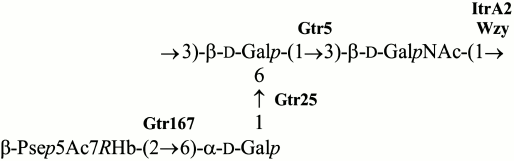Structure and Gene Cluster of the K93 Capsular Polysaccharide of Acinetobacter baumannii B11911 Containing 5-N-Acetyl-7-N-[(R)-3-hydroxybutanoyl]pseudaminic Acid
A. A. Kasimova1,2, M. M. Shneider3, N. P. Arbatsky1, A. V. Popova4,5, A. S. Shashkov1, K. A. Miroshnikov3, Veeraraghavan Balaji6, Indranil Biswas7, and Yu. A. Knirel1*
1Zelinsky Institute of Organic Chemistry, Russian Academy of Sciences, 119991 Moscow, Russia; E-mail: yknirel@gmail.com2Higher Chemical College of the Russian Academy of Sciences, Dmitry Mendeleev University of Chemical Technology of Russia, 125047 Moscow, Russia
3Shemyakin–Ovchinnikov Institute of Bioorganic Chemistry, Russian Academy of Sciences, 119997 Moscow, Russia; E-mail: mm_shn@mail.ru
4Moscow Institute of Physics and Technology, 141701 Dolgoprudny, Moscow Region, Russia; E-mail: popova_nastya86@mail.ru
5State Research Center for Applied Microbiology and Biotechnology, 142279 Obolensk, Moscow Region, Russia
6Department of Clinical Microbiology, Christian Medical College, Vellore, 632004 Tamil Nadu, India; E-mail: vbalaji@cmcvellore.ac.in
7Department of Microbiology, Molecular Genetics and Immunology, University of Kansas Medical Center, Kansas City, KS 66160, USA; E-mail: ibiswas@kumc.edu
* To whom correspondence should be addressed.
Received October 13, 2016; Revision received November 5, 2016
Capsular polysaccharide (CPS) assigned to the K93 type was isolated from the bacterium Acinetobacter baumannii B11911 and studied by sugar analysis along with one- and two-dimensional 1H and 13C NMR spectroscopy. The CPS was found to contain a derivative of pseudaminic acid, and the structure of the branched tetrasaccharide repeating unit was established. Genes in the KL93 capsule biosynthesis locus were annotated and found to be consistent with the CPS structure established. The K93 CPS has the α-d-Galp-(1→6)-β-d-Galp-(1→3)-d-GalpNAc trisaccharide fragment in common with the K14 CPS of Acinetobacter nosocomialis LUH 5541 and A. baumannii D46. It also shares the β-d-Galp-(1→3)-d-GalpNAc disaccharide fragment and the corresponding predicted Gal transferase Gtr5, as well as the initiating GalNAc-1-P transferase ItrA2, with a number of A. baumannii strains.
KEY WORDS: Acinetobacter baumannii, capsular polysaccharide structure, capsule biosynthesis locus, pseudaminic acid, glycosyltransferaseDOI: 10.1134/S0006297917040101
Abbreviations: COSY, correlation spectroscopy; CPS, capsular polysaccharide; HMBC, heteronuclear multiple-bond correlation; HSQC, heteronuclear single-quantum coherence; Pse, pseudaminic acid; RHb, (R)-3-hydroxybutanoyl; ROESY, rotating-frame nuclear Overhauser effect spectroscopy; TOCSY, total correlation spectroscopy.
Recently, Acinetobacter baumannii has become one of the most
widespread agents causing health-care associated infections. Their
treatment is complicated owing to the ability of the bacteria to
acquire and to accumulate various antibiotic resistance mechanisms [1]. Acinetobacter baumannii has a short-chain
lipooligosaccharide without any O-polysaccharide chain [2] but possesses a capsular polysaccharide (CPS),
which consists of many oligosaccharide repeats (K units). The CPS forms
a thick layer around the bacterial cell and protects A.
baumannii from the action of immune system components and
penetration of antimicrobial agents into the cell [3].
Acinetobacter baumannii strain B11911 was isolated from a patient with bloodstream infection in 2014 in Vellore, India. Its genome has been sequenced previously [4] (GenBank accession number NZ_LFYX00000000.1) and was found to contain a novel capsule biosynthesis cluster at the K locus. It was designated KL93, as suggested by J. J. Kenyon (personal communication), and, accordingly, the CPS of this strain was assigned to the K93 type. In this work, we established the structure of this CPS and assigned putative functions to the genes present in the KL93 locus.
MATERIALS AND METHODS
Cultivation of the bacterium and isolation of CPS. Acinetobacter baumannii B11911 was cultivated in 2× TY medium for 24 h. Cells were collected by centrifugation at 10,000g per 20 min, washed with a 7 : 3 (v/v) acetone–water mixture, and dried.
CPS was isolated by phenol–water extraction [5] of bacterial mass (450 mg), and then the crude extract was dialyzed without layer separation and freed from insoluble contaminations by centrifugation. Proteins and nucleic acids were removed by precipitation with aqueous 50% CCl3CO2H at 0°C; after centrifugation, the supernatant was dialyzed against distilled water and lyophilized.
CPS preparation (85 mg) was heated with aqueous 2% HOAc (100°C, 3 h), lipid precipitate was removed by centrifugation (13,000g per 20 min), and the purified CPS sample (33 mg) was isolated from the supernatant by gel-permeation chromatography on a column (56 × 2.6 cm) of Sephadex G-50 Superfine (Amersham Biosciences, Sweden) in 0.05 M pyridinium acetate buffer (pH 4.5) monitored using a differential refractometer (Knauer, Germany).
Composition analysis. A CPS sample (0.5 mg) was hydrolyzed with 2 M CF3CO2H (120°C, 2 h). Monosaccharides were analyzed by GLC of the alditol acetates on a Maestro (Agilent 7820) chromatograph (Interlab, Russia) equipped with a HP-5 column (0.32 mm × 30 m) using a temperature program of 160°C (1 min) to 290°C at 7°C/min. The absolute configurations of monosaccharides and 3-hydroxybutanoic acid were determined by GLC of the acetylated (S)-2-octyl glycosides [6] and trifluoroacetylated (S)-2-octyl ester as described [7].
NMR spectroscopy. Samples were deuterium-exchanged by lyophilization from 99.9% D2O and then examined as solutions in 99.95% D2O at 65°C using internal sodium 3-trimethylsilylpropanoate-2,2,3,3-d4 (δH 0.0, δC —1.6) as a reference for calibration. The 13C NMR spectra were recorded on a Bruker DRX-500 instrument, and two-dimensional NMR spectra were run on a Bruker Avance II 600 MHz spectrometer (Germany) using standard Bruker software. Relaxation delay was set to 3 s in the 13C NMR experiment. A spin-lock time of 60 ms and a mixing time of 150 ms were used in TOCSY and ROESY experiments, respectively. A 60-ms delay was applied for evolution of long-range coupling to optimize the 1H,13C HMBC experiment. Other NMR parameters were set as previously described [8]. The BrukerTopSpin 2.1 program was used to acquire and process the NMR data.
Bioinformatics. The K locus sequence for A. baumannii B11911 was obtained from GenBank (accession number NZ_LFYX00000000.1, base position range: 3339095 to 3366570). The sequence was annotated using the established nomenclature system [2], and novel genes were assigned names by J. J. Kenyon (personal communication). Protein functions were predicted using the BLASTp bioinformatics tool [9].
RESULTS AND DISCUSSION
Structure elucidation of the CPS. A crude CPS preparation was isolated from cells of A. baumannii B11911 by phenol–water extraction and freed from contaminations by heating under mild acidic conditions followed by gel-permeation chromatography on Sephadex G-50. The 1H NMR and 13C NMR spectra of the purified CPS sample compared to those of the crude preparation demonstrated that the acid treatment introduced no structural modification.
Sugar analysis of the CPS using GLC of the alditol acetates derived after full acid hydrolysis revealed Gal and GalN in the ratio 1 : 0.25 (detector response). Further studies showed that the CPS also contained a derivative of pseudaminic acid (Pse), namely 5-acetamido-3,5,7,9-tetradeoxy-7-(3-hydroxybutanoylamino)-l-glycero-l-manno-non-2-ulosonic acid (Pse5Ac7Hb). GLC analysis of the acetylated (S)-2-octyl glycosides indicated that Gal and GalN had the d-configuration. 3-Hydroxybutanoic acid was released by acid hydrolysis and identified as the R isomer by GLC of the trifluoroacetylated (S)-2-octyl ester.
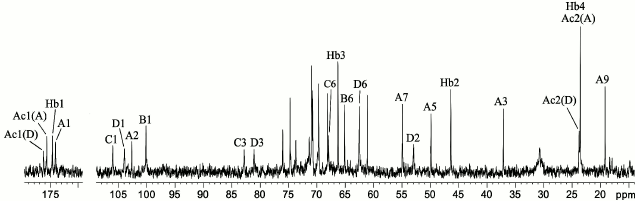
The 1H NMR spectrum showed signals for three anomeric protons at δ 4.48-4.97 (table), two C-CH2-C groups at δ 1.57, 2.50 (Pse H-3ax, H-3eq) and δ 2.35 (2 H, NHb H-2), two CH3-C groups at δ 1.22 (Pse H-9 and NHb H-4), and two NAc groups at δ 2.02 and 2.03. The 13C NMR spectrum of the CPS (Fig. 1) contained signals for four anomeric carbons at δ 100.1-106.0, two C-CH2-C groups at δ 37.1 (Pse C-3) and δ 46.4 (NHb C-2), four CH3-C groups at δ 19.2, 23.6 (2 C), and 23.8 (Pse C-9, NHb C-4, and 2 NAc C-2), three C-CH2O groups at δ 62.5, 65.1, and 67.9 (C-6 of GalN and 2 Gal, data of the attached proton test), one C-CHOH-C group at δ 66.3 (NHb C-3), three CH-NH groups at δ 49.9, 54.9, and 53.0 (Pse C-5 and C-7, GalN C-2), four CO groups at δ 174.1-176.2 (C-1 of Pse, NHb, and 2 NAc), and other sugar carbons at δ 68.1-82.7 (table).
1H and 13C NMR chemical shifts (δ, ppm) of
the K93 CPS of A. baumannii B11911
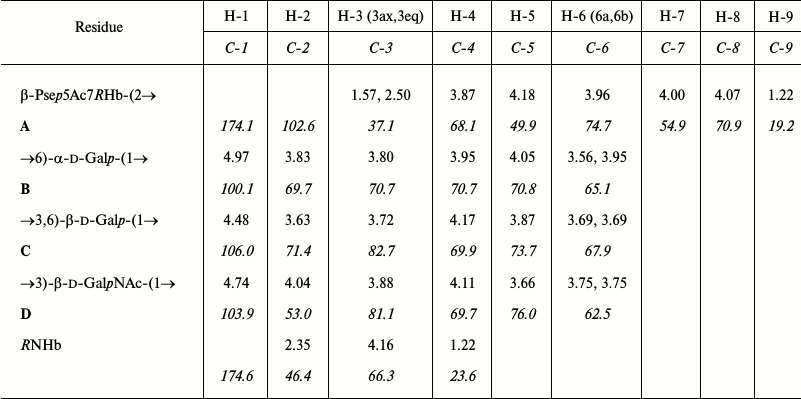
Note: 13C NMR chemical shifts are italicized. Chemical shifts
for the N-acetyl groups are δH 2.02 and 2.03,
δC 23.6 and 23.8 (both Me), 175.7 and 176.2 (both
CO).
Fig. 1. 13C NMR spectrum of the K93 CPS of A. baumannii B11911. Numbers refer to carbons in sugar residues denoted by letters as shown in the table.
These data together indicated that the CPS is composed of tetrasaccharide K units containing two d-Gal residues and one residue each of d-GalN and Pse. Two amino groups of the amino sugars are N-acetylated, and one is N-acylated with a (R)-3-hydroxybutanoyl group.
The 1H NMR spectrum of the CPS was assigned using two-dimensional shift-correlated 1H,1H COSY, 1H,1H TOCSY, and 1H,1H ROESY experiments. Spin-systems for four sugar residues were revealed, which were designated as units A (β-Psep), B (α-Galp), C (β-Galp), and D (β-GalpN), all being in the pyranose form (table). Particularly, the monosaccharides were identified by the following cross-peaks in the TOCSY spectrum: H-3ax/H-4,5, H-3eq/H-4,5, and H-9/H-8,7,6 for unit A, H-1/H-2,3,4 for units B, C, and D, combined with correlations between neighboring protons within each monosaccharide residues that were demonstrated by the COSY spectrum. H-1/H-5 correlations for units C and D in the 1H,1H ROESY spectrum (figure; see Supplement to this paper on the site of the journal (http://protein.bio.msu.ru/biokhimiya) and Springer site (Link.springer.com)) enabled assignment of the H-5 signals and defined the β configuration of these units. The α configuration of unit B was inferred by a relatively low 3J1,2 coupling constant of <3 Hz. With the 1H NMR spectrum assigned, the 13C NMR spectrum was assigned using two-dimensional 1H,13C HSQC and 1H,13C HMBC experiments (table).
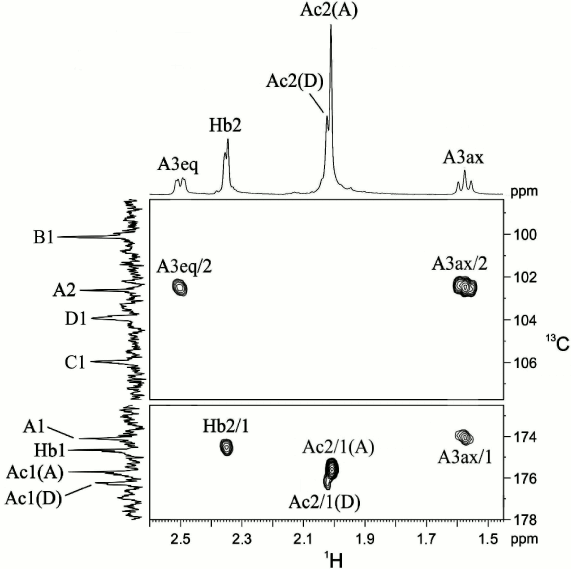
That unit A was a 3-deoxynonulosonic acid was confirmed by H-3ax/C-1, H-3ax/C-2, and H-3eq/C-2 correlations at δ 1.57/174.1, 1.57/102.6, and 2.50/102.6, respectively, in the 1H,13C HMBC spectrum (Fig. 2). The 1H and 13C NMR chemical shifts of unit A and 3JH,H coupling constants estimated from the two-dimensional NMR spectra were consistent with a non-substituted 5,7-diamino-3,5,7,9-tetradeoxy-l-glycero-β-l-manno-non-2-ulopyranosonic (pseudaminic) acid and differed from the spectral parameters of the other known stereoisomers [10-13]. Particularly, relatively large J3ax,4 (~12.5 Hz) and small J4,5 and J5,6 (<5 Hz) coupling constants indicated the axial orientation of H-4 and the equatorial orientation of H-5. Combined with the J6,7 coupling constant of ~10 Hz, these values suggested the l-glycero-l-manno configuration of unit A, which is thus a derivative of Pse. A relatively large difference between the H-3ax and H-3eq chemical shifts (0.93 ppm) indicated the axial orientation of the carboxyl group, i.e. the β configuration of Pse [10-13]. The absolute configuration of Pse was confirmed by analysis of the gene cluster for the CPS biosynthesis (see below).
Fig. 2. Part of a two-dimensional 1H,13C HMBC spectrum of the K93 CPS of A. baumannii B11911. The corresponding parts of the 1H and 13C NMR spectra are shown along the horizontal and vertical axes, respectively. Numbers before and after oblique stroke refer to protons and carbons, respectively, in sugar residues denoted by letters as shown in the table.
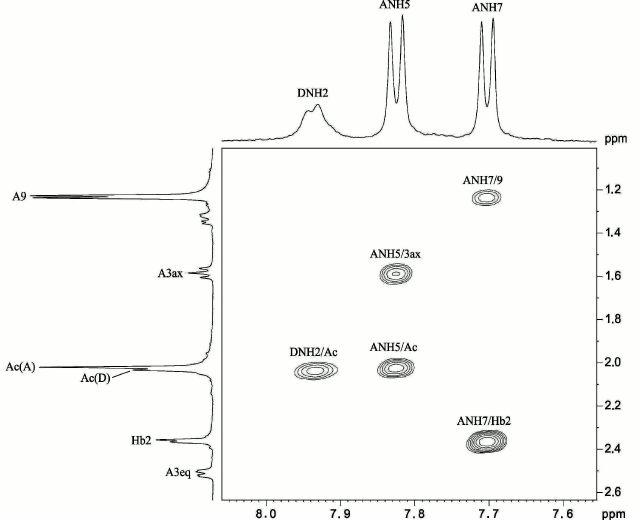
To determine positions of the N-acyl substituents, the NMR spectra of the CPS were measured in a 9 : 1 H2O–D2O mixture, which enabled detection of the nitrogen-linked protons. Signals for three NH protons were observed at δ 7.70, 7.82, and 7.93 and were assigned to Pse NH-7 and NH-5 and GalN NH-2, respectively, by correlations with CH protons of Pse in the 1H,1H TOCSY spectrum: NH-5/H-3ax,3eq,4,5 and NH-7/H-6,7,8,9. The 1H,1H ROESY spectrum (Fig. 3) showed correlations of GalN NH-2 and Pse NH-5 with the NAc groups at δ 7.93/2.03 and 7.82/2.02, respectively, and Pse NH-7 with H-2 of the NHb group at δ 7.70/2.35. The position of the NHb group was confirmed by a correlation between Pse H-7 and NHb C-1 (CO group) at δ 4.00/174.6 in the 1H,13C HMBC spectrum. Therefore, Pse5Ac7RHb was present in the CPS.
Fig. 3. Part of a two-dimensional 1H,1H ROESY spectrum of the K93 CPS of A. baumannii B11911 measured in 9 : 1 H2O–D2O mixture. The corresponding parts of the 1H NMR spectrum are shown along the axes. Numbers refer to protons in sugar residues denoted by letters as shown in the table.
Low-field positions of the signals for C-3 of unit D, C-3 and C-6 of unit C, and C-6 of unit B at δ 81.1, 82.7, 67.9, and 65.1, respectively, compared with their positions in the corresponding non-substituted monosaccharides at δ 72.4, 74.1 (both C-3), 62.4 and 62.2 (both C-6), respectively [14], showed that the CPS is branched and defined the glycosylation pattern in the K unit. The 1H,1H ROESY experiment (figure in Supplement) showed C H-1/D H-3, D H-1/C H-2,3,4, and B H-1/C H-6 interresidue correlations. The 1H,13C HMBC spectrum showed a correlation between C-2 of unit A (Pse) and H-6b of unit B at δ 102.6/3.95. These data were consistent with the 13C NMR chemical shift data of the linkage carbons and defined the monosaccharide sequence in the CPS.
Based on these data, we concluded that the K93 CPS of A. baumannii B11911 has the structure shown in Fig. 4. The CPS contains β-Pse5Ac7RHb that belongs to a class of higher acidic monosaccharides that are rather common in A. baumannii strains. Particularly, α-Pse5Ac7RHb has been found in K42 [15], α-Pse5Ac7Ac in K2 [16, 17] and K33 [18], and β-Pse5Ac7Ac in K6 [19]. The presence of genes for synthesis of a nucleotide precursor of Pse in the K locus of some other A. baumannii strains [17] suggests that there are more CPS K types that include a Pse derivative. Although, to our knowledge, the K93 CPS has a unique structure among known bacterial polysaccharides, it shares the α-d-Galp-(1→6)-β-d-Galp-(1→3)-d-GalpNAc trisaccharide fragment with K14 of A. nosocomialis LUH 5541 and A. baumannii D46 [20, 21]. CPS of a number of other A. baumannii strains, including K2 [16, 17], K6 [19], K27 [22], K33 [18], and K44 [22], have the β-d-Galp-(1→3)-d-GalpNAc disaccharide fragment in common with K93.
Fig. 4. Structure of the K93 CPS of A. baumannii B11911. Glycosyltransferases are shown next to the linkage each is assigned to.
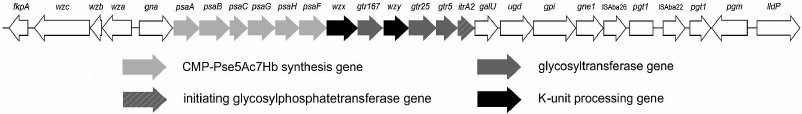
Characterization of the CPS biosynthesis gene locus. The gene locus for synthesis and export of the K93 CPS, KL93, has the common genetic organization for A. baumannii, with a central region containing genes for synthesis of a specific sugar component (Pse5Ac7RHb), glycosyltransferases for the K-unit assembly, Wzx translocase, and Wzy polymerase (Fig. 5). This CPS-specific portion is flanked by a block of wz genes for capsule export on one side and a block of genes for synthesis of common sugars on the other side [2]. In KL93, the latter is distinguished by the occurrence of an insertion sequence (IS) immediately adjacent to the pgt1 gene for phosphoglycerol transferase, which is interrupted by another IS (Fig. 5).
Fig. 5. Organization and content of the KL93 locus of A. baumannii B11911. The figure is drawn to scale from GenBank accession number NZ_LFYX00000000.1 (base position range: 3339095 to 3366570).
In KL93, there is a set of six psa genes that are close homologs of the genes involved with the synthesis of Pse5Ac7RHb, which are present in KL42 [15]. The same pseudaminic acid derivative is present in the K93 CPS, and hence the psa genes were assigned to its synthesis. Other monosaccharides found in K93 (Gal and GalNAc) are common sugars in A. baumannii and, accordingly, no other monosaccharide synthesis module is present in KL93.
KL93 encodes an initiating glycosylphosphate transferase having 99 and 97% sequence identity with ItrA2 of KL2 [17] and KL14 [21], respectively. This enzyme initiates the K-unit synthesis by transfer of d-GalpNAc-1-P from UDP-d-GalpNAc to the undecaprenol phosphate carrier in A. baumannii ATCC 17978 (KL3) [23], which is consistent with the presence of GalNAc in K93 CPS and shows that this monosaccharide is the first in the K unit. The Wzy polymerase would then catalyze the formation of the β-d-GalpNAc-(1→3)-d-Galp linkage between the K93 units.
In accordance with the presence of three internal linkages in the K93 unit, KL93 includes three glycosyltransferases. Two of them are 98 and 90% identical to the Gtr5 and Gtr25 predicted glycosyltransferases, which have been assigned to the β-d-Galp-(1→3)-d-GalpNAc and α-d-Galp-(1→6)-d-Galp linkages in the K14 CPS [21], respectively. Both linkages are present in the K93 unit, and the two Gtr proteins were named accordingly (Fig. 5). The remaining glycosyltransferase gene is evidently responsible for formation of the β-Psep5Ac7RHb-(2→6)-d-Galp linkage. It was named gtr167, as suggested by J. J. Kenyon (personal communication). The predicted Gtr167 protein is 42% identical and 62% similar to the Gtr16 inverting glycosyltransferase of A. baumannii RBH4 (KL6), which was assigned to the β-Psep5Ac7Ac-(2→4)-d-Galp linkage [19].
Therefore, the genes in the KL93 capsule biosynthesis locus of A. baumannii B11911 are consistent with the K93 CPS structure established.
Acknowledgements
We thank J. J. Kenyon for her help and critical reading of the manuscript.
This work was supported by the Russian Foundation for Basic Research (grant No. 14-04-00657).
REFERENCES
1.Roca, I., Espinal, P., Vila-Farres, X., and Vila,
J. (2012) The Acinetobacter baumannii oxymoron: commensal
hospital dweller turned pan-drug-resistant menace, Front.
Microbiol., 3, article 148.
2.Kenyon, J. J., and Hall, R. M. (2013) Variation in
the complex carbohydrate biosynthesis loci of Acinetobacter
baumannii genomes, PLoS One, 3, e62160.
3.Russo, T. A., Luke, N. R., Beanan, J. M., Olson,
R., Sauberan, S. L., MacDonald, U., Schultz, L. W., Umland, T. C., and
Campagnari, A. A. (2010) The K1 capsular polysaccharide of
Acinetobacter baumannii strain 307-0294 is a major virulence
factor, Infect. Immun., 78, 3993-4000.
4.Balaji, V., Rajenderan, S., Anandan, S., and
Biswas, I. (2015) Genome sequences of two multidrug-resistant
Acinetobacter baumannii clinical strains isolated from Southern
India, Genome Announc., 3, e01010-e01015.
5.Westphal, O., and Jann, K. (1965) Bacterial
lipopolysaccharides. Extraction with phenol–water and further
applications of the procedure, Methods Carbohydr. Chem.,
5, 83-91.
6.Leontein, K., and Lönngren, J. (1993)
Determination of the absolute configuration of sugars by gas-liquid
chromatography of their acetylated 2-octylglycosides, Methods
Carbohydr. Chem., 9, 87-89.
7.Shashkov, A. S., Paramonov, N. A., Veremeychenko,
S. N., Grosskurth, H., Zdorovenko, G. M., Knirel, Y. A., and Kochetkov,
N. K. (1998) Somatic antigens of pseudomonads: structure of the
O-specific polysaccharide of Pseudomonas fluorescens biovar B,
strain IMV 247, Carbohydr. Res., 306, 297-303.
8.Senchenkova, S. N., Shashkov, A. S., Popova, A. V.,
Shneider, M. M., Arbatsky, N. P., Miroshnikov, K. A., Volozhantsev, N.
V., and Knirel, Y. A. (2015) Structure elucidation of the capsular
polysaccharide of Acinetobacter baumannii AB5075 having the KL25
capsule biosynthesis locus, Carbohydr. Res., 408,
8-11.
9.Altschul, S. F., Gish, W., Miller, W., Myers, E.
W., and Lipman, D. J. (1990) Basic local alignment search tool, J.
Mol. Biol., 215, 403-410.
10.Knirel, Y. A., Shashkov, A. S., Tsvetkov, Y. E.,
Jansson, P.-E., and Zähringer, U. (2003)
5,7-Diamino-3,5,7,9-tetradeoxynon-2-ulosonic acids in bacterial
glycopolymers: chemistry and biochemistry, Adv. Carbohydr. Chem.
Biochem., 58, 371-417.
11.Knirel, Y. A., Kocharova, N. A., Shashkov, A. S.,
Dmitriev, B. A., Kochetkov, N. K., Stanislavsky, E. S., and Mashilova,
G. M. (1987) Somatic antigens of Pseudomonas aeruginosa. The
structure of the O-specific polysaccharide chains of the
lipopolysaccharide from P. aeruginosa O5 (Lanyi) and immunotype
6 (Fisher), Eur. J. Biochem., 163, 639-652.
12.Knirel, Y. A., Vinogradov, E. V., Shashkov, A.
S., Dmitriev, B. A., Kochetkov, N. K., Stanislavsky, E. S., and
Mashilova, G. M. (1986) Somatic antigens of Pseudomonas
aeruginosa. The structure of O-specific polysaccharide chains of
P. aeruginosa O10 (Lányi) lipopolysaccharides, Eur. J.
Biochem., 157, 129-138.
13.Knirel, Y. A., Vinogradov, E. V., Shashkov, A.
S., Dmitriev, B. A., Kochetkov, N. K., Stanislavsky, E. S., and
Mashilova, G. M. (1987) Somatic antigens of Pseudomonas
aeruginosa. The structure of the O-specific polysaccharide chain of
the lipopolysaccharide from P. aeruginosa O13 (Lányi),
Eur. J. Biochem., 163, 627-637.
14.Lipkind, G. M., Shashkov, A. S., Knirel, Y. A.,
Vinogradov, E. V., and Kochetkov, N. K. (1988) A computer-assisted
structural analysis of regular polysaccharides on the basis of
13C-n.m.r. data, Carbohydr. Res., 175,
59-75.
15.Senchenkova, S. N., Popova, A. V., Shashkov, A.
S., Shneider, M. M., Mei, Z., Arbatsky, N. P., Liu, B., Miroshnikov, K.
A., Volozhantsev, N. V., and Knirel, Y. A. (2015) Structure of a new
pseudaminic acid-containing capsular polysaccharide of Acinetobacter
baumannii LUH5550 having the KL42 capsule biosynthesis locus,
Carbohydr. Res., 407, 154-157.
16.Senchenkova, S. N., Shashkov, A. S., Shneider, M.
M., Arbatsky, N. P., Popova, A. V., Miroshnikov, K. A., Volozhantsev,
N. V., and Knirel, Y. A. (2014) Structure of the capsular
polysaccharide of Acinetobacter baumannii ACICU containing
di-N-acetylpseudaminic acid, Carbohydr. Res., 391,
89-92.
17.Kenyon, J. J., Marzaioli, A. M., Hall, R. M., and
De Castro, C. (2014) Structure of the K2 capsule associated with the
KL2 gene cluster of Acinetobacter baumannii,
Glycobiology, 24, 554-563.
18.Arbatsky, N. P., Shneider, M. M., Shashkov, A.
S., Popova, A. V., Miroshnikov, K. A., Volozhantsev, N. V., and Knirel,
Y. A. (2016) Structure of the N-acetylpseudaminic
acid-containing capsular polysaccharide of Acinetobacter
baumannii NIPH67, Russ. Chem. Bull., 588-591.
19.Kenyon, J. J., Marzaioli, A. M., Hall, R. M., and
De Castro, C. (2015) Structure of the K6 capsular polysaccharide from
Acinetobacter baumannii isolate RBH4, Carbohydr. Res.,
409, 30-35.
20.Haseley, S. R., and Wilkinson, S. G. (1996)
Structural studies of the putative O-specific polysaccharide of
Acinetobacter baumannii O11, Eur. J. Biochem.,
237, 266-271.
21.Kenyon, J. J., Hall, R. M., and De Castro, C.
(2015) Structural determination of the K14 capsular polysaccharide from
an ST25 Acinetobacter baumannii isolate, D46, Carbohydr.
Res., 417, 52-56.
22.Shashkov, A. S., Kenyon, J. J., Senchenkova, S.
N., Shneider, M. M., Popova, A. V., Arbatsky, N. P., Miroshnikov, K.
A., Volozhantsev, N. V., Hall, R. M., and Knirel, Y. A. (2016)
Acinetobacter baumannii K27 and K44 capsular polysaccharides
have the same K unit but different structures due to the presence of
distinct wzy genes in otherwise closely related K gene clusters,
Glycobiology, 26, 501-508.
23.Lees-Miller, R. G., Iwashkiw, J. A., Scott, N.
E., Seper, A., Vinogradov, E., Schild, S., and Feldman, M. F. (2103) A
common pathway for O-linked protein-glycosylation and synthesis of
capsule in Acinetobacter baumannii, Mol. Microbiol.,
89, 816-830.
Supplementary Figure (PDF)