (Gibco) at 37°C in an atmosphere with 5% CO2 and at 95% humidity. To induce apoptosis, the cells were cultured in the presence of cisplatin (Sigma, USA) at the concentration of 40 mM during 24 h. To determine the number of apoptotic cells, the tissue culture was treated with trypsin solution containing EDTA (Gibco), resuspended in phosphate-buffered saline (PBS; Gibco), and incubated with annexin V-FITC and propidium iodide from an FITC Annexin V/Dead Cell Apoptosis kit (Invitrogen, USA) according to the producer’s protocol. About 5·104 cells were analyzed using a FACSCalibur flow cytometer (Becton Dickinson, USA).
Suppression of EndoG gene expression. The expression of the EndoG gene was suppressed using small interfering RNAs (siRNAs) as described in the work by Apostolov et al. [20]. The cells were cultured to the state of 70-80% confluent monolayer and transfected with a pair of siRNAs (direct 5′-AUGCCUGGAACAACCUGGAdTdT-3′ and reverse 3′-UCCAGGUUGUUCCAGGCAUdTdT-5′; IDT, USA) or with a pair of control nonspecific siRNAs: Non-Targeting siRNA No. 1 (Dharmacon, USA) in serum-free medium containing 50 nM siRNA and a TransIT-TKO transfection reagent (Mirus, USA) for 48 h. The cells were washed and incubated with cisplatin for 24 h.
Isolation of cell organelles. Mitochondria were isolated as described in the work of Justo et al. [21]. The cells were homogenized on ice with an Ultra-Turrax T25 rotor homogenizer (IKA, USA) in 1 ml of buffer (220 mM D-mannitol, 70 mM sucrose, 20 mM Tris, 1 mM EDTA, pH 7.4) and centrifuged at 700g for 10 min at 4°C. The supernatant was centrifuged for 10 min at 1000g to precipitate the mitochondria. The quality of the mitochondria was assessed using a Cytochrome c Oxidase Assay Kit (Sigma) according to the producer’s protocol. The cytoplasm was prepared as described in the work of Laukova et al. [22]. The cells were homogenized on ice with the rotor homogenizer in 300 µl of buffer (10 mM Hepes, 10 mM KCl, 1.5 mM MgCl2, 0.1 mM EGTA, pH 7.4) and centrifuged at 2000g for 3 min at 4°C. The supernatant was taken as the cytoplasm fraction. The nuclei were isolated as described by Pravdenkova et al. [23]. The cells were homogenized on ice in 300 µl of buffer (50 mM Tris-HCl, pH 7.9, 0.25 M sucrose, 10 mM CaCl2), and then the suspension was centrifuged using an Optima XE ultracentrifuge (Beckman Coulter, USA) in a gradient solution (2 M sucrose, 50 mM Tris-HCl, 10 mM CaCl2, 5 mM β-mercaptoethanol) at 70,000g for 90 min. The nuclei were washed twice in TBE buffer (89 mM Tris-HCl, 89 mM H3BO3, 2 mM EDTA).
Real-time extraction of RNA and RT-PCR. Total RNA was isolated from the cells with an RNeasy Mini Kit (Qiagen, USA) according to the producer’s protocol. The reverse transcription (RT) and PCR were performed in real time as described in the work of Basnakian et al. [24]. To perform the RT, 5 µg of total RNA was placed in 25 µl of reaction mixture (Invitrogen). As the reaction mixture for real time PCR, we used Platinum SYBR Green qPCR Supermix-UDG (Invitrogen). The primer structures (Syntol, Russia) are presented in Table 1. Amplification was performed using the CFX96 Touch™ Real-Time PCR Detection System (Bio-Rad, USA) and a two-temperature reaction regimen (primer annealing/elongation). The quantity of amplificates was determined by fluorescence at the end of the elongation cycle. The amplification quality was assessed by analyzing the melting curve from 60 to 95°C at the reaction end (after the 35th cycle). Standard curves of PCR efficiency were obtained by serial dilutions (1 : 40, 1 : 80, 1 : 160, and 1 : 320) of total cDNAs. The relative concentration of RNA was calculated using the Qbase PLUS (Bio-Rad) program. Reference genes were as follows: 18S (the “housekeeping gene” for cytoplasm), Lamin B1 (the “housekeeping gene” for nuclei), and NADH dehydrogenase, subunit 4 (the “housekeeping gene” for mitochondria). Data are presented as normalization of mRNA levels of the studied genes using the averaged expression values of three reference genes. Amplification products were visualized in 2% agarose gel by staining with ethidium bromide and photographed with a ChemiDoc™ XRS imaging system (Bio-Rad).
Table 1. Primers used for real-time
RT-PCR

Western blotting. Cells or cellular organelles were lysed by ultrasonication in 1 ml of TBE buffer for 2 min at the power of 50 W using a Model 50 Sonic Dismembrator (Fisher Scientific, USA) and centrifuged for 10 min at 12,000g for removal of debris. The total protein concentration was determined in specimens by the method of Bradford [25] using a Bradford Protein Assay (Pierce, USA) with BSA for the calibration curve. The lysate (calculated per 50 µg total protein) was diluted in 50 mM Tris-HCl (pH 6.8) containing 1% SDS, 2 mM EDTA, 1% β-mercaptoethanol, and 7.5% glycerol, denatured by heating at 100°C for 10 min, and separated by electrophoresis in a gradient polyacrylamide gel [26] (100 V, 2 h) using NuPAGE® Novex® 4-12% Bis-Tris Protein Gels (Life Technologies, USA). Then the proteins were transferred onto a nitrocellulose membrane in Novex transferring buffer (Invitrogen) at 40 V during 3 h, and then the membranes were stained with Ponceau S (Sigma) as described by Hofnagel et al. [27]. Then the membranes were blocked with Blotting-Grade Blocker (Bio-Rad) and incubated for 2 h with primary monoclonal antibodies anti-GAPDH (glyceraldehyde-3-phosphate dehydrogenase, the reference protein for cytoplasm), anti-Lamin B1 (the reference protein for nuclei), anti-COX IV (cytochrome c oxidase IV, the reference protein for mitochondria), and anti-hTERT (all antibodies were from Abcam, USA) diluted 1 : 1000, or with polyclonal anti-EndoG (Millipore, USA) antibodies diluted 1 : 500. Then the primary antibodies were washed in PBS (pH 7.6) with 0.1% Tween-20, and the membranes were incubated with secondary antibodies conjugated with horseradish peroxidase (Cell Signaling, USA). For visualization, the SuperSignal chemiluminescent kit (Pierce Biotechnology, USA) was used with subsequent documentation in a ChemiDoc™ XRS imaging system (Bio-Rad). The protein was determined quantitatively by densitometry using the GelAnalyzer 2010a program.
Telomerase activity was determined using the TRAP method [5, 28]. The cells or cell organelles were lysed in buffer containing 10 mM Tris-HCl (pH 7.5), 1 mM MgCl2, 1 mM EGTA, 0.1 mM phenylmethylsulfonyl fluoride, 5 mM β-mercaptoethanol, 0.5% CHAPS, and 10% glycerol, taking 1 µl of the buffer per 103 cells. The lysates were centrifuged at 12,000g for 30 min at 4°C, and the supernatant was taken and stored at —80°C. The oligonucleotide substrate-primer was elongated with subsequent amplification in 30 µl of reaction mixture containing 67 mM Tris-HCl (pH 8.8), 16.6 mM (NH4)2SO4, 0.01% Tween 20, 1.5 mM MgCl2, 1 mM EGTA, 0.25 mM of each dNTP, 0.1 µg TS-primer (5′-AATCCGTCGAGCAGAGTT-3′), and 2 µl of the cell extract, diluted with the lysing buffer and equivalent to 2000 cells, and incubated at 37°C for 25 min. After incubation, the mixture was maintained for 10 min at 96°C for inactivation of telomerase. Then the mixture was supplemented with 0.1 µg CX-primer (5′-CCCTTACCCTTACCCTTACCCTAA-3′) and 2.5 units of Taq-polymerase. The reaction mixture was subjected to PCR as follows: 94°C – 2 min; 30 cycles: 94°C – 30 s, 50°C – 30 s, 72°C – 40 s; 72°C – 5 min. The amplification products were separated by PAGE in 12% nondenaturing polyacrylamide gel and TBE buffer. The specimens in volume of 10 µl were placed into the gel wells. The separated products were visualized with the ChemiDoc™ XRS imaging system after the gel was maintained for 30 min in a solution of the SYBR Green I dye (Invitrogen). Telomerase activity was assessed by densitometry of the TRAP results using the GelAnalyzer 2010a program.
Immunocytochemistry. Cells were cultured in 8-well chambers of a Nunc Lab-Tek Chamber Slide System (Thermo Scientific, USA), fixed with 10% formaldehyde (Sigma), and studied by methods of cytochemistry as described by Apostolov et al. [20]. The cells were incubated with rabbit antibodies to EndoG (Millipore, USA) and with goat antibodies to hTERT (Abcam, USA) diluted 1 : 200 in buffer BSA-T-PBS (0.5% BSA, 0.05% Tween 20, PBS). The primary antibodies were detected using secondary antibodies to rabbit antibodies IgG-AlexaFlour 647 (Invitrogen) or to goat antibodies IgG-AlexaFluor 594. For control, cells were used that were incubated only with the secondary antibodies. After the secondary antibodies were washed off, the cell nuclei were stained with 4,6-diamidino-2-phenylindole (DAPI). The cells were covered with Prolong Antifade kit cover glasses (Invitrogen) and analyzed by fluorescent microscopy with an Olympus IX-51 inverted microscope (USA) equipped with a Hamamatsu ORCA-ER monochromatic photo camera (Japan). The images were analyzed using the SlideBook 4.2 program. The average fluorescence intensity was measured in each channel. Red color was used for the signal from EndoG, green color was used for hTERT, and blue color for DAPI. For each cell-containing well, 20 images were analyzed. The intracellular colocalization and the pixel correlation of EndoG and hTERT were evaluated using the Pearson–Manders algorithm in the SlideBook 4.2 program [29].
Statistics. Results were analyzed statistically with Student’s t-test and presented as mean values ± standard deviation. Values are considered significant at p ≤ 0.05.
RESULTS
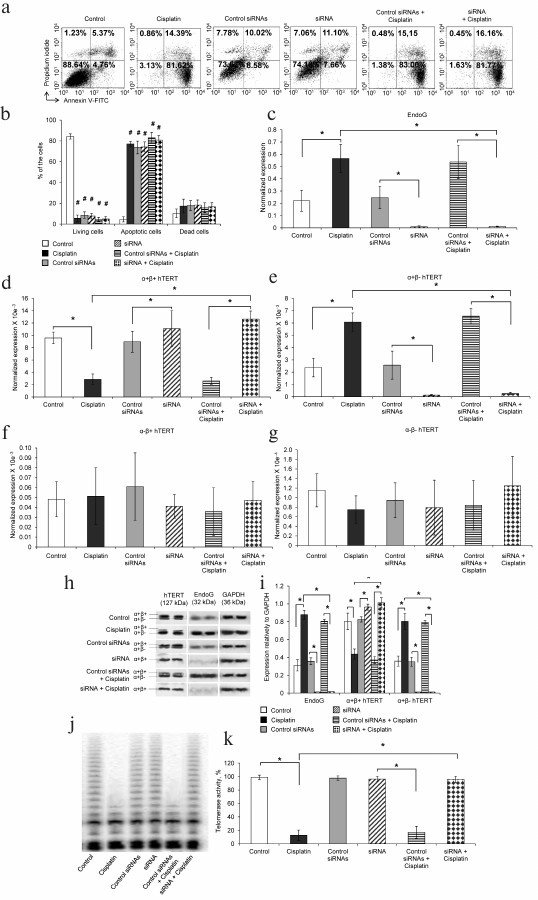
Changes in expression of hTERT splice-variants in apoptotic cells and under suppressed expression of EndoG. To study relations between expression of EndoG and hTERT splice-variants, the amounts of their mRNAs were evaluated in normal and apoptotic cells of human intestine cancer CaCo-2 and in normal and apoptotic cells with the suppressed expression of EndoG. Apoptosis was induced by incubation of the cells with cisplatin. The expression of EndoG was suppressed by RNA interference. Cell viability was assessed by flow cytometry on staining with propidium iodide and annexin V-FITC, and it was revealed that after 24 h of incubation with cisplatin or after transfection of the cells and subsequent incubation with cisplatin more than 81% of the cells were in the state of apoptosis (Fig. 1, a and b). There was no significant difference in the number of dead cells. Therefore, this population of cells was considered homogenous and consisting of apoptotic cells. In the group of cells transfected with siRNAs or with control siRNAs, the level of apoptotic cells was not higher than in the control group. Total RNA was isolated from the cells, and expression was studied by real-time RT-PCR. In the cells incubated with cisplatin, the expression of EndoG was significantly increased (Fig. 1c), the amount of mRNA of the full-size α+β+ splice-variant of hTERT was significantly decreased (Fig. 1d), and the amount of mRNA of the α+β– splice-variant of hTERT was significantly increased (Fig. 1e). Transfection of the cells with siRNAs induced suppression of the EndoG expression in both normal and apoptotic cells. The suppression of EndoG expression was accompanied by increase in the amount of mRNA of the hTERT α+β+ splice-variant, whereas mRNA of the hTERT α+β– splice-variant was virtually absent. Apoptosis induced by cisplatin, as well as suppression of EndoG expression, did not lead to changes in the amount of mRNA of α–β+ (Fig. 1f) and α–β– (Fig. 1g) splice-variants of hTERT.
Fig. 1. Changes in hTERT splice-variants and in telomerase activity during cisplatin-induced apoptosis of cells and suppression of the EndoG siRNA expression. a) Results of flow cytometry of CaCo-2 cells transfected with siRNA incubated with cisplatin and labeled with annexin V-FITC and propidium iodide. Shown are percent of living cells (lower left quadrants), apoptotic cells (lower right quadrants), and dead cells (upper quadrants); b) histograms of the living CaCo-2 cells, apoptotic cells, and dead cells incubated with cisplatin or transfected with siRNA; N = 4, # p ≤ 0.05 relative to the control cells. Levels of mRNA of (c) EndoG, (d) hTERT α+β+ splice-variant, (e) hTERT α+β– splice-variant, (f) hTERT α–β+ splice-variant, and (g) hTERT α–β– splice-variant measured by real-time RT-PCR. Levels of gene mRNAs are normalized relative to rRNA of the 18S gene; N = 4, * p ≤ 0.05; h) Western blotting of hTERT splice-forms, EndoG, and reference protein GAPDH in CaCo-2 cells incubated with cisplatin or transfected with siRNA; i) results of determination of amounts of hTERT splice-forms and EndoG relative to GAPD; * p ≤ 0.05; j) gel electrophoresis of TRAP cells incubated with cisplatin; k) results of determination of telomerase activity measured by the TRAP method; N = 4, * p ≤ 0.05.
We have studied the influence of apoptosis and, consequently, of changes in the ratio of hTERT splice-variants on telomerase activity. Western blotting with antibodies to the studied proteins revealed that in the apoptotic cells EndoG significantly increased, the full-size α+β+ form of hTERT decreased, and the hTERT α+β– form increased (Fig. 1, h and i). The suppression of EndoG expression was accompanied by an increase in the amount of the hTERT α+β+ form and by absence of synthesis of the hTERT α+β– form in both normal and apoptotic cells. We did not observe changes in the pool of proteins of the hTERT α–β+ and α–β– forms because their amount in the cell was too small and could not be detected by Western blotting.
Telomerase activity was analyzed in cell extracts by the TRAP method. Telomerase activity in apoptotic cells was significantly decreased (Fig. 1, j and k). These data are in a good agreement with results of measurement of the amount of mRNA of the hTERT splice-variants by real-time RT-PCR and of the amount of the hTERT splice-variants determined by Western blotting. The decrease in telomerase activity more likely was due to decrease in the amount of the hTERT full-size α+β+ form of hTERT, because just this form had the catalytic activity, and due to increase in the amount of the dominant negative α+β– form [8]. Suppression of EndoG expression and, therefore, absence of the hTERT α+β– form did not lead to inhibition of telomerase activity in the apoptotic cells. The transfection of cells with control siRNAs did not cause suppression of EndoG expression and failed to influence the change in the pool of hTERT splice-variants in both the normal and apoptotic cells.
Changes in intracellular localization of hTERT splice-variants during apoptosis and under conditions of suppressed expression of EndoG. It is known that telomerase displays enzymatic activity in the nucleus, synthesizing telomeric repeats [1], whereas the apoptotic endonuclease EndoG is normally localized in the intermembrane space of mitochondria and is internalized into the nucleus on development of apoptosis [18]. Alternative splicing of pre-mRNA of proteins is also localized in nuclei [19]. Using real-time RT-PCR, we determined amounts of mRNA of EndoG and of hTERT splice-variants in the cytoplasm and nuclei of normal cells, apoptotic cells, and in the organelles of cells with suppressed expression of EndoG. We did not observe a significant difference between amounts of EndoG mRNA in the cytoplasm and nuclei of the normal cells (Fig. 2a). The mRNA amount was significantly increased in both cytoplasm and nuclei of cells incubated with cisplatin. As expected, suppression of EndoG expression by RNA-interference caused a decrease in the amount of EndoG mRNA in the cytoplasm and nuclei of normal and apoptotic cells to the detection limit.
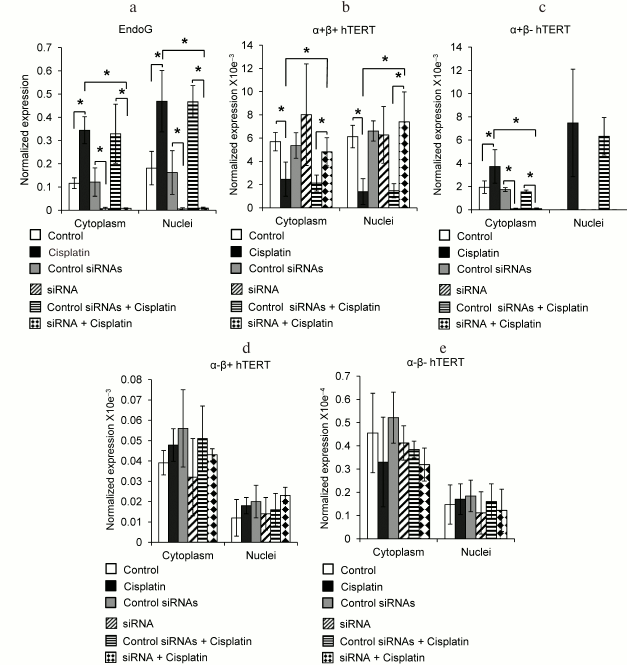
Fig. 2. Intracellular localization of mRNA of EndoG and of hTERT splice-variants during apoptosis and on suppression of the EndoG expression with siRNA. mRNA levels of (a) EndoG, (b) hTERT α+β+ splice-variant, (c) hTERT α+β– splice-variant, (d) hTERT α–β+ splice-variant, and (e) hTERT α–β– splice-variant measured by real-time RT-PCR. Levels of gene mRNAs are normalized with respect to mRNA levels of reference genes; N = 4, * p ≤ 0.05.
We did not find a significant difference between the levels of mRNA of the full-size hTERT α+β+ splice-variant in the cytoplasm and nuclei of normal cells (Fig. 2b). The significant decrease in the amount of mRNA of this splice-variant in both the cytoplasm and nuclei was found during the incubation of cells with cisplatin. Suppression of EndoG expression led to a significant increase in the amount of mRNA of the hTERT α+β+ splice-variant in the cytoplasm and nuclei of the apoptotic cells, whereas its increase in these compartments of normal cells was not significant (Fig. 2b).
Regardless of expectations, we failed to detect mRNA of the hTERT α+β– splice-variant in the nuclei of normal cells, whereas in the cytoplasm the level of this variant mRNA was rather high (Fig. 2c). Incubation of cells with cisplatin resulted in a significant increase in the amount of mRNA of the hTERT α+β– splice-variant in the cytoplasm and in appearance of this variant mRNA in the nucleus. Suppression of EndoG expression caused a significant decrease in the amount of mRNA of the hTERT α+β– splice-variant in the cytoplasm of normal and apoptotic cells. The amount of mRNA of the α+β– splice-variant in the nuclei of apoptotic cells with suppressed expression of EndoG was below the detection limits (Fig. 2c).
Induction of apoptosis did not lead to changes in the amounts of mRNA of the hTERT α–β+ (Fig. 2d) and α–β– (Fig. 2e) splice-variants in all organelles studied in normal cells and in cells with suppressed expression of EndoG. Transfection of cells with control siRNAs did not cause a decrease in the level of EndoG mRNA and failed to influence changes in the mRNA pool of the hTERT splice-variants in the cytoplasm and nuclei of normal and apoptotic cells.
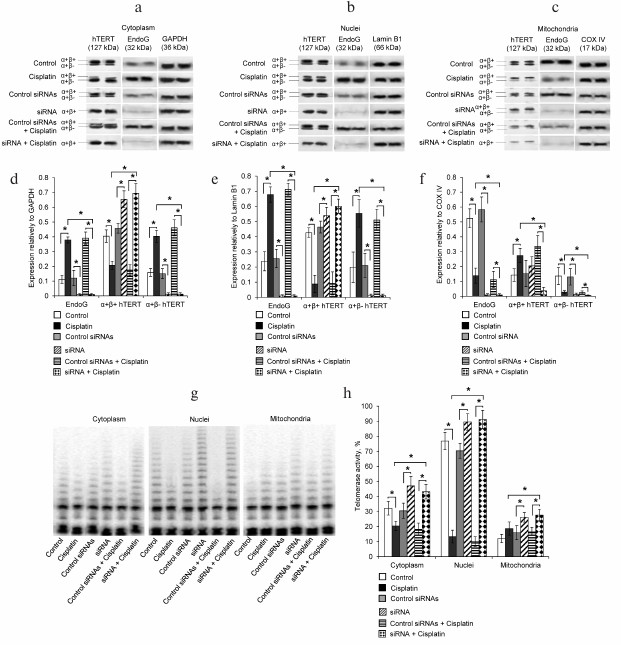
Decrease in telomerase activity in cell organelles with changes in the ratio of hTERT splice-variants during apoptosis and on suppression of EndoG expression. We tried to elucidate the influence of changes in the ratio of the hTERT splice-variants on telomerase activity in the cytoplasm, nuclei, and mitochondria. The Western blotting results revealed that in cytoplasm of apoptotic cells, the amount of the hTERT full-size α+β+ variant was significantly decreased, whereas the amount of the hTERT α+β– variant was increased (Fig. 3, a and d). These data are in correlation with the increase in EndoG in these cells and with results of determination of the mRNA amount by real-time RT-PCR. The study using the TRAP method revealed that telomerase activity in normal cell cytoplasm was about 30% of the total telomerase activity of the cell (Fig. 3, g and h). On the incubation of cells with cisplatin, telomerase activity in the cytoplasm significantly decreased to the level of 20%.
Fig. 3. Decrease in telomerase activity with increase in the amount of the hTERT α+β– splice-variant. a) Western blotting of hTERT splice-forms, EndoG, and of the reference GAPDH in cytoplasm of CaCo-2 cells incubated with cisplatin and on suppressed EndoG expression by siRNA; b) Western blotting of the hTERT splice-forms, EndoG, and of the reference protein Lamin B1 in nuclei of cells; c) Western blotting of the hTERT splice-forms, EndoG, and of the reference protein COX IV in mitochondria of cells. Results of determination of amounts of the hTERT splice-forms and EndoG with respect to the reference proteins in (d) cytoplasm, (e) nuclei, and (f) mitochondria of cells; * p ≤ 0.05; g) gel electrophoresis TRAP in organelles of cells incubated with cisplatin or transfected with siRNA; h) results of determination of telomerase activity in organelles of cells by the TRAP method; N = 4, * p ≤ 0.05.
Incubation of cells with cisplatin significantly increased the amount of EndoG in nuclei and decreased the amount of the hTERT full-size α+β+ splice-variant (Fig. 3, c and d). Although mRNA of the hTERT α+β– splice-variant was not localized in nuclei of normal cells, we detected its presence by Western blotting. Incubation of cells with cisplatin significantly decreased the amount of α+β+ and increased the amount of α+β– proteins of the hTERT splice-variant. Telomerase activity in nuclei of normal cells was about 75% of the total telomerase activity of the cell. On the incubation of cells with cisplatin, telomerase activity in the cytoplasm significantly decreased to the level of 15%.
The effect of cisplatin on the amounts of EndoG and hTERT splice-variants in mitochondria was the opposite. The development of apoptosis led to a significant decrease in the amount of EndoG, an increase in the amount of the full-size α+β+, and a decrease in the amount of the hTERT α+β– splice-variant (Fig. 3, e and f). Telomerase activity in mitochondria of normal cells was about 10% of the total telomerase activity of the cell. No significant changes in telomerase activity were observed in mitochondria on the incubation of the cells with cisplatin.
As expected, the suppression of EndoG expression by RNA interference led to the absence of this protein in the cytoplasm, nuclei, and mitochondria of normal and apoptotic cells (Fig. 3, a-f). The absence of EndoG was accompanied by an increase in the amount of the hTERT full-size α+β+ and by virtually full absence of the hTERT α+β– splice-variant in all studied cell compartments (Fig. 3, a-f). In apoptotic and normal cells with suppressed expression of EndoG, telomerase activity was significantly increased, which correlated with increase in the amount of mRNA of the hTERT α+β+ variant (Fig. 3, g and h).
Transfection of cells with control siRNAs did not decrease the level of mRNA of EndoG and did not influence changes in the pool of mRNA of the hTERT splice-variants in cytoplasm, nuclei, and mitochondria of normal and apoptotic cells.
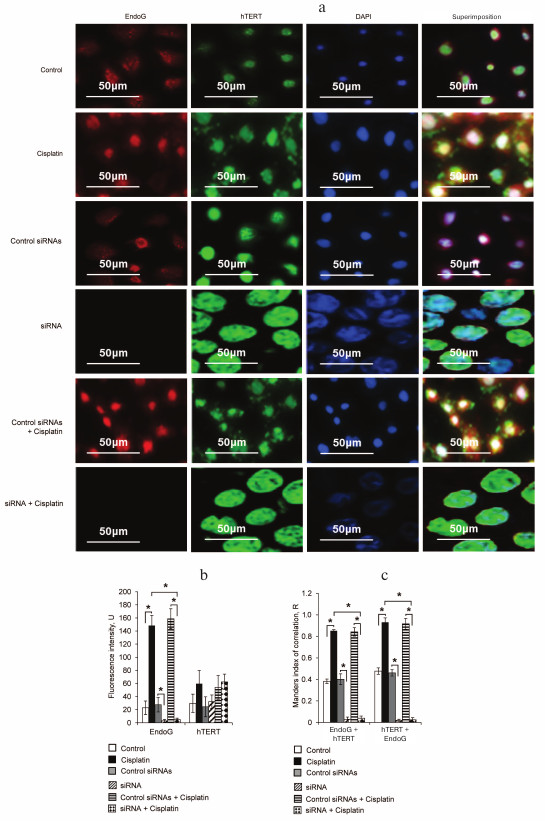
Endonuclease EndoG is colocalized with hTERT normally and during apoptosis. The intracellular localization of EndoG and hTERT in the normal and apoptotic cells was studied using fluorescence microscopy. Incubation of cells with cisplatin significantly increased the amount of EndoG, whereas the amount of hTERT was unchanged (Fig. 4, a and b).
Fig. 4. Increase in intracellular colocalization of EndoG and hTERT during apoptosis. a) Fluorescence microscopy of cells labeled with antibodies to EndoG, hTERT, and DAPI; b) results of quantitative determination of EndoG and hTERT by fluorescence intensity; N = 4, * p ≤ 0.05; c) Manders index of colocalization of EndoG with hTERT and of hTERT with EndoG; * p ≤ 0.05.
The intracellular colocalization of EndoG and hTERT was analyzed using two algorithms, those proposed by Pearson and by Manders. The results of the Manders test indicated a high degree of EndoG and hTERT colocalization in normal cells (Fig. 4c). The colocalization degree significantly increased in apoptotic cells on incubation with cisplatin. The pixel analysis using Pearson’s test also revealed a high degree of EndoG and hTERT colocalization in normal and apoptotic cells (Table 2).
Table 2. Pearson’s correlation
coefficients of EndoG and hTERT colocalization (N = 4)
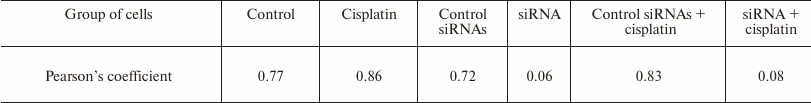
DISCUSSION
Most the hTERT splice-variants were identified rather long ago [30], but their biological function and mechanism of AS are studied insufficiently. It is known that some spice-variants function as dominant negative, which results in inhibition of telomerase activity [8, 31, 32]. We showed earlier that transfection of human intestine carcinoma cells with gene EndoG led to changes in the amount of mRNA of hTERT splice-variants [16, 17]. Changes in the ratio of hTERT splice-variants caused inhibition of telomerase activity, shortening of telomeres, and death of the cells. It is known that EndoG expression increases in response to damage of DNA [33]; moreover, during apoptosis EndoG is internalized into the cell nuclei from the intermembrane space of mitochondria [18]. To induce apoptosis, we incubated cells with cisplatin, and 24 h later we obtained a virtually homogenous population of apoptotic cells. In the apoptotic cells, EndoG expression was increased, which agreed with data of Yin et al. [33], the amount of the α+β– variant was increased, and the amount of the full-size α+β+ variant was decreased. Changes in the ratio of the hTERT splice-variants resulted in decrease in telomerase activity that was shown earlier by us [15, 17] and by other authors [8, 34, 35]. Then we followed changes in the amounts of mRNAs of EndoG and of hTERT splice-variants in cell organelles in the norm and during apoptosis. The increase in the amount of EndoG mRNA in the cytoplasm and nuclei of cells was a consequence of the damaging effect of cisplatin on their DNA. Changes in the amount of mRNA of the α+β+ and α+β– splice-variants in the cytoplasm and nuclei of the apoptotic cells were results of increased expression of EndoG.
In most cases, AS of mRNA is localized in the cell nucleus [19]. The absence of mRNA of the α+β– splice-variant in nuclei of normal cells was given special attention and is likely due to a rapid export of mRNA into the cytoplasm from the nucleus, to rapid degradation, or to AS localization outside the nucleus. Results of this work do not clearly answer why the α+β– splice-variant is absent from the nuclei of normal cells. We found mRNA of the α+β– splice-variant in nuclei of apoptotic cells. We supposed that EndoG should be involved in AS of hTERT mRNA and that the internalization of EndoG into the nucleus during apoptosis should lead to appearance of mRNA of the α+β– splice-variant. Determination of hTERT level in organelles of normal and apoptotic cells by Western blotting revealed that in the nuclei of normal cells, the hTERT α+β– splice-variant is present, and that its amount increases during apoptosis as expected. In mitochondria, normally both splice-variants of hTERT are found in approximately equal amounts. The presence of hTERT in mitochondria agrees with data of other authors [8, 36, 37]. During apoptosis, the amounts of the hTERT α+β– splice-variant and of EndoG are decreased. The decrease in the amount of EndoG in mitochondria and the increase in nuclei of apoptotic cells can be explained by translocation of EndoG into the nucleus under conditions of apoptosis [18]. The increase in α+β– amount and decrease in α+β+ amount of splice-variants of hTERT agree with the decrease in telomerase activity and with the increase in EndoG in nuclei and cytoplasm. We also found telomerase activity in mitochondria, which did not change during apoptosis.
To confirm the participation of EndoG in AS of hTERT mRNA, we suppressed EndoG expression by RNA interference. The absence of EndoG synthesis was accompanied by changes in the pool of hTERT splice-variants: amounts of both mRNA and proteins of the full-size α+β+ variant increased, whereas the amount of the α+β– splice-variant decreased. These changes were accompanied by an increase in telomerase activity in cytoplasm, nuclei, and mitochondria of normal and apoptotic cells.
Studies on the cellular localization of EndoG and hTERT by the cytochemistry method confirmed a high degree of their colocalization. The colocalization degree increased with development of apoptosis.
Apoptotic endonuclease EndoG is involved in the regulation of telomerase activity through induction of AS of hTERT. It has been shown in the present work that the increase in expression and amount of EndoG protein in cytoplasm, nuclei, and mitochondria leads to increase in the amount of mRNA of the inactive splice-variant of hTERT and to inhibition of telomerase activity. Induction of apoptosis on incubation of cells with cisplatin leads to changes in the intracellular localization of EndoG and, consequently, to increase in AS of hTERT and inhibition of telomerase activity in the cellular organelles. The high degree of intracellular colocalization of EndoG and hTERT indirectly confirms the involvement of EndoG in regulation of hTERT activity. Obviously, the participation of EndoG in AS of hTERT is a fundamental process that needs to be studied more deeply.
Acknowledgments
The work was supported by the State Academies Program of Basic Scientific Researches for 2013-2020.
REFERENCES
1.Blackburn, E. H. (2001) Switching and signaling at
the telomere, Cell, 106, 661-673.
2.Wright, W. E., Brasiskyte, D., Piatyszek, M. A.,
and Shay, J. W. (1996) Experimental elongation of telomeres extends the
lifespan of immortal x normal cell hybrids, EMBO J., 15,
1734-1741.
3.Harley, C. B. (1991) Telomere loss: mitotic clock
or genetic time bomb? Mutation Res., 256, 271-282.
4.Kaszubowska, L. (2008) Telomere shortening and
ageing of the immune system, J. Physiol. Pharmacol., 59,
169-186.
5.Kim, N. W., Piatyszek, M. A., Prowse, K. R.,
Harley, C. B., West, M. D., Ho, P. L., and Shay, J. W. (1994) Specific
association of human telomerase activity with immortal cells and
cancer, Science, 266, 2011-2015.
6.Blackburn, E. H. (2000) Telomere states and cell
fates, Nature, 408, 53-56.
7.Meyerson, M., Counter, C. M., Eaton, E. N.,
Ellisen, L. W., Steiner, P., Caddle, S. D., and Weinberg, R. A. (1997)
hEST2, the putative human telomerase catalytic subunit gene, is
up-regulated in tumor cells and during immortalization, Cell,
90, 785-795.
8.Listerman, I., Sun, J., Gazzaniga, F. S., Lukas, J.
L., and Blackburn, E. H. (2013) The major reverse
transcriptase-incompetent splice variant of the human telomerase
protein inhibits telomerase activity but protects from apoptosis,
Cancer Res., 73, 2817-2828.
9.Krams, M., Claviez, A., Heidorn, K., Krupp, G.,
Parwaresch, R., Harms, D., and Rudolph, P. (2001) Regulation of
telomerase activity by alternate splicing of human telomerase reverse
transcriptase mRNA in a subset of neuroblastomas, Am. J.
Pathol., 159, 1925-1932.
10.Daniel, M., Peek, G. W., and Tollefsbol, T. O.
(2012) Regulation of the human catalytic subunit of telomerase (hTERT),
Gene, 498, 135-146.
11.Saeboe-Larssen, S., Fossberg, E., and Gaudernack,
G. (2006) Characterization of novel alternative splicing sites in human
telomerase reverse transcriptase (hTERT): analysis of expression and
mutual correlation in mRNA isoforms from normal and tumour tissues,
BMC Mol. Biol., 7, 26.
12.Hrdlickova, R., Nehyba, J., and Bose, H. R.
(2012) Alternatively spliced telomerase reverse transcriptase variants
lacking telomerase activity stimulate cell proliferation, Mol. Cell.
Biol., 32, 4283-4296.
13.Ulaner, G. A., Hu, J. F., Vu, T. H., Giudice, L.
C., and Hoffman, A. R. (1998) Telomerase activity in human development
is regulated by human telomerase reverse transcriptase (hTERT)
transcription and by alternate splicing of hTERT transcripts, Cancer
Res., 58, 4168-4172.
14.Ulaner, G. A., Hu, J. F., Vu, T. H., Oruganti,
H., Giudice, L. C., and Hoffman, A. R. (2000) Regulation of telomerase
by alternate splicing of human telomerase reverse transcriptase (hTERT)
in normal and neoplastic ovary, endometrium and myometrium, Int. J.
Cancer, 85, 330-335.
15.Zhdanov, D. D., Vasina, D. A., Orlova, E. V.,
Orlova, V. S., Pokrovskaya, M. V., Aleksandrova, S. S., and
Sokolov, N. N. (2016) Apoptotic endonuclease EndoG regulates
alternative splicing of human telomerase catalytic subunit hTERT,
Biomed. Khim., 62, 544-554.
16.Zhdanov, D. D., Vasina, D. A., Orlova, V. S.,
Gotovtseva, V. Y., Bibikova, M. V., Pokrovsky, V. S., and Sokolov,
N. N. (2016) Apoptotic endonuclease EndoG induces alternative splicing
of telomerase catalytic subunit hTERT and death of tumor cells,
Biomed. Khim., 62, 239-250.
17.Vasina, D. A., Zhdanov, D. D., Orlova, E. V.,
Orlova, V. S., Pokrovskaya, M. V., Aleksandrova, S. S., and Sokolov, N.
N. (2017) Apoptotic endonuclease EndoG inhibits telomerase activity and
induces malignant transformation of human CD4+ T cells,
Biochemistry (Moscow), 82, 24-37.
18.Ohsato, T., Ishihara, N., Muta, T., Umeda, S.,
Ikeda, S., Mihara, K., and Kang, D. (2002) Mammalian mitochondrial
endonuclease G. Digestion of R-loops and localization in intermembrane
space, Eur. J. Biochem., 269, 5765-5770.
19.Montes, M., Becerra, S., Sanchez-Alvarez, M., and
Sune, C. (2012) Functional coupling of transcription and splicing,
Gene, 501, 104-117.
20.Apostolov, E. O., Ray, D., Alobuia, W. M.,
Mikhailova, M. V., Wang, X., Basnakian, A. G., and Shah, S. V.
(2011) Endonuclease G mediates endothelial cell death induced by
carbamylated LDL, Am. J. Physiol. Heart Circ. Physiol.,
300, 1997-2004.
21.Justo, R., Alcolea, M. P., Colom, B., Riera, A.
N., Gianotti, M., and Garcia-Palmer, F. J. (2002) Morphofunctional
changes in the mitochondrial subpopulations of conceptus tissues during
the placentation process, Cell. Mol. Life Sci., 59,
2199-2209.
22.Laukova, M., Alaluf, L. G., Serova, L. I.,
Arango, V., and Sabban, E. L. (2014) Early intervention with intranasal
NPY prevents single prolonged stress-triggered impairments in
hypothalamus and ventral hippocampus in male rats,
Endocrinology, 155, 3920-3933.
23.Pravdenkova, S. V., Basnakian, A. G., James, S.
J., and Andersen, B. J. (1996) DNA fragmentation and nuclear
endonuclease activity in rat brain after severe closed head injury,
Brain Res., 729, 151-155.
24.Basnakian, A. G., Apostolov, E. O., Yin, X.,
Abiri, S. O., Stewart, A. G., Singh, A. B., and Shah, S. V. (2006)
Endonuclease G promotes cell death of non-invasive human breast cancer
cells, Exp. Cell Res., 312, 4139-4149.
25.Bradford, M. M. (1976) A rapid and sensitive
method for the quantitation of microgram quantities of protein
utilizing the principle of protein–dye binding, Anal.
Biochem., 72, 248-254.
26.Laemmli, U. K. (1970) Cleavage of structural
proteins during the assembly of the head of bacteriophage T4,
Nature, 227, 680-685.
27.Hofnagel, O., Luechtenborg, B., Stolle, K.,
Lorkowski, S., Eschert, H., Plenz, G., and Robenek, H. (2004)
Proinflammatory cytokines regulate LOX-1 expression in vascular smooth
muscle cells, Arterioscler. Thromb. Vasc. Biol., 24,
1789-1795.
28.Kovalenko, N. A., Zhdanov, D. D., Bibikova, M.
V., and Gotovtseva, V. I. (2011) The influence of compound aITEL1296 on
telomerase activity and the growth of cancer cells, Biomed.
Khim., 57, 501-510.
29.Manders, E. M., Stap, J., Brakenhoff, G. J., van
Driel, R., and Aten, J. A. (1992) Dynamics of three-dimensional
replication patterns during the S-phase, analysed by double labelling
of DNA and confocal microscopy, J. Cell Sci., 103,
857-862.
30.Kilian, A., Bowtell, D. D., Abud, H. E., Hime, G.
R., Venter, D. J., Keese, P. K., and Jefferson, R. A. (1997) Isolation
of a candidate human telomerase catalytic subunit gene, which reveals
complex splicing patterns in different cell types, Hum. Mol.
Genet., 6, 2011-2019.
31.Colgin, L. M., Wilkinson, C., Englezou, A.,
Kilian, A., Robinson, M. O., and Reddel, R. R. (2000) The hTERTalpha
splice variant is a dominant negative inhibitor of telomerase activity,
Neoplasia, 2, 426-432.
32.Yi, X., White, D. M., Aisner, D. L., Baur, J. A.,
Wright, W. E., and Shay, J. W. (2000) An alternate splicing variant of
the human telomerase catalytic subunit inhibits telomerase activity,
Neoplasia, 2, 433-440.
33.Yin, X., Apostolov, E. O., Shah, S. V., Wang, X.,
Bogdanov, K. V., Buzder, T., and Basnakian, A. G. (2007) Induction of
renal endonuclease G by cisplatin is reduced in DNase I-deficient mice,
J. Am. Soc. Nephrol., 18, 2544-2553.
34.Lincz, L. F., Mudge, L.-M., Scorgie, F. E.,
Sakoff, J. A., Hamilton, C. S., and Seldon, M. (2008) Quantification of
hTERT splice variants in melanoma by SYBR green real-time polymerase
chain reaction indicates a negative regulatory role for the beta
deletion variant, Neoplasia, 10, 1131-1137.
35.Liu, Y., Wu, B., Zhong, H., Tian, X., and Fang,
W. (2012) Quantification of alternative splicing variants of human
telomerase reverse transcriptase and correlations with telomerase
activity in lung cancer, PLoS One, 7, e38868.
36.Eitan, E., Braverman, C., Tichon, A., Gitler, D.,
Hutchison, E. R., Mattson, M. P., and Priel, E. (2015) Excitotoxic and
radiation stress increase TERT levels in the mitochondria and cytosol
of cerebellar purkinje neurons, Cerebellum, 15,
509-517.
37.Li, P., Tong, Y., Yang, H., Zhou, S., Xiong, F.,
Huo, T., and Mao, M. (2014) Mitochondrial translocation of human
telomerase reverse transcriptase in cord blood mononuclear cells of
newborns with gestational diabetes mellitus mothers, Diabetes Res.
Clin. Pract., 103, 310-318.