REVIEW: Rpf Proteins Are the Factors of Reactivation of the Dormant Forms of Actinobacteria
V. D. Nikitushkin*, G. R. Demina, and A. S. Kaprelyants
Bach Institute of Biochemistry, Federal Research Center “Fundamentals of Biotechnology” of the Russian Academy of Sciences, 119071 Moscow, Russia; E-mail: vadimchemist@gmail.com* To whom correspondence should be addressed.
Received June 10, 2016; Revision received September 20, 2016
As the response to unfavorable growth conditions, nonsporulating mycobacteria transform into the dormant state with the concomitant formation of the specialized dormant forms characterized by low metabolic activity and resistance to antibiotics. Such dormant cells can be reactivated under the influence of several factors including proteins of Rpf (Resuscitation promoting factor) family, which possess peptidoglycan hydrolase activity and were considered to belong to the group of the autocrine growth factors of the bacteria. Remarkable interest toward Rpf family is determined by its participation in resuscitation of the dormant forms of Mycobacterium tuberculosis, what in turn is the key element in resuscitation of the latent tuberculosis – an infectious disease that affects one third of the World’s population. Experiments with Rpf mutant forms and with strains deleted in these proteins revealed a relationship between the enzymatic activity of this protein and its ability to resuscitate mycobacteria both in vitro and in vivo. This review discusses possible mechanisms of Rpf action including those related to possible participation of the products of mycobacterial Rpf-mediated cell wall hydrolysis (muropeptides) as signaling molecules. The unique ability of Rpf proteins to resuscitate the dormant forms of mycobacteria and to stimulate their proliferation would allow these proteins to occupy their niche in medicine – in diagnostics and in creation of antituberculosis subunit vaccines.
KEY WORDS: Resuscitation promoting factor (Rpf), actinobacteria, dormancy, tuberculosis, lytic enzymes, bacterial cell wall, muropeptides, MALDI, vaccines, diagnosticsDOI: 10.1134/S0006297916130095
Abbreviations: DAP, diaminopimelic acid; GlcNAc, N-acetylglucosamine; MALDI, matrix-assisted laser desorption/ionization; MurNAc, N-acetylmuramic acid; PG, peptidoglycan; RipA, resuscitation promoting factor interacting protein A; Rpf, resuscitation promoting factor (reactivation factor of dormant actinobacteria forms).
Bacteria occupy virtually all ecological niches on the planet, whereby
their unique ability to endure unfavorable environmental conditions is
an important factor of their survival, dissemination, and
pathogenicity. In addition to that, under unfavorable conditions
bacteria can enter a specific physiologic state – a state of
dormancy, which is characterized by low metabolic activity, changes in
morphology, thickening of the cell wall, and by the absence of division
on the typical nutrition media as well as inability to form colonies.
Since the cells in this state display poor metabolic activity, such
dormant bacteria may develop resistance to antibiotics and may
represent a source of chronic infection that is difficult to treat.
Notably, it is generally thought that latent tuberculosis is associated
with the ability of the tuberculosis causative agent Mycobacterium
tuberculosis to enter the specific physiological state – the
dormant state, similar to sporulation. It is supposed that dormant
M. tuberculosis can reactivate in the host tissues producing
actively dividing cells.
The dormant state in nonsporulating bacteria is often accompanied by the appearance of the state of “non-culturability” (NC), i.e. transient inability of the bacterial cells to form colonies on solid nutritional media. These cells however retain proliferative potential, i.e. the ability to grow on both liquid and solid media, which develops after the implementation of the resuscitation procedure in the liquid medium [1]. Studies on the gram-positive bacterium Micrococcus luteus belonging to the same order as mycobacteria Actinomycetales revealed that the bacteria grown on the poor nutrition media entered the dormant state, and 99.99% of these cells lost the ability to grow on solid nutrition media [2-4]. However, the cells could be resuscitated (their ability for division was recovered) by addition of a sterile filtered supernatant obtained from the culture of actively growing M. luteus cells [5, 6]. In subsequent studies, it was found that the M. luteus cells released into the cultural medium a protein capable of resuscitating dormant cells and of stimulating multiplication of the normal cells under certain conditions. This protein was also found to stimulate growth of some other (G+C)-rich gram-positive bacteria: Mycobacterium avium, Mycobacterium bovis (BCG), Mycobacterium kansasii, Mycobacterium smegmatis and M. tuberculosis [6]. This protein was active in picomolar concentrations and was named as a factor of reactivation of the dormant form of microorganisms, or Resuscitation promoting factor (Rpf) [7, 8].
Rpf secreted by M. luteus cells is a thermolabile trypsin-sensitive ~19-kDa protein [7]. Due to its activity in picomolar concentrations, Rpf was initially assigned to the group of bacterial cytokines [6], but relatively high concentration of Rpf in the cultural medium of M. luteus and the absence of hypothetical receptors to it on the cell surface failed to confirm this initial hypothesis. Full-genomic sequencing revealed that M. luteus contains only one copy of this protein [8]. The appearance of many sequenced genomes of various bacteria made clear that Rpf of M. luteus was a member of a large family of the secreted bacterial Rpf-like proteins among many gram-positive (G+C)-rich bacteria belonging to the phylum Actinobacteria. This phylum includes representatives of about 20 families from six orders (Actinomycetales, Streptomycetales, Corynebacteriales, Micrococcales, Pseodonocardiales, Propionobacteriales) including the representatives of Mycobacterium, Corynebacterium and Nocardia genera. The number of the rpf gene orthologs for each species varies from 1 to 5. All these proteins have in common a conservative domain with a similar amino acid sequence (about 75 amino acid residues). In the Fig. 1, the amino acid sequences of Rpf catalytic domains of different bacteria (M. luteus, M. tuberculosis and M. smegmatis) are aligned.
Fig. 1. Comparison of the amino acid sequences of the Rpf catalytic domains of actinobacteria: Micrococcus luteus, Mycobacterium tuberculosis, Mycobacterium smegmatis. Bioinformatic analysis was performed on the UniProt portal (www.uniprot.org).
Figure 2 presents a cladogram based on the similarity (>60%) of the Rpf conservative domain in different species of actinobacteria, which illustrates wide distribution of Rpf proteins in bacteria of various orders. There are also some reports on the presence of Rpf-like proteins among Firmicutes [9] as well as among gram-negative bacterium Salmonella typhimurium, although their amino acid sequences are significantly different from those of Rpf proteins in actinobacteria [10]. Wide distribution of the Rpf proteins and their important role in bacterial physiology have stimulated intense studies on this protein family. The current work is aimed to acquaint readers with modern views on the structure, biological functions, and application of the proteins from this family.
Fig. 2. Cladogram of the conservative Rpf domain distribution among different species of actinobacteria. Homology lower than 60% has not been considered. Bioinformatic analysis was performed on the UniProt portal (www.uniprot.org).
STRUCTURE OF THE Rpf PROTEINS
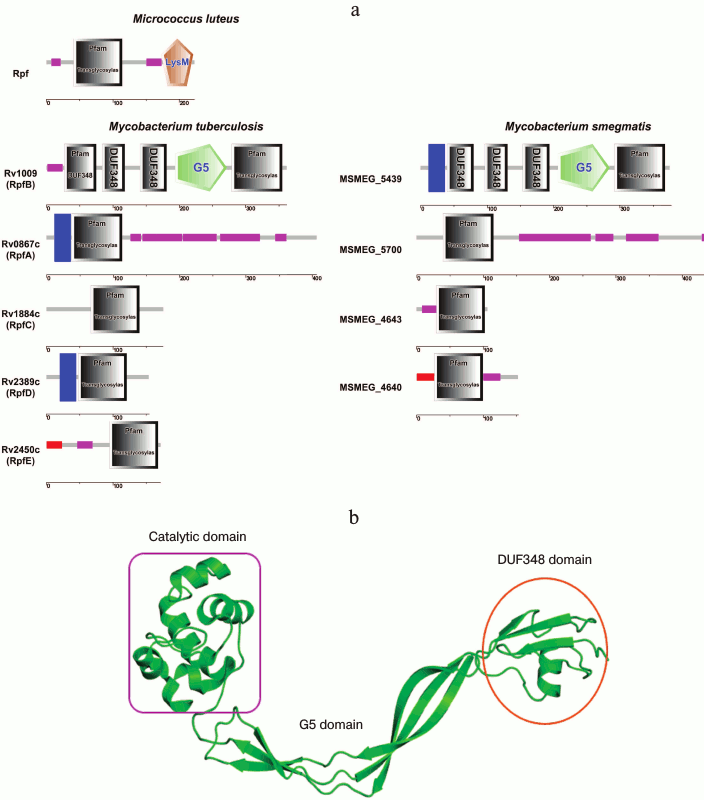
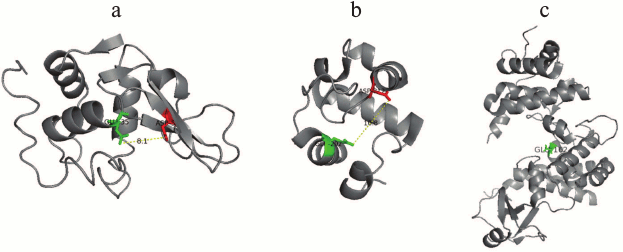
The studies on the Rpf structure revealed that the protein from M. luteus consists of a conservative domain located at the N-end of the molecule and the LysM domain (which is known to be a component of enzymes involved in cell wall metabolism) located at the C-end [7]. The LysM domain was established to participate in the enzyme interaction with peptidoglycan components [11]. Later on, the domain organization of the Rpf proteins for other representatives of the Mycobacterium genus was also confirmed (Fig. 3). As opposed to the M. luteus Rpf, mycobacterial Rpf proteins do not contain LysM domain, however alongside with catalytic domain the other intrinsic domains could be distinguished, such as DUF and G5 domains, as well as the domains responsible for binding of some proteins with the cytoplasmic membrane (Fig. 3a). According to the model of the Rpf protein tertiary structure, its conservative domain has spatial organization similar to the tertiary structure of lysozyme C. This has been found in study on the structure of the conservative domain RpfB from Mycobacterium tuberculosis by high resolution NMR [14, 15] and later confirmed by X-ray crystallographic analysis [16, 17]. The conservative domain of Rpf has six α-helices and one β-sheet composed of three strands spatially disposed identically to α-helical parts of lysozyme C and of some lytic enzymes involved in bacterial cell wall metabolism [14, 15]. According to the data on the crystalline structure studies, it has been established that RpfB can be considered a “mini-lysozyme” [16]. Similarly to lysozyme, the protein catalytic center consists of six peptidoglycan-binding sites (A-F) [16]. On the other hand, RpfB is also homologous to some lytic transglycosylases from E. coli (Slt35, Slt70) [13], what is shown in Fig. 4. Thus, structural studies suggest that the conservative domain of Rpf proteins may be considered as a hybrid of the well-known enzyme lysozyme (of animal origin) with the bacterial lytic transglycosylases. In turn, this structure implies that Rpf proteins should have enzymatic features.
Fig. 3. Domain organization of the Rpf products from mycobacteria M. tuberculosis and M. smegmatis. a) Schematic comparison of the Rpf proteins’ domains in M. luteus and mycobacteria (cytoplasmic membrane-binding domains are shown in blue; signaling sequences are shown in red; Pro/Ala-enriched regions are shown in violet). b) Spatial organization of the domains in the RpfB molecule (PDB 3EO5).
Fig. 4. Structures of lysozyme (PDB 1LZE), Rpf (PDB 3EO5), lytic transglycosylase Slt35 – E. coli (PDB 1D0K). Attention should be paid to the similarity in organization of the active center of these enzymes as well as to the spatial location of the Asp and Glu residues. Glu is common for all three cases.
In comparison with the rather simply organized structure of Rpf from M. luteus, RpfB protein from M. tuberculosis consisting of 362 a.a. has more complicated structure, i.e. besides the catalytic domain consisting of 75 a.a., it also contains additionally a G5 domain and three Domains of Unknown Function (DUF348) [18-20]. The G5 domain (it is named because of five conservative glycine residues in its structure) is an important component for many proteins involved in cell wall degradation and in formation of biological films; its functional role is close to that of the LysM domain of Rpf from M. luteus [18]. Detailed studies of the crystal structure of the ΔDUFRpfB+G5 protein established the accurate topology and mutual locations of the G5 β-strands allowing to ascribe this domain to the β-TH-β motif structure [16]. However, the most interesting observation was that this domain was adhesive to mycobacterial peptidoglycan, and its bended shape was favorable for sterically advantageous localization of the Rpf protein in peptidoglycan cells [16].
It is not so long ago that the structure and functions of three DUF348 domains in the RpfB structure were unknown. However, just recently this domain was shown to consist of one α-helix packed with four β-strands [19]; moreover, the spatial organization of this domain was found to be similar to that of the eukaryotic protein ubiquitin [19]. The role of DUF-domains in the Rpf proteins still remains unclear. It should be emphasized that the main feature of ubiquitin in eukaryotic cells is its ability to interact with a large numbers of macromolecules [19]. It seems that the existence of the ubiquitin-like domain in the Rpf molecule allows this protein to interact with other proteins, as a variant for such interaction may serve a partner of the Rpf protein – RipA (see below).
Thus, Rpf structural studies unambiguously evidence that these proteins belong to the class of peptidoglycan hydrolases suggesting that they could possess enzymatic activity that could be important for the observed physiological effects.
ENZYMATIC FEATURES OF THE Rpf PROTEINS
The assumption that the Rpf protein belongs to the class of hydrolytic enzymes which, similarly to lysozyme and lytic transglycosylases, cleave 1→4 glycosidic bonds between the residues of N-acetylglucosamine and N-acetyl(glycolyl)muramic acid of bacterial peptidoglycan [15, 23] can be tested experimentally.
The presence of enzymatic activity in the Rpf proteins was established both in experiments with the application of the artificial substrate 4-methylumbelliferyl-β-D-N,N′,N′′-acetylglucosamine (MUF-3-NAG), a synthetic analog of the cell wall component, and directly on isolated peptidoglycan. In both cases, the capability of the recombinant Rpf protein from M. luteus to hydrolyze peptidoglycan was shown [24, 25].
To confirm the enzymatic activity of Rpf proteins, site-directed mutagenesis was performed. It is well known that in the active center of lysozyme-like proteins there is a conservative residue of glutamic acid involved in catalysis. In lysozyme, the glutamate is in position 35 (Glu35), in lytic transglycosylase Slt35 it is in position 162, and in transglycosylase Slt70 it is in position 478 [23]. In Rpf proteins from M. luteus, the catalytic glutamate is supposed to be in position 54 [13].
It has been shown that substitution of the hypothetical catalytic glutamate in Rpf protein from M. luteus by glutamine (E54Q) resulted only in partial loss of activity (as also described in the case of lysozyme and lytic transglycosylases) [15, 26, 27], whereas substitution by alanine (E54A) or lysine (E54K) essentially suppressed the enzymatic activity [21]. The activity was inhibited maximally in a mutant Rpf with two substituted cysteine residues that could participate in formation of a functionally important intramolecular disulfide bond [13]. In fact, the substitution of either Cys53 or Cys114 separately resulted only in partial loss of the activity, whereas substitution of both amino acids caused virtually complete inactivation of Rpf [21]. Recent studies on the RpfC protein using NMR have shown the importance of disulfide bond formation between the conservative cysteine residues for supporting and modulating the catalytic domain conformation during catalysis [15]. These findings have confirmed the catalytic activity of Rpf proteins and their similarity with muramidases.
FUNCTIONAL FEATURES OF THE Rpf PROTEINS
In the earlier studies, it was shown that introduction of recombinant Rpf protein from M. luteus in picomolar concentrations stimulated the growth of M. luteus cells on a poor nutrition medium [6]. Later, a similar effect was found for M. smegmatis cells [25]. In general, in these and other experiments, it was found that the growth dependence of bacterial cells on exogenous Rpf was manifested only under exposure of the cells to the stress conditions, whereas the active cells grown on the corresponding balanced media did not display such sensitivity [7, 28]. Particularly, the pronounced effect of the recombinant Rpf A-D proteins from M. tuberculosis was manifested in stimulation of the bacterial growth during the lag phase at the low initial concentrations of the proteins. In this case, the cross-activity of the Rpf proteins from M. tuberculosis was observed toward M. luteus and M. smegmatis cells [26].
In a number of publications it has also been shown that the recombinant Rpf proteins were able to stimulate effectively resuscitation of the NC, dormant forms of M. tuberculosis, Rhodococcus rodochrous [27], and M. smegmatis [28] cells in vitro. The resuscitation ability of the Rpf protein from M. luteus was also shown on the dormant mycobacteria isolated from the MTB infected macrophages [29]. Exogenous addition of the recombinant Rpf protein could be replaced by hyperexpression of this protein in M. smegmatis strain (pAGR strain); such modified cells displayed the ability for spontaneous resuscitation from the nonculturable state without addition of the exogenous Rpf [28].
It was found that, synthesis of Rpf in M. luteus cells is maximal at the end of the logarithmic growth phase, and Rpf molecules can be found not only in the cultural medium but also on the surface of the cells [7, 9]. Rpf proteins have also been found in the cultural medium of M. tuberculosis, but their concentrations were significantly less in comparison with the concentrations produced by M. luteus cells [26].
The biological function of Rpf proteins seems to be associated with their necessity for growth of the producing bacteria. Synthesis of Rpf seems to be necessary for M. luteus cells division, since the mutant cells with an inactivated rpf gene appeared to be nonviable [7]. Antibodies to Rpf suppressed bacterial growth in vitro [7], what confirmed the essential role of the rpf genes for growth of the culture. As distinct from M. luteus null-mutants, the null-mutants in the rpf genes of M. tuberculosis were viable both in vitro and in vivo. Mutation in one of the genes encoding the synthesis of Rpf in M. tuberculosis did not influence resuscitation, did not stop cell growth, and did not result in bacterial death, however such mutations could cause changes in cell morphology [33, 34]. Deletion of the certain genes was accompanied with the slight increase in expression of the other genes. The fact, that the inactivation of individual rpf genes did not influence the resuscitation, demonstrates certain interchangeability of the Rpf proteins in the resuscitation process. However, concurrent mutations in the genes encoding RpfA/C/D and RpfB/D/E resulted in a significant decrease in the virulence of M. tuberculosis strains in mice and in the inability of these mutants for resuscitation from the dormant state in vivo [32]. Thus, M. tuberculosis displays redundancy of the rpf genes that is obviously responsible for the evolutionary resistance of this pathogen.
Mutant M. tuberculosis strains with the concurrently deleted four and completely knocked-out five rpf (A-E) genes were analyzed. These strains were studied under various conditions both in vitro and in vivo. Quadruple mutants could grow in vitro both in liquid and on solid media, whereas the quintuple mutant cells delayed in colony formation on solid medium [33]. Moreover, the studied mutants were incapable of resuscitation from the dormant state, but introduction of the rpf genes into such cells partially recovered this ability [35, 37]. On the mice model, the quadruple mutants suppressed division of the aerogenic chronic tuberculosis, and one of the mutants with the deleted rpf ACBE genes virtually completely lost the ability to grow in vivo and thus its virulence for animals [33]. The absence of both rpf genes in Corynebacterium glutamicum did not influence on the culture growth, however the repeated growth got impaired after long-term storage [34]. It should also be noted that one of the Rpf proteins (Rpf2) from C. glutamicum was glycosylated (the protein molecule contained mannose and galactose). However, there are no reports about the presence of glycosylated forms among mycobacterial Rpf proteins.
Although the Rpf proteins are dispensable for growth of M. tuberculosis, they determine the transition of the M. tuberculosis dormant forms to the active state both in vitro [33] and in vivo [37, 39] in the model of chronic TB on animals. The expression of all five M. tuberculosis H37Rv rpf genes was studied by RT-PCR during different growth stages (from the exponential phase to the dormancy state with the subsequent Rpf-mediated resuscitation). Analysis of the MTB rpf genes revealed that all of them (rpfA, rpfB, rpfC, rpfD, rpfE) were differently expressed at the early exponential phase [31]. Exponentially growing cells retained the high expression of rpfB only [34, 40], whereas the data on expression of the other rpf genes are differing [34, 40]. During the stationary growth phase, general tendency for decrease in the expression of all M. tuberculosis rpf genes was observed , what is similar to the rpf expression from M. luteus (except for the rpfC gene) [36].
Analysis of the changes in rpf gene expression during resuscitation of the NC M. smegmatis cells revealed that only RpfA (MSMEG_5700) homolog manifested an elevated expression during the late resuscitation stages before the beginning of cell division [37].
The expression levels of rpf genes were analyzed under various stress conditions (starvation, hypoxia, lowering pH values). During starvation (incubation for 24 and 96 h in phosphate buffer at pH 7.2), expression of all rpf genes was increased after 24 h comparing with the control (the cells before the exposure to the stress conditions), and further incubation up to 96 h resulted in a significant decrease in the expression of rpfA, rpfB and rpfE, whereas the expressions of rpfC and rpfD were unchanged. Under hypoxic conditions, the expression of rpfC and rpfE increased. As a response to pH lowering to the values of 5.5-4.5, the expression of rpfD and rpfE increased. The transcription analysis data revealed that proteins encoded by rpf genes are being expressed differently depending on conditions and growth phases; therefore, they could be regulated separately. Thus, the biosynthesis of RpfA is regulated by cAMP-dependent transcriptional receptor protein (CRP) Rv3676, which is responsible for regulation of the expression of the genes involved in the persistence of MTB and/or resuscitation from the dormant state [38]; RpfC was positively regulated by the sigma-factor SigD [39] and negatively regulated by the two-component regulatory system MprAB influencing many genes in the SigD regulon [43-45].
It seems that Rpf orthologs do not functionally differ in M. tuberculosis, but their expression is regulated differently, what may indicate on “association” of different Rpf proteins with different stress factors of the environment.
MECHANISM OF ACTION
Data presented in the previous sections (“Structure of the Rpf proteins” and “Enzymatic Features of the Rpf Proteins”) indicate that Rpf proteins possess enzymatic activity and therefore, we supposed that the enzymatic hydrolysis of peptidoglycan under the influence of Rpf could be important for stimulation of growth and resuscitation of the dormant mycobacteria. In this regard, the question of how two processes – the hydrolysis of the cell wall peptidoglycan and the resuscitation process – can be related arises. The relation of these two processes has been shown in the experiments on resuscitation of the dormant M. smegmatis cells transformed by the plasmid pAG carrying the rpf gene (strain pAGR) and the rpf mutant genes (mutant AGX strains). The findings were very similar to the data on the enzymatic activity of the recombinant proteins carrying the corresponding amino acid substitutions (see “Enzymatic Features of the Rpf Proteins” section). Thus, the substitution of glutamate by glutamine (E54Q) in the active center of the RpfC protein from M. tuberculosis (Rv1884c) led only to a slight decrease in resuscitation, whereas substitution of glutamate by alanine and especially by lysine resulted in sharp decrease in resuscitation. The substitution of each cysteine (C53K and C114T) separately resulted in a partial decrease in resuscitation, whereas the substitution of both cysteines completely suppressed the resuscitating ability of the protein. Thus, in addition to the glutamate important for catalysis, the stabilizing function of two cysteine residues in the molecule is also crucial for resuscitation [24, 25]. These results directly confirm the relation between the enzymatic activity of the Rpf proteins and resuscitation of the dormant NC bacteria.
Nevertheless, the question about the nature of the relationship between the hydrolytic and resuscitating abilities of Rpf was unclear for a long time. In our opinion, three hypotheses for mechanism of action of the Rpf protein are worth being discussed:
1) Rpf may act as a hydrolase causing restricted hydrolysis of the modified peptidoglycan in the dormant cells and thus promoting synthesis and growth of the cell wall and stimulating the beginning of division process in the NC cells.
Peptidoglycan of mycobacteria consists of the interchangingly bound residues of N-acetylglucosamine and muramic acid (either N-acetyl or N-glycolyl) connected with pentapeptide side chains [46-49]. On transition into the dormant state, the number of intermolecular bonds in the structure of peptidoglycan increases and PG becomes denser and thicker [46, 47, 50, 51]. At the same time, mycobacterial peptidoglycan of the actively growing cells is a very dynamical structure that is constantly increasing and being remodeled during growth [48, 52]. Therefore, a restricted hydrolysis of peptidoglycan can be critical for starting cell division. However, although being the target for exogenous Rpf, peptidoglycan is located under the outer mycobacterial membrane layer [44], and therefore its availability for exogenous Rpf in the dormant cells is problematic. However, endogenous Rpf may adopt this function. This hypothesis has been partially confirmed by the fact that expression of the Rpf molecules was observed not at the beginning of resuscitation, but somewhat later when transcriptional and translational activities appear in the cells, i.e. immediately before the beginning of cell division [45]. This also contradicts the initial hypothesis that Rpf is a cytokine responsible for triggering early stage of reactivation process [6].
2) The second hypothesis about the Rpf action mechanism is based on the ability of recombinant Rpf to disperse bacterial aggregations, which seems to be due to its hydrolytic activity [46]. It was found earlier that aggregation of M. luteus cells was important for initiation of the growth of the culture at the lag-phase [47] as well as during the initial steps of reactivation of M. tuberculosis [48]. It seems that Rpf proteins can participate in dispersion of these aggregates before the beginning of cell division.
3) Finally, according to the “signaling hypothesis”, in the process of PG hydrolysis, Rpf can release low molecular weight molecules transmitting the signal onto the cell as it is and onto the neighboring cells acting on a surface cellular receptor.
As discussed above, Rpf proteins hydrolyze 1→4 glycosidic bonds of the bacterial peptidoglycan. However, up to the present it was not clear what products are generated during the hydrolysis, since it was unknown to which group of the enzymes Rpf should be ascribed – to the group of lysozymes or to the group of lytic transglycosylases. The known peptidoglycan hydrolases are classified based on their specificity to cleave the structural sites of bacterial peptidoglycan [42]: thus, N-acetylmuramyl-L-alanine amidase hydrolyzes the amide bond between N-acetylmuramic acid and L-alanine releasing the glycan chain from the peptide stem, whereas carboxy- and endopeptidases destroy LD-, DD-, and DL-bonds in the peptide chain. Three types of glycosidases that cleave the glycan strands by residues of N-acetylmuramic acid and N-acetylglucosamine have been described so far: N-acetylglucosaminidase, lysozymes, and lytic transglycosylases. The latter two enzymes destroy one β-(1→4)-glycosidic bond of glycan; therefore, lysozymes and lytic transglycosylases are termed together as N-acetyl-β-D-muramidases. It should also be noted that hydrolases are specific toward a glycan type, in particular, toward the presence or absence of glycan modifications, the number of cross-links, etc. [23, 48].
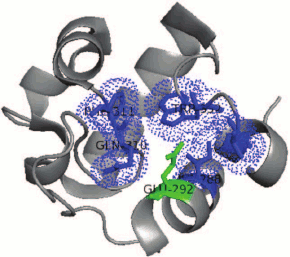
Detailed comparison of the structure of catalytic centers of proteins reveals a conservative residue of glutamic acid in RpfB from M. tuberculosis (Glu292 RpfB corresponds to Glu35 of lysozyme C). The catalytic glutamate of RpfB is surrounded by a “hydrophobic pocket” similarly to lysozyme. The hydrophobic surrounding is created by the side chain of amino acids Ile288, Gln310, Phe311, Trp352, and Val354, which stabilize the protonated uncharged state of Glu292 at the optimum of the enzyme activity (pH 5.0) that makes glutamate to serve a “strong acid” during the catalysis (Fig. 5). Recently, it has been shown that a similar structure is characteristic for both the hydrophobic pocket and RpfC, which may indicate on a common organization and structure of all the Rpf proteins [15]. It should be noted that RpfB, as opposed to lysozyme, does not have an equivalent of Asp52 from C-type lysozyme (in its place Tyr305 is located in RpfB). Therefore, it has been supposed that protein Rpf functionally belongs to lytic transhydrolases, i.e. the group of enzymes cleaving bacterial peptidoglycan with the release of anhydro-products of the reaction [57, 58]. However, the experimental proofs of formation of these products were obtained later (see below).
Fig. 5. Organization of the “hydrophobic pocket” of the RpfB protein catalytic center. Hydrophobic surrounding is created by the side chains of the amino acids Ile288, Gln310, Phe311, Trp352, and Val354 (blue) that stabilize the protonated uncharged state of Glu292 (green).
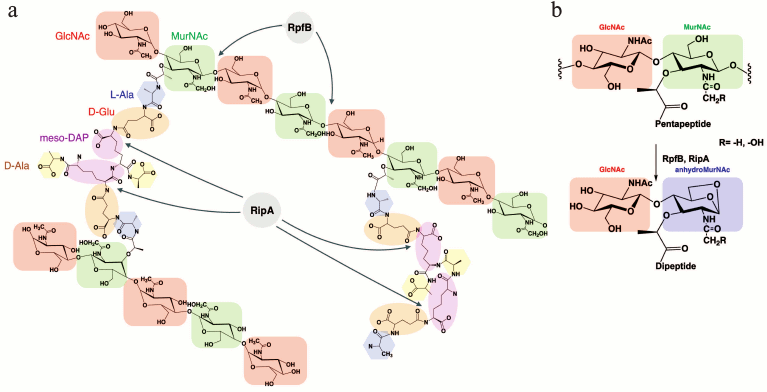
There are many enzymes involved in synthesis and degradation of peptidoglycan that function in combination with other enzymes. By means of screening in a yeast two-hybrid system it has been found that RpfB of M. tuberculosis had a partner — endopeptidase RipA (Resuscitation promoting factor interacting protein) [59-61]. RipA, or L,D-endopeptidase, is a proteolytic enzyme capable of cleaving peptide bonds within the peptide chain of peptidoglycan (it hydrolyzes D-glutamine-meso-diaminopimelic acid to isoglutamine (D-iGln) and meso-diaminopimelic acid (m-DAP)). RipA can also interact with RpfE [51]. RipA is a secreted protein that has been detected in many mycobacteria including pathogenic forms and it is also found among Rhodococcus and Corynebacterineae. The catalytic domain of RipA contains in its “core” 70 amino acids (385-445) that have the 35% similarity with a cysteine protease of the NlpC/P60 family. The localization of the two proteins (RpfB and RipA) on the septum of the dividing cells suggests that the mode of action of these enzymes in process of cell division is coordinated [59-63]. It was also shown that deletion of the gene encoding RipA leads to decrease of the growth rate and to anomalous morphology of the M. tuberculosis and M. smegmatis cells (branching and formation of chains). Formation of the long branched chains could be explained due to the incomplete formation of the septum, which increases cell sensitivity to β-lactam antibiotics. Therefore, RipA is supposed to play an important role during the final stage of the cell division. Since RpfB can hydrolyze the glycoside bond between the residues of N-acetylglucosamine (GlcNAc) and N-acetyl(glycolyl)muramic acid and the endopeptidase RipA is active at the D-Glu-meso-DAP sites of the peptide chain of the mycobacterial peptidoglycan, their interaction should result in a synergistic hydrolysis of the bacterial cell wall (Fig. 6), what has been shown experimentally [54].
Fig. 6. Structural organization of the mycobacterial peptidoglycan and its hydrolysis. a) The hydrolysis sites of the mycobacterial peptidoglycan under the influence of RpfB and RipA. b) Expected product of hydrolysis – anhydro-disaccharide-dipeptide.
It has also been experimentally shown that the combined effect of RpfB and RipA proteins on the hydrolysis of M. smegmatis fluorescently labeled peptidoglycan was stronger than the effects of individual RpfB and RipA proteins [64, 65]. Similarly, addition of both proteins also demonstrated synergistic effect in the procedure of reactivation of the dormant NC M. smegmatis cells [55].
Theoretically, one could suggest that the combined action of RpfB and RipA could result in release of the disaccharide-peptide [55]. Application of the method of mass-spectrometry (MALDI-TOF) allowed us to establish the structure of the product of mycobacterial peptidoglycan synergistic hydrolysis under the influence of two enzymes, RpfB and RipA (Fig. 6b). As it turned out, the main product of the enzymatic reaction appeared to be N-acetylglucosaminyl-β(1→4)-N-glycolyl-1,6-anhydromuramoyl-L-alanyl-D-isoglutamate (anhydroMurNAc), what confirmed the hypothesis of the protein Rpf belonging to the class of lytic transglycosylases [55].
Detection of the low molecular weight products generated under the influence of Rpf and Rip combination suggested us to consider anhydromuropeptides as potential intermediate signaling molecules. In fact, application of a synthetic analog (N-acetylglucosaminyl-β(1→4)-N-acetyl-1,6-anhydromuramoyl-L-alanyl-D-glutamate) on resuscitation assays confirmed the ability of this compound (in concentrations of about 100 ng/ml) to stimulate the resuscitation of NC mycobacteria cells [55].
Muropeptides are considered as potential signaling molecules involved in organization of molecular “parasite–host” cascades [43]. Recently, in the work of Shah et al. [56], muropeptides releasing in the process of mutanolysine-induced hydrolysis of peptidoglycan were shown to play a role of signaling molecules in germination of Bacillus subtilis endospores. According to the scheme proposed by the authors, muropeptides interacted with a specific exogenous receptor – PASTA domain of the membrane serine-threonine protein kinase PrkC; muropeptides activated it thus inducing subsequent phosphorylation of regulatory cellular proteins [56]. Only meso-DAP-containing muropeptides released during the active growth of the bacteria promoted germination of B. subtilis spores [60]. Noteworthy that prkC gene controls an expression of a supposed homolog of the Rpf protein, YocH from B. subtilis and, correspondingly, can regulate release of muropeptides under the influence of YocH activity [57]. Mycobacterium tuberculosis has a PrkC homolog – serine-threonine kinase PknB, which is one of the 11 serine-threonine protein kinases [65, 68]. Therefore, it is reasonable to suppose that muropeptides produced during enzymatic hydrolysis under the influence of RipA and RpfB can interact similarly with the given protein kinase. The PknB regulator is essential for growth of the pathogenic M. tuberculosis and the saprophytic M. smegmatis [65]. Overexpression of the PASTA-domain of PknB (a structural analog of the PASTA-domain of PrkC) delays the growth of both bacteria [66]. Both PknB and PknA regulate the shape of the cells and, possibly, cell division of mycobacteria as well, associated with the reversible phosphorylation of intracellular proteins [67].
PknB controls the main metabolic pathways through phosphorylation of protein substrates: the protein regulator GarA, which in the non-phosphorylated state inhibits α-ketoglutarate decarboxylase in TCA cycle [69], KasA/B proteins involved in biosynthesis of mycolic acids [70], virulence factors RshA and SigH, Wag31 protein participating in the cell division [68, 71], and the cell wall biosynthesis enzymes GlmU [72] and PBPA [73]. It seems that the PASTA domain recognizes growing chains of peptidoglycan and regulates distribution of the proteins involved in division and formation of the septum. Interaction of the PASTA-domain of PknB with the cell wall is an important process that can serve as a mechanism for control of cell wall integrity and growth of the bacteria [59].
Among known substrates of phosphorylation by mycobacterial PknB, proteins involved in cell division Wag31 and PpbA are interesting, which seem to be involved in the early stages of reactivation of the dormant cells [60].
A molecular model of PknB activation based on extracellular domain dimerization under the influence of muropeptides was proposed [58], and binding of the synthesized muropeptides with the PknB extracellular domain was also studied by the method of surface plasmon resonance [50]. However, the serine-threonine protein kinase PrkC from S. aureus was found to be unable to dimerize even in the presence of high concentrations of muropeptides [61]. The authors, however, did not exclude this possibility in vivo; in this case, the other molecular players might be involved [61].
Although the involvement of protein kinase PknB in signal transduction through muropeptides is an interesting but not confirmed hypothesis, the signaling action mechanism of Rpf proteins mediated by muropeptides is plausible.
In fact, one cannot rule out simultaneous implementation of several proposed mechanisms, for instance, disintegration of bacterial aggregations under the influence of Rpf [46] can precede the subsequent generation of muropeptides triggering the resuscitation (Fig. 7). However, these processes might occur in parallel. Thus, peptidoglycan fragments that are always present in bacterial cultures can serve as substrate for Rpf proteins [72, 73]. It is obvious now that resuscitation of the dormant bacterial forms is a complex phenomenon that includes several stages. It is likely that, the first stage (the metabolic activation stage) seems to be associated with an increase in the level of intracellular cAMP as a result of adenylate cyclase activation. The increase in cAMP level and activation of metabolism preceded cell proliferation and rpfA expression, which grew by the beginning of cell division [37]. Thus, endogenous Rpf proteins can directly stimulate the division of mycobacterial cells and concurrently promote the generation of low molecular weight products (muropeptides) capable of inducing early stages of resuscitation [62].
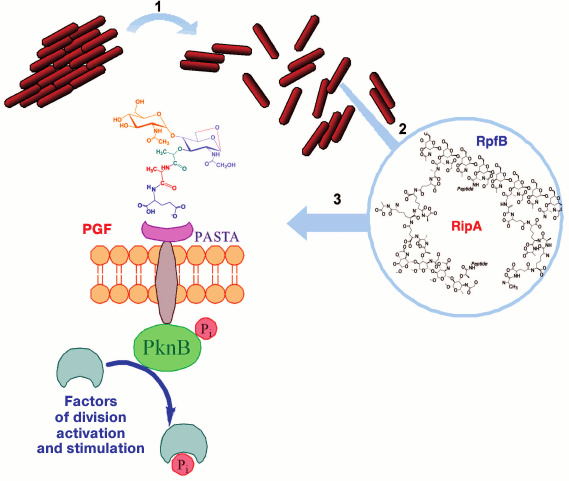
Fig. 7. A possible pathway of resuscitation of the dormant mycobacteria. 1) The Rpf protein induces disintegration of cellular aggregates; 2) under the combined action of Rpf and Rip proteins on mycobacterial peptidoglycan, peptidoglycan fragments, muropeptides, are released; 3) peptidoglycan fragments (PGF) bind with the surface receptor PASTA-domain of serine-threonine protein kinase PknB.
PROSPECTS FOR THE Rpf PROTEINS IN MEDICINE
Antituberculosis drugs development. In a number of publications, Rpf proteins are associated with the latent form of tuberculosis and its further transition into active tuberculosis [29, 34, 37, 74, 75]. The problem of latent tuberculosis is urgent in case of immunosuppressive therapy, when resuscitation of the latent form could appear as a side effect [76, 77]. In connection with the above-described role of Rpf proteins in resuscitation of the dormant forms, approaches for inhibition of Rpf activity seem to be promising for the struggle against activation of the latent TB. Such approaches include usage of monoclonal antibodies against the RpfB domain, what inhibit the stimulatory effect of Rpf on M. tuberculosis division in vivo [65].
The enzymatic activity of Rpf protein allows us to select compounds with potential inhibitory activity that can be considered as potential antituberculosis drugs. There are earlier reports on the existence of two inhibitors of lytic transglycosylases, bulgecin and NAG-thiazoline; none of them, however, has ever been tested on gram-positive bacteria [79, 80]. Moreover, NAG-thiazolines did not have a pronounced inhibitory effect but were active only in high concentrations [66]. From this perspective, using nitrophenyl thiocyanates (NPT) appeared to be promising because they could inhibit germination of fungal spores. The knowledge on involvement of hydrolytic enzymes, in particular, lytic transglycosylases and chitinases, performing key functions in germination process, suggested a new class of potential inhibitors of such hydrolases – nitrophenyl thiocyanates [81-83]. It was supposed that NPTs could be active toward Rpfs. Studies on the interaction of some NPTs with the proteins by the method of protein autofluorescence quenching revealed that such inhibitors interacted with residues of aromatic amino acids (Trp352, Tyr305) and prevented the binding of Rpf’s substrate (peptidoglycan fragments) with its active center [69]. It was also found that this class of compounds could inhibit Rpf hydrolysis of both the synthetic substrate 4-MUF-3-NAG and the fluorescently labeled mycobacterial peptidoglycan [69]. Studies on biological effect of NPTs on Rpf-producing microorganisms (M. luteus, M. smegmatis, and M. tuberculosis) revealed that they could inhibit growth of the bacteria and suppress their resuscitation from the dormant state, increasing the resuscitation time and lowering the number of viable cells at the last stage of the resuscitation (M. smegmatis, M. tuberculosis) [57, 83]. Moreover, NPTs suppressed reactivation of latent tuberculous infection in an in vivo mouse model after immunosuppression with aminoguanidine [49]. Although these inhibitory effects were observed at rather high concentrations of the compounds tested and some cells still could be resuscitated from the nonculturable state, NPTs can be considered as a promising class of molecules which modification would provide effective medicines preventing activation of latent tuberculosis.
Because it is difficult to eradicate dormant forms of TB within a host (common antibiotics do not act on such forms [84, 85]), it has been proposed to use Rpf for transition of nonculturable M. tuberculosis cells into the active state inside the host’s organs to make them sensitive to antibiotics [86, 87]. This idea directly was published in 2013, but it has not been realized so far. In particular, it is unclear how the unstable recombinant protein could be introduced into the host organism. There is also another danger – an antibiotic-resistant M. tuberculosis strain can be resuscitated that cannot be predicted beforehand.
Immunological aspects. In M. tuberculosis, five Rpf orthologs (Rpf A-E) have been described, and, according to predictions, four of them are secreted proteins. Therefore, it was supposed that these proteins could be recognized by the host’s immune system and serve as protectors in development of new vaccines. Usage of the Rpf proteins for stimulation of immune response against latent tuberculosis and its activation seemed especially attractive. In fact, in the works following the discovery of Rpf, mRNAs corresponding to five orthologs of Rpf were detected in pulmonary tissues of infected mice, rabbits, and humans [34, 88, 89]. The corresponding protein was detected in the intestinal lymph nodes of TB patients using antibodies against RpfB. In this case, the protein was localized in the necrosis centers, giant cells, and macrophages [74].
Immunological studies have revealed that two of the five Rpf proteins (Rpf A and Rpf D) from M. tuberculosis can stimulate production of γ-interferon by T-cells of patients with tuberculosis, i.e. their immune system can recognize these antigens. It should be noted that the immune response was expressed more strongly in patients with latent tuberculosis than in patients with its active form [75]. These findings seem promising for using Rpf proteins for serodiagnosis, as well as for TB diagnostics in cell response reactions.
Use of Rpf proteins as prophylactic subunit vaccines was studied on a model of mouse tuberculosis. In immunized animals, the count of bacteria after infection decreased [76]. On a similar model, it was shown that vaccination of animals with DNA vaccines encoding synthesis of the RpfB and RpfD proteins had the maximal protective effect manifested as a decrease in bacterial count in the lungs after infection [77]. Application of four of the five recombinant Rpf proteins from M. tuberculosis as subunit vaccines revealed their ability to induce in mice both cellular and humoral immune response. Vaccination of mice with these proteins resulted in significant protection on infecting with a virulent strain of M. tuberculosis: the survival of the animals increased and the pathogen proliferation in the lungs and spleen was significantly decreased [78]. Another approach was constructing attenuated strains of M. tuberculosis for using them as live vaccines. In this case, knocked-out strains of M. tuberculosis were created with deletions of two-to-four Rpf genes. As a result of these deletions, the strains lost virulence but could protect immunized mice infected with a virulent strain [79]. Certainly, for the development and approbation of new antituberculosis vaccines, a long time is required, and the above-presented experiments are only the first steps in this line. In particular, the known biotechnological company AERAS specialized in development of new anti-tuberculous vaccines reported an experimental study of a new vaccine based on recombinant BCG expressing three proteins of the M. tuberculosis Rpf along with other antigens [80].
Rpf for diagnosis of tuberculosis in patient samples. Resuscitating features of Rpf relative to dormant mycobacteria have been applied in diagnostic studies. In microbiological examination of sputum samples taken from patients with tuberculosis, false-negative results were observed rather frequently, i.e. there was no growth of the pathogen on standard media. Now it is clear that this can be caused by the presence in sputum of M. tuberculosis cells in the nonculturable state. Such population of the dormant cells was first detected by Mukamolova et al. in 2010 [81]. It was shown that a significant fraction of the cells in the sputum could be resuscitated by addition of Rpf into the medium [81]. Later a similar effect was found on resuscitation of sputum samples in liquid medium upon addition of supernatant obtained from the growing cells of M. tuberculosis [82] or M. luteus [83]. Note that the maximal resuscitating effect of supernatant presumably containing Rpf was manifested on slowly growing cultures, i.e. on the deeply dormant cells [82]. Unfortunately, in these works the active bacterial supernatants required for the maximal result were not studied in detail (first, the culture age and the medium composition), although this could be important for such experiments (M. O. Shleeva, personal communication). A very similar result was obtained in the examination of patients’ sputum with recombinant Rpf proteins E and B, which stimulated dormant bacteria growth in the samples, which decreased the time necessary for diagnosis of the pathogen [84].
Thus, the Rpf family proteins can be used for improvement of tuberculosis diagnostics and decreasing the number of false-negative results associated with difficulties in cultivation of the pathogen.
CONCLUSIONS
Eighteen years have passed since the time of the first publication about the Rpf protein in M. luteus as a released factor stimulating the resuscitation of dormant forms of this nonsporulating bacterium [6]. During this time, significant progress has been achieved in understanding of the distribution of such proteins, their nature, structure, and functions. The Rpf proteins are now known to be a large family widely represented in gram-positive bacteria of the Actinobacteria phylum. Based on structural studies, it was supposed that the conservative domain in the proteins of this family should be a hybrid of lysozyme and bacterial lytic transglycosylase, which suggested the presence of enzymatic features in the Rpf proteins. This hypothesis was confirmed later in several experimental studies, which resulted in the assignment of these proteins to the class of peptidoglycan hydrolases – lytic transglycosylases, i.e. to enzymes involved in bacterial wall remodeling. The Rpf proteins in many bacteria were shown to exist as several orthologs, which, at least partially, were functionally interchangeable in vitro. In many publications, the resuscitating role of Rpf proteins was shown in vitro on the dormant and stressed cells of various actinobacteria. Special attention was paid to the role of this family of proteins in mycobacteria, in particular in M. tuberculosis. In these works, an association was established between the resuscitation of the tuberculosis agent from the dormant state in vitro and in the host organism and the expression of Rpf proteins.
Notwithstanding the obvious progress, mechanisms of Rpf protein action stimulating growth and resuscitation are not clear in detail. Is the enzymatic activity of the Rpf proteins as it is sufficient for the resuscitating effect? Alternatively, do products of the enzymatic activity of Rpf (muropeptides) act as intermediate signaling molecules? If the first hypothesis is correct, a question arises whether a sequence of events exists between the peptidoglycan hydrolysis and the stimulation of proliferation. If the second hypothesis is correct, what are the mechanisms of the signal transmission? These questions have no answers and are awaiting experimental studies.
Obviously, proteins of the Rpf family are important for medicine. First, this is associated with studies on mechanisms of latent tuberculosis, which is a widely distributed disease potentially dangerous because of uncontrolled resuscitation and transition to the active form. Experiments with M. tuberculosis strains knocked-out in their rpf genes have shown that Rpf proteins determine the ability of such strains to establish the latent state in animals and to be resuscitated with development of an active tuberculosis process. Thus, the Rpf family is an important factor in the pathogenesis of latent tuberculosis.
On the other hand, the finding that Rpf proteins can be recognized by the host immune system suggests the use of these proteins as diagnostic antibodies and as a basis for creating subunit prophylactic vaccines. Finally, the resuscitating function of the Rpf proteins makes possible their use for improvement of microbiological diagnostics of tuberculosis in biological specimens (detection of nonculturable forms of the mycobacteria). The above-mentioned possibilities of using the Rpf proteins are already realized, at least in experimental works. These studies may result in the creation of new vaccines and diagnostic tests for application in medical practice.
Acknowledgements
This work was supported by the Russian Scientific Foundation (project No. 16-15-00245): sections “Functional Features of the Rpf Proteins”, “Mechanism of Action”, “Prospects of Using of the Rpf Proteins in Medicine”; and by the Program “Molecular and Cell Biology” of the Presidium of the Russian Academy of Sciences: introductory part and following sections “Structure of the Rpf proteins” and “Enzymatic Features of the Rpf Proteins”.
REFERENCES
1.Mukamolova, G. V., Kaprelyants, A. S., Kell, D. B.,
and Young, M. (2003) Adoption of the transiently non-culturable state
– a bacterial survival strategy? Adv. Microb. Physiol.,
47, 65-129.
2.Kaprelyants, A. S., Gottschal, J. C., and Kell, D.
B. (1993) Dormancy in non-sporulating bacteria, FEMS Microbiol.
Lett., 104, 271-286.
3.Votyakova, T. V., Kaprelyants, A. S., and Kell, D.
B. (1994) Influence of viable cells on the resuscitation of dormant
cells in Micrococcus luteus cultures held in an extended
stationary phase: the population effect, Appl. Environ.
Microbiol., 60, 3284-3291.
4.Kaprelyants, A. S., and Kell, D. B. (1993) Dormancy
in stationary-phase cultures of Micrococcus luteus: flow
cytometric analysis of starvation and resuscitation, Appl. Environ.
Microbiol., 59, 3187-3196.
5.Kaprelyants, A. S., Mukamolova, G. V., and Kell, D.
B. (1994) Estimation of dormant Micrococcus luteus cells by
penicillin lysis and by resuscitation in cell-free spent culture medium
at high dilution, FEMS Microbiol. Lett., 115,
347-352.
6.Mukamolova, G. V., Kaprelyants, A. S., Young, D.
I., and Kell, D. B. (1998) A bacterial cytokine, Proc. Natl. Acad.
Sci. USA, 95, 8916-8921.
7.Mukamolova, G. V., Turapov, O. A., Kazarian, K.,
Telkov, M., Kaprelyants, A. S., Kell, D. B., and Young, M. (2002) The
rpf gene of Micrococcus luteus encodes an essential
secreted growth factor, Mol. Microbiol., 46, 611-621.
8.Young, M., Artsatbanov, V., Beller, H. R., Chandra,
G., Chater, K. F., Dover, L. G., Goh, E. B., Kahan, T., Kaprelyants, A.
S., Kyrpides, N., Lapidus, A., Lowry, S. R., Lykidis, A., Mahillon, J.,
Markowitz, V., Mavromatis, K., Mukamolova, G. V., Oren, A., Rokem, J.
S., Smith, M. C., Young, D. I., and Greenblatt, C. L. (2010) Genome
sequence of the Fleming strain of Micrococcus luteus, a simple
free-living actinobacterium, J. Bacteriol., 192,
841-860.
9.Ravagnani, A., Finan, C. L., and Young, M. (2005) A
novel firmicute protein family related to the actinobacterial
resuscitation-promoting factors by non-orthologous domain displacement,
BMC Genomics, 6, 39.
10.Panutdaporn, N., Kawamoto, K., Asakura, H., and
Makino, S. I. (2006) Resuscitation of the viable but non-culturable
state of Salmonella enterica serovar Oranienburg by recombinant
resuscitation-promoting factor derived from Salmonella
typhimurium strain LT2, Int. J. Food Microbiol.,
106, 241-247.
11.Bateman, A., and Bycroft, M. (2000) The structure
of a LysM domain from E. coli membrane-bound lytic murein
transglycosylase D (MltD), J. Mol. Biol., 299,
1113-1119.
12.Cohen-Gonsaud, M., Keep, N. H., Davies, A. P.,
Ward, J., Henderson, B., and Labesse, G. (2004) Resuscitation-promoting
factors possess a lysozyme-like domain, Trends Biochem. Sci.,
29, 7-10.
13.Cohen-Gonsaud, M., Barthe, P., Bagneris, C.,
Henderson, B., Ward, J., Roumestand, C., and Keep, N. H. (2005) The
structure of a resuscitation-promoting factor domain from
Mycobacterium tuberculosis shows homology to lysozymes, Nat.
Struct. Mol. Biol., 12, 270-273.
14.Ruggiero, A., Tizzano, B., Geerlof, A., Pedone,
E., Pedone, C., Wilmanns, M., and Berisio, R. (2007) Expression,
purification, crystallization and preliminary X-ray crystallographic
analysis of a resuscitation-promoting factor from Mycobacterium
tuberculosis, Acta Crystallogr. Sect. F Struct. Biol. Cryst.
Commun., 63, 870-873.
15.Maione, V., Ruggiero, A., Russo, L., De Simone,
A., Pedone, P. V., Malgieri, G., Berisio, R., and Isernia, C. (2015)
NMR structure and dynamics of the resuscitation promoting factor RpfC
catalytic domain, PLoS One., 10, e0142807.
16.Ruggiero, A., Tizzano, B., Pedone, E., Pedone,
C., Wilmanns, M., and Berisio, R. (2009) Crystal structure of the
resuscitation-promoting factor (DeltaDUF)RpfB from M.
tuberculosis, J. Mol. Biol., 385, 153-162.
17.Ruggiero, A., Marchant, J., Squeglia, F.,
Makarov, V., De Simone, A., and Berisio, R. (2013) Molecular
determinants of inactivation of the resuscitation promoting factor B
from Mycobacterium tuberculosis, J. Biomol. Struct. Dyn.,
31, 195-205.
18.Bateman, A., Holden, M. T., and Yeats, C. (2005)
The G5 domain: a potential N-acetylglucosamine recognition domain
involved in biofilm formation, Bioinformatics, 21,
1301-1303.
19.Ruggiero, A., Squeglia, F., Romano, M.,
Vitagliano, L., De Simone, A., and Berisio, R. (2016) The structure of
resuscitation promoting factor B from M. tuberculosis reveals
unexpected ubiquitin-like domains, Biochim. Biophys. Acta,
1860, 445-451.
20.Vollmer, W. (2008) Structural variation in the
glycan strands of bacterial peptidoglycan, FEMS Microbiol. Rev.,
32, 287-306.
21.Mukamolova, G. V., Murzin, A. G., Salina, E. G.,
Demina, G. R., Kell, D. B., Kaprelyants, A. S., and Young, M. (2006)
Muralytic activity of Micrococcus luteus Rpf and its
relationship to physiological activity in promoting bacterial growth
and resuscitation, Mol. Microbiol., 59, 84-98.
22.Telkov, M. V., Demina, G. R., Voloshin, S. A.,
Salina, E. G., Dudik, T. V., Stekhanova, T. N., Mukamolova, G. V.,
Kazaryan, K. A., Goncharenko, A. V., Young, M., and Kaprelyants, A. S.
(2006) Proteins of the Rpf (resuscitation promoting factor) family are
peptidoglycan hydrolases, Biochemistry, 71, 414-422.
23.Thunnissen, A. M., Dijkstra, A. J., Kalk, K. H.,
Rozeboom, H. J., Engel, H., Keck, W., and Dijkstra, B. W. (1994)
Doughnut-shaped structure of a bacterial muramidase revealed by X-ray
crystallography, Nature, 367, 750-753.
24.Grutter, M. G., Weaver, L. H., and Matthews, B.
W. (1983) Goose lysozyme structure: an evolutionary link between hen
and bacteriophage lysozymes? Nature, 303, 828-831.
25.Mukamolova, G. V., Kormer, S. S., Kell, D. B.,
and Kaprelyants, A. S. (1999) Stimulation of the multiplication of
Micrococcus luteus by an autocrine growth factor, Arch.
Microbiol., 172, 9-14.
26.Mukamolova, G. V., Turapov, O. A., Young, D. I.,
Kaprelyants, A. S., Kell, D. B., and Young, M. (2002) A family of
autocrine growth factors in Mycobacterium tuberculosis, Mol.
Microbiol., 46, 623-635.
27.Shleeva, M. O., Bagramyan, K., Telkov, M. V.,
Mukamolova, G. V., Young, M., Kell, D. B., and Kaprelyants, A. S.
(2002) Formation and resuscitation of “non-culturable”
cells of Rhodococcus rhodochrous and Mycobacterium
tuberculosis in prolonged stationary phase, Microbiology,
148, 1581-1591.
28.Shleeva, M., Mukamolova, G. V., Young, M.,
Williams, H. D., and Kaprelyants, A. S. (2004) Formation of
“non-culturable” cells of Mycobacterium smegmatis in
stationary phase in response to growth under suboptimal conditions and
their Rpf-mediated resuscitation, Microbiology, 150,
1687-1697.
29.Biketov, S., Mukamolova, G. V., Potapov, V.,
Gilenkov, E., Vostroknutova, G., Kell, D. B., Young, M., and
Kaprelyants, A. S. (2000) Culturability of Mycobacterium
tuberculosis cells isolated from murine macrophages: a bacterial
growth factor promotes recovery, FEMS Immunol. Med. Microbiol.,
29, 233-240.
30.Downing, K. J., Betts, J. C., Young, D. I.,
McAdam, R. A., Kelly, F., Young, M., and Mizrahi, V. (2004) Global
expression profiling of strains harboring null mutations reveals that
the five rpf-like genes of Mycobacterium tuberculosis
show functional redundancy, Tuberculosis, 84,
167-179.
31.Tufariello, J. M., Jacobs, W. R., and Chan, J.
(2004) Individual Mycobacterium tuberculosis
resuscitation-promoting factor homologues are dispensable for growth
in vitro and in vivo, Infect. Immun., 72,
515-526.
32.Downing, K. J., Mischenko, V. V., Shleeva, M. O.,
Young, D. I., Young, M., Kaprelyants, A. S., Apt, A. S., and Mizrahi,
V. (2005) Mutants of Mycobacterium tuberculosis lacking three of
the five rpf-like genes are defective for growth in vivo
and for resuscitation in vitro, Infect. Immun.,
73, 3038-3043.
33.Kana, B. D., Gordhan, B. G., Downing, K. J.,
Sung, N., Vostroktunova, G., Machowski, E. E., Tsenova, L., Young, M.,
Kaprelyants, A., Kaplan, G., and Mizrahi, V. (2008) The
resuscitation-promoting factors of Mycobacterium tuberculosis
are required for virulence and resuscitation from dormancy but are
collectively dispensable for growth in vitro, Mol.
Microbiol., 67, 672-684.
34.Hartmann, M., Barsch, A., Niehaus, K., Puhler,
A., Tauch, A., and Kalinowski, J. (2004) The glycosylated cell surface
protein Rpf2, containing a resuscitation-promoting factor motif, is
involved in intercellular communication of Corynebacterium
glutamicum, Arch. Microbiol., 182, 299-312.
35.Tufariello, J. M., Mi, K., Xu, J., Manabe, Y. C.,
Kesavan, A. K., Drumm, J., Tanaka, K., Jacobs, W. R., Jr., and Chan, J.
(2006) Deletion of the Mycobacterium tuberculosis
resuscitation-promoting factor Rv1009 gene results in delayed
resuscitation from chronic tuberculosis, Infect. Immun.,
74, 2985-2995.
36.Gupta, R. K., Srivastava, B. S., and Srivastava,
R. (2010) Comparative expression analysis of rpf-like genes of
Mycobacterium tuberculosis H37Rv under different physiological
stress and growth conditions, Microbiology, 156,
2714-2722.
37.Shleeva, M., Goncharenko, A., Kudykina, Y.,
Young, D., Young, M., and Kaprelyants, A. (2013) Cyclic AMP-dependent
resuscitation of dormant Mycobacteria by exogenous free fatty
acids, PLoS One, 8, e82914.
38.Rickman, L., Scott, C., Hunt, D. M., Hutchinson,
T., Menendez, M. C., Whalan, R., Hinds, J., Colston, M. J., Green, J.,
and Buxton, R. S. (2005) A member of the cAMP receptor protein family
of transcription regulators in Mycobacterium tuberculosis is
required for virulence in mice and controls transcription of the
rpfA gene coding for a resuscitation promoting factor, Mol.
Microbiol., 56, 1274-1286.
39.Raman, S., Hazra, R., Dascher, C. C., and Husson,
R. N. (2004) Transcription regulation by the Mycobacterium
tuberculosis alternative sigma factor SigD and its role in
virulence, J. Bacteriol., 186, 6605-6616.
40.Keep, N. H., Ward, J. M., Cohen-Gonsaud, M., and
Henderson, B. (2006) Wake up! Peptidoglycan lysis and bacterial
non-growth states, Trends Microbiol., 14, 271-276.
41.Typas, A., Banzhaf, M., Gross, C. A., and
Vollmer, W. (2011) From the regulation of peptidoglycan synthesis to
bacterial growth and morphology, Nat. Rev. Microbiol.,
10, 123-136.
42.Vollmer, W., Joris, B., Charlier, P., and Foster,
S. (2008) Bacterial peptidoglycan (murein) hydrolases, FEMS
Microbiol. Rev., 32, 259-286.
43.Boneca, I. G. (2005) The role of peptidoglycan in
pathogenesis, Curr. Opin. Microbiol., 8, 46-53.
44.Bansal-Mutalik, R., and Nikaido, H. (2014)
Mycobacterial outer membrane is a lipid bilayer and the inner membrane
is unusually rich in diacyl phosphatidylinositol dimannosides, Proc.
Natl. Acad. Sci. USA, 111, 4958-4963.
45.Gupta, R. K., Srivastava, B. S., and Srivastava,
R. (2010) Comparative expression analysis of rpf-like genes of
Mycobacterium tuberculosis H37Rv under different physiological
stress and growth conditions, Microbiology, 156,
2714-2722.
46.Nikitushkin, V. D., Demina, G. R., and
Kaprelyants, A. S. (2011) Effect of secreted Rpf protein on
intracellular contacts in Micrococcus luteus and
Mycobacterium smegmatis cultures, Microbiology,
80, 143-149.
47.Voloshin, S. A., and Kapreliants, A. S. (2005)
Cell aggregation in cultures of Micrococcus luteus studied by
dynamic light scattering, Prikl. Biokhim. Mikrobiol., 41,
647-651.
48.Shleeva, M. O., Mukamolova, G. V., Telkov, M. V.,
Berezinskaia, T. L., Syroeshkin, A. V., Biketov, S. F., and
Kapreliants, A. S. (2003) Formation of non-culturable Mycobacterium
tuberculosis and their regeneration, Mikrobiologiya,
72, 76-83.
49.Kaprelyants, A. S., Mukamolova, G. V., Ruggiero,
A., Makarov, V. A., Demina, G. R., Shleeva, M. O., Potapov, V. D., and
Shramko, P. A. (2012) Resuscitation-promoting factors (Rpf): in search
of inhibitors, Protein Pept. Lett., 19, 1026-1034.
50.Scheurwater, E., Reid, C. W., and Clarke, A. J.
(2008) Lytic transglycosylases: bacterial space-making autolysins,
Int. J. Biochem. Cell Biol., 40, 586-591.
51.Hett, E. C., Chao, M. C., Steyn, A. J., Fortune,
S. M., Deng, L. L., and Rubin, E. J. (2007) A partner for the
resuscitation-promoting factors of Mycobacterium tuberculosis,
Mol. Microbiol., 66, 658-668.
52.Ruggiero, A., Squeglia, F., Esposito, C.,
Marasco, D., Pedone, E., Pedone, C., and Berisio, R. (2010) Expression,
purification, crystallization and preliminary X-ray crystallographic
analysis of the resuscitation promoting factor interacting protein RipA
from M. tuberculosis, Protein Pept. Lett., 17,
70-73.
53.Kana, B. D., and Mizrahi, V. (2010)
Resuscitation-promoting factors as lytic enzymes for bacterial growth
and signaling, FEMS Immunol. Med. Microbiol., 58,
39-50.
54.Hett, E. C., Chao, M. C., Deng, L. L., and Rubin,
E. J. (2008) A mycobacterial enzyme essential for cell division
synergizes with resuscitation-promoting factor, PLoS Pathog.,
4, e1000001.
55.Nikitushkin, V. D., Demina, G. R., Shleeva, M.
O., Guryanova, S. V., Ruggiero, A., Berisio, R., and Kaprelyants, A. S.
(2015) A product of RpfB and RipA joint enzymatic action promotes the
resuscitation of dormant mycobacteria, FEBS J., 282,
2500-2511.
56.Shah, I. M., Laaberki, M. H., Popham, D. L., and
Dworkin, J. (2008) A eukaryotic-like Ser/Thr kinase signals bacteria to
exit dormancy in response to peptidoglycan fragments, Cell,
135, 486-496.
57.Shah, I. M., and Dworkin, J. (2010) Induction and
regulation of a secreted peptidoglycan hydrolase by a membrane Ser/Thr
kinase that detects muropeptides, Mol. Microbiol., 75,
1232-1243.
58.Barthe, P., Mukamolova, G. V., Roumestand, C.,
and Cohen-Gonsaud, M. (2010) The structure of PknB extracellular PASTA
domain from Mycobacterium tuberculosis suggests a
ligand-dependent kinase activation, Structure, 18,
606-615.
59.Dasgupta, A., Datta, P., Kundu, M., and Basu, J.
(2006) The serine/threonine kinase PknB of Mycobacterium
tuberculosis phosphorylates PBPA, a penicillin-binding protein
required for cell division, Microbiology, 152,
493-504.
60.Molle, V., and Kremer, L. (2010) Division and
cell envelope regulation by Ser/Thr phosphorylation: mycobacterium
shows the way, Mol. Microbiol., 75, 1064-1077.
61.Ruggiero, A., Squeglia, F., Marasco, D.,
Marchetti, R., Molinaro, A., and Berisio, R. (2011) X-ray structural
studies of the entire extracellular region of the serine/threonine
kinase PrkC from Staphylococcus aureus, Biochem. J.,
435, 33-41.
62.Nikitushkin, V. D., Demina, G. R., Shleeva, M.
O., and Kaprelyants, A. S. (2013) Peptidoglycan fragments stimulate
resuscitation of ”non-culturable” mycobacteria, Antonie
van Leeuwenhoek, Int. J. Gen. Mol. Microbiol., 103,
37-46.
63.Orlicka, K., Barnes, E., and Culver, E. L. (2013)
Prevention of infection caused by immunosuppressive drugs in
gastroenterology, Ther. Adv. Chronic Dis., 4,
167-185.
64.Mohan, V. P., Scanga, C. A., Yu, K., Scott, H.
M., Tanaka, K. E., Tsang, E., Tsai, M. M., Flynn, J. L., and Chan, J.
(2001) Effects of tumor necrosis factor alpha on host immune response
in chronic persistent tuberculosis: possible role for limiting
pathology, Infect. Immun., 69, 1847-1855.
65.Fan, A., Jian, W., Shi, C., Ma, Y., Wang, L.,
Peng, D., Bai, Y., An, Q., Hao, X., and Xu, Z. (2010) Production and
characterization of monoclonal antibody against Mycobacterium
tuberculosis RpfB domain, Hybridoma (Larchmt), 29,
327-332.
66.Reid, C. W., Blackburn, N. T., Legaree, B. A.,
Auzanneau, F. I., and Clarke, A. J. (2004) Inhibition of membrane-bound
lytic transglycosylase B by NAG-thiazoline, FEBS Lett.,
574, 73-79.
67.Templin, M. F., Edwards, D. H., and Holtje, J. V.
(1992) A murein hydrolase is the specific target of bulgecin in
Escherichia coli, J. Biol. Chem., 267,
20039-20043.
68.Atrih, A., and Foster, S. J. (1999) The role of
peptidoglycan structure and structural dynamics during endospore
dormancy and germination, Antonie van Leeuwenhoek, Int. J.
Gen. Mol. Microbiol., 75, 299-307.
69.Demina, G. R., Makarov, V. A., Nikitushkin, V.
D., Ryabova, O. B., Vostroknutova, G. N., Salina, E. G., Shleeva, M.
O., Goncharenko, A. V., and Kaprelyants, A. S. (2009) Finding of the
low molecular weight inhibitors of resuscitation promoting factor
enzymatic and resuscitation activity, PLoS One, 4,
e8174.
70.Zhang, Y. (2004) Persistent and dormant tubercle
bacilli and latent tuberculosis, Front. Biosci., 9,
1136-1156.
71.Salina, E., Ryabova, O., Kaprelyants, A., and
Makarov, V. (2014) New 2-thiopyridines as potential candidates for
killing both actively growing and dormant Mycobacterium
tuberculosis cells, Antimicrob. Agents Chemother.,
58, 55-60.
72.Seidi, K., and Jahanban-Esfahlan, R. (2013) A
novel approach to eradicate latent TB: based on resuscitation promoting
factors, J. Med. Hypotheses Ideas, 7, 69-74.
73.Gan, Y., Yao, Y., and Guo, S. (2015) The dormant
cells of Mycobacterium tuberculosis may be resuscitated by
targeting-expression system of recombinant mycobacteriophage-Rpf:
implication of shorter course of TB chemotherapy in the future, Med.
Hypotheses, 84, 477-480.
74.Davies, A. P., Dhillon, A. P., Young, M.,
Henderson, B., McHugh, T. D., and Gillespie, S. H. (2008)
Resuscitation-promoting factors are expressed in Mycobacterium
tuberculosis-infected human tissue, Tuberculosis, 88,
462-468.
75.Riano, F., Arroyo, L., Paris, S., Rojas, M.,
Friggen, A. H., Van Meijgaarden, K. E., Franken, K. L., Ottenhoff, T.
H., Garcia, L. F., and Barrera, L. F. (2012) T cell responses to DosR
and Rpf proteins in actively and latently infected individuals from
Colombia, Tuberculosis (Edinb.), 92, 148-159.
76.Radaeva, T. V., Nikonenko, B. V., Kapina, M. A.,
Mishchenko, V. V., and Apt, A. S. (2006) Experimental approaches to
designing vaccines against tuberculous infection resuscitation,
Probl. Tuberk. Bolezn. Legk., 5, 45-48.
77.Romano, M., Aryan, E., Korf, H., Bruffaerts, N.,
Franken, C. L., Ottenhoff, T. H., and Huygen, K. (2012) Potential of
Mycobacterium tuberculosis resuscitation-promoting factors as
antigens in novel tuberculosis subunit vaccines, Microbes
Infect., 14, 86-95.
78.Yeremeev, V. V., Kondratieva, T. K., Rubakova, E.
I., Petrovskaya, S. N., Kazarian, K. A., Telkov, M. V., Biketov, S. F.,
Kaprelyants, A. S., and Apt, A. S. (2003) Proteins of the Rpf family:
immune cell reactivity and vaccination efficacy against tuberculosis in
mice, Infect. Immun., 71, 4789-4794.
79.Kondratieva, T., Rubakova, E., Kana, B. D.,
Biketov, S., Potapov, V., Kaprelyants, A., and Apt, A. (2011)
Mycobacterium tuberculosis attenuated by multiple deletions of
rpf genes effectively protects mice against TB infection,
Tuberculosis (Edinb.)., 91, 219-223.
80.Zvi, A., Ariel, N., Fulkerson, J., Sadoff, J. C.,
and Shafferman, A. (2008) Whole genome identification of
Mycobacterium tuberculosis vaccine candidates by comprehensive
data mining and bioinformatic analyses, BMC Med. Genomics,
1, 18.
81.Mukamolova, G. V., Turapov, O., Malkin, J.,
Woltmann, G., and Barer, M. R. (2010) Resuscitation-promoting factors
reveal an occult population of tubercle Bacilli in Sputum, Am. J.
Respir. Crit. Care Med., 181, 174-180.
82.Kolwijck, E., Friedrich, S. O., Karinja, M. N.,
Van Ingen, J., Warren, R. M., and Diacon, A. H. (2014) Early stationary
phase culture supernatant accelerates growth of sputum cultures
collected after initiation of anti-tuberculosis treatment, Clin.
Microbiol. Infect., 20, 418-420.
83.Freeman, R., Dunn, J., Magee, J., and Barrett, A.
(2002) The enhancement of isolation of mycobacteria from a rapid liquid
culture system by broth culture supernate of Micrococcus luteus,
J. Med. Microbiol., 51, 92-93.
84.Huang, W., Qi, Y., Diao, Y., Yang, F., Zha, X.,
Ren, C., Huang, D., Franken, K. L., Ottenhoff, T. H., Wu, Q., and Shen,
J. (2014) Use of resuscitation-promoting factor proteins improves the
sensitivity of culture-based tuberculosis testing in special samples,
Am. J. Respir. Crit. Care Med., 189, 612-614.