REVIEW: Role of Proteolytic Enzymes in the Interaction of Phytopathogenic Microorganisms with Plants
T. A. Valueva*, B. Ts. Zaichik, and N. N. Kudryavtseva
Federal Research Center for Fundamental Basics of Biotechnology, Bach Institute of Biochemistry, Russian Academy of Sciences, 119071 Moscow, Russia; E-mail: valueva@inbi.ras.ru* To whom correspondence should be addressed.
Received August 29, 2016; Revision received September 19, 2016
Various forms of participation of proteolytic enzymes in pathogenesis and defense in plants are reviewed. Along with extracellular proteinases, phytopathogenic microorganisms produce specific effectors having proteolytic activity and capable of acting on proteins inside plant cells. In turn, for defense against pathogens, plants use both extracellular and intracellular proteinases.
KEY WORDS: phytopathogenic microorganisms, proteolytic enzymes, pathogenesis-related proteinsDOI: 10.1134/S0006297916130083
Abbreviations: Avr proteins, avirulence pathogen proteins; CBD, cellulose binding domain of plant chitinase; HR, hypersensitive response in plants; PR proteins, plant pathogenesis-related proteins; R genes, resistance genes of the host plant; ROS, reactive oxygen species; SAR, systemic acquired resistance response in plants; TTSS, type three secretion system.
Both plants and their pathogens have developed strategies during mutual
evolution that allow them to survive and win in their struggle for
existence. Taking into consideration that most paths used by either
plants or pathogenic organism are based on the synthesis and activation
of defense proteins (pathogenesis-related, PR) participating in the
plant response to the pathogen action [1, 2], investigation of their proteomes contributes
significantly to understanding of these paths.
For example, increase in relative protein content in the leaf blade of the rice plant (Oryza sativa L.), which participates in energy exchange, photosynthesis, protective reactions, and demonstrates antioxidative activity, was shown following infection with the Rhizoctonia solani fungus, and concurrently it was shown that the molecules of some proteins unfolded and were degraded [3]. It is necessary to establish during investigation of the interaction of host plant with pathogen the synthesis of which plant proteins is activated in response to the pathogen invasion, whether this activation occurs as a result of new transcription/translation or posttranslational changes of preformed inactive products, and whether it depends on the nature of the pathosystem, as well as what bioagents affect protective functions of the plant.
The plant cell wall comprises an external interface between the plant and its abiotic or biotic environment, and it provides the major protective barrier against pathogenic microorganisms that try to colonize a new host plant. The plant cell wall and its apoplastic environment can be considered as a molecular battlefield where interaction occurs between secreted plant and pathogen proteomes or secretomes (coevolution) required for the attack and defense of both plant and pathogen. The phytopathogens form and secrete an excess of effectors following invasion into the plant, which facilitate the successful infection process. These effectors include extracellular proteinases (that destroy plant proteinases) and their inhibitors (forming the plant defense mechanism). On the other hand, they can be identified as pathogen proteinases and inhibitors that results in induction of plant protective reactions [4, 5].
Formation of proteinases and their inhibitors in plants as well as proteinases in phytopathogens resulted in coevolutionary diversification and adaptation of the pathogen to the certain host plant. On one hand, successful infection suggests availability of mechanisms in the phytopathogen ensuring plant invasion and colonization [6]. On the other hand, plants also possess a certain defense system, which allows identification of the pathogen and protection from its actions.
Various forms of participation of proteolytic enzymes in pathogenesis have been considered. It was shown that phytopathogenic microorganisms produced specific effectors with proteolytic activities that can act upon the proteins inside the plant cell.
PROTEOLYTIC ENZYMES FROM PATHOGENIC MICROORGANISMS
The plant cell secretes a wide spectrum of proteins and peptides either constitutively expressed or induced, which are related to plant defense [5, 6]. Likewise, the phytopathogens secrete many peptides and proteins in the process of infection, which can be either attached to a hypha wall or penetrate the plant apoplast [7]. These proteins are referred to as effectors that change the structure of the host plant cell or affect the functions of the pathogen. The enzymes from the pathogen usually include those that destroy the plant wall structure [8, 9] as well as suppressors of extracellular plant protective compounds [8, 10, 11]. Depending on the character of interaction of the host plant and the pathogen in the case of resistant organisms, the specific secreted proteins termed avirulence (Avr) proteins interact with the products of the respective resistance gene of the host (R). At the same time, these proteins facilitate the development of disease in plants that do not possess resistance genes. It was shown that different proteins were secreted into the plant apoplast in the process of infection, which was likely associated with biotrophic or necrotrophic phases of pathogenesis [3].
Gram-negative bacteria developed the “type three-secretion system” (TTSS) for transfer of virulence proteins including proteolytic enzymes through the plasma membrane directly into the host plant cytosol [12, 13]. Representatives of Pseudomonas, Xanthomonas, Ralstonia, Erwinia, and Pantoea genera belong to the group of Gram-negative plant pathogens with TTSS. Recognition of threat by the plant cell is achieved at the point of contact with the microorganism. Certain signaling entities from the infected cells can induce a systemic acquired response (SAR) in the healthy non-infected plant tissues, which helps the whole plant to prepare for the fight against an invader. The SAR is manifested by the change of flow of ions including calcium across the membrane and hormonal status, as well as by oxidative burst, protein phosphorylation, and hypersensitive response (HR) accompanied by necrosis at the point of infection that prevents the spread of the pathogen, and by accumulation of pathogenesis related proteins and antimicrobial compounds – phytoalexins [10, 12, 14]. Even though most Avr proteins from eukaryotic phytopathogens are apoplastic [15], it was shown that similarly to the case of bacterial phytopathogens, some Avr proteins from fungi are translocated into the plant cells [16-18]. These data confirm the complexity of interactions of the secretomes of pathogens with the secretomes of their host plants.
It was established that the activity of protein effectors is based on the availability of enzymatic (including proteolytic) activity [19]. A list of known protein effectors with proteolytic properties is presented in the table. They can be separated into four families of related proteins. The representatives of the YopJ and XopD families belong to clan CE of cysteine proteinases, and the representatives of the YopT and AvrRpt2 families – to clan CA.
Bacterial effector proteins having properties of cysteine proteinases
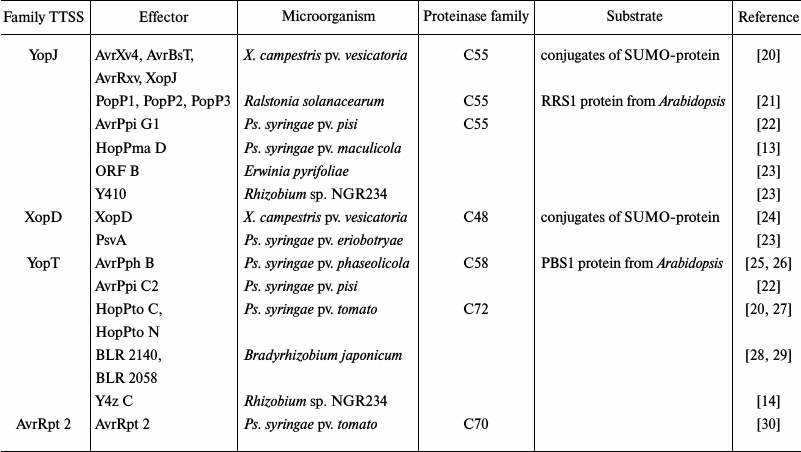
The 35-kDa avirulence protein AvrPphB from the Pseudomonas syringae pv. phaseolicola bacterium is subjected to autoactivation in the bacterial cell, forming two fragments with molecular masses of 7 and 28 kDa. The 28-kDa fragment is acylated by a fatty acid residue following transfer into the plant cell, and this acylated form is transferred through the cytoplasmic membrane. The AvrPphB protein causes hypersensitive response in Arabidopsis plants expressing the RPS5 protein [24]. The 3D structure of AvrPphB protein and location of the Cys98, His212, and Asp227 residues forming the catalytic triad in the active center of the proteinase is similar to papain [31]. Mutants with replaced Cys, His, or Asp residues in the active center lose to the same degree their biological activity and ability for autoactivation [25, 26]. Interaction of AvrPphB with RPS5 is not direct. The presence of a third component – PBS1 protein – is required, which is the true substrate of the proteinase. PBS1 protein exhibiting properties of serine/threonine kinase is cleaved into two fragments and is then subjected to phosphorylation, and it acts as a trigger activating signal transfer with participation of RPS5 [24, 26]. It has been suggested that all three proteins participating in this process remain bound to the plasma membrane [32].
The effector protein AvrRpt2 found in P. syringae pv. tomato belongs to another family and does not have any known analogs (table). The AvrRpt2 protein induces hypersensitive response in resistant Arabidopsis plants expressing the RPS2 protein. On the contrary, the AvrRpt2 effector facilitates disease development in plants lacking the RPS2 protein [33]. The effector protein with molecular mass of 28 kDa is proteolytically cleaved at its Gly71–Gly72 bond inside the plant cell forming two fragments with molecular masses of 21 and 7 kDa. The C-terminal fragment with molecular mass of 21 kDa acts as a trigger of protective response [34]. Even though the processing of AvrRpt2 is autocatalytic in nature, the presence of a certain additional factor contained in the extract of animal and plant tissues is necessary [35]. The structure of the AvrRpt2 effector molecule and location of amino acid residues in the active center was found to be similar to the cysteine proteinase from Staphylococcus aureus – staphopain [22]. RIN4 was identified as a substrate of AvrRpt2 in Arabidopsis cells [30, 36]. RIN4 proteolysis serves as a signal for hypersensitive response, in which the RPS2 protein also participates. At the same time, the purified AvrRpt2 effector does not affect RIN4 protein in vitro. The presence of a eukaryotic cofactor is necessary for the interaction, which is likely identical to the one participating in the processing of the effector itself [22]. The nature of this factor was established, the cyclophilin protein playing this role [37].
Different representatives of bacteria of the Yersinia genus are animal and plant pathogens, and they produce effectors known as Yop (Yersinia outer proteins). When they are introduced in the plant or animal host cell, these effectors prevent the development of immune response. Some of the effectors belonging to the YopJ protein family, such as AvrBsT and AvrXv4 from Xanthomonas campestris pv. vesicatoria that bears similarities to the ULP1 proteinase from Saccharomyces cerevisiae yeast [38, 39]. The ULP1 enzyme controls the SUMO (small ubiquitin-like modifier) process of interaction with different proteins in yeasts. This type of proteinases contains either the His/Glu/Cys or His/Asp/Cys triad in the catalytic center, and it belongs to the CE clan of cysteine proteinases [38]. Modification of the residues in the enzyme active center results in the loss of ability to initiate the hypersensitive response in plants [24, 38, 39].
Expression of the X. campestris effectors XopD and AvrXv4 in leaves of Nicotiana benthamiana (L.) results in a decrease in the content of SUMO protein conjugates. It was concluded based on this that the effectors exhibited isopeptidase activity in vivo [24, 39]. Further confirmation was obtained with purified XopD protein. It was shown that the effector acted as an isopeptidase in vitro, cleaving the bond between the carboxyl group of the SUMO N-terminal residue and the ε-amino group of a Lys residue in the protein substrate. Moreover, it can act as a peptidase capable of transforming the SUMO precursor into the active form containing Gly-Gly N-terminal sequence [24, 40]. The XopD action is very specific; it exhibits activity exclusively towards SUMO forms of plant origin [24].
Modification of SUMO proteins plays an important role in regulation of various biological processes in plants. Furthermore, according to available data, proteins subjected to modification are concentrated mainly in the nucleus [41]. The fact that the XopD and PopP2 effectors entering the plant cell are also concentrated in the nuclei is intriguing in this connection [21, 24]. It can be suggested that the availability of proteinases in phytopathogenic microorganisms that can imitate the intrinsic plant enzymes provides them significant advantage. On the other hand, mechanisms could exist in plants that allow neutralizing activity of the SUMO-proteinases from bacteria. The product of the resistance gene in Arabidopsis – RRS1 protein – ensures resistance to Ralstonia solanacearum strains expressing the PopP2 effector [42]. The RRS1 protein directly interacts with the bacterial proteinase [21].
Products of avirulence genes exhibiting proteolytic activity were also found in phytopathogenic fungi. A major rice pathogen – Magnaporthe grisea fungus – produces the Avr-Pita protein, which is a metalloproteinase belonging to the family of zinc-containing proteinases M35 [43]. The mature form of this enzyme can initiate the hypersensitivity reaction in rice plants. It was established that Avr-Pita interacts directly with the leucine-rich domain of the product of the resistance gene – Pi-ta protein [44]. A mutant form of the Avr-Pita proteinase with the Glu residue in active center replaced by Asp loses its ability to interact with the Pi-ta protein and, respectively, loses its biological activity [45]. These data indicate that the product of the resistance gene – Pi-ta protein – is a proteinase substrate. This is the first example described in the literature of a direct interaction between the products of either avirulence or resistance gene [33].
To introduce the products of avirulence genes having proteolytic activity into cells of the host plant, the availability of invasive effectors comprising proteins carrying the RxLR or other conserved motifs is necessary. These effectors introduce the virulence factors into the plant cells, which, in turn, results in infection expression [2, 46]. Effectors with lysine-containing motifs (LysM) participate in protection of the chitin layer of the fungus cell wall, which allows maintaining its integrity and are defensive [47-49]. As suggested, these two types of effectors occur widely in fungi, especially in biotrophic ones. It was shown that positive selection of C-terminal amino acid residues of these effectors contributes to their evolution and diversity [50]. This is consistent with the suggestion that the N-terminal domain participates in their secretion, while the C-terminal domain – in suppression of the plant cell protective response. Hence, the posttranslational modification of proteins directly inside the plant cell can serve as an important means of phytopathogen action upon plant defense systems.
There appears to be evolutionary strategies for coexistence of pathosystems that allow adaptation of pathogens to possibilities emerging during their trophic interaction with the host plant. Because of this, most biotrophs have managed to adapt themselves to such plant defense mechanisms involving detoxifying phytoalexins and other metabolites related to cell defense, while the enhancement of their own defense means in response to the protective reaction of the host plant was characteristic for both hemibiotrophs and necrotrophs [2]. For example, such necrotrophic fungi as Sclerotinia sclerotiorum suppress the signaling cascade of reactive oxygen species (ROS) of the host plant in planta using catalase, superoxide dismutase, and mannite or oxalic acid, which results in local enhancement of the hypersensitive reaction and/or prevention of signal transduction in remote healthy parts [2]. At the same time, the Botrytis cinerea fungus forces its host plant to produce ROS, which destroys tissues, thus providing easier colonization. Jasmonate is secreted during the action of the Fusarium oxysporum fungus on Arabidopsis thaliana (L.), which facilitates the development of infection [51]. The same use of signaling paths was reported also for attack of the potato plant (Solanum tuberosum L.) by the phytopathogenic fungus Verticillium dahliae and of the tomato (Solanum lycopersicum L.) by the B. cinerea fungus [2, 33, 52, 53].
Hence, the pathogens adapted to certain plant families acquire the ability to overcome their immunity to microbial infections by secreting effector molecules that suppress or decrease the immune response. For example, the Cladosporium fulvum (syn. Passalora fulva) fungus secretes the chitin-binding protein CfAvr4 into the intercellular space of tomato leaf for protection from the action of the plant apoplastic chitinase [54]. Functional homologs of CfAvr4 were found also in other ascomycetes [49, 55, 56]. In cases when there are no homologs of the CfAvr4 gene in the phytopathogen genome, they secrete proteinases that cleave the chitinase cellulose-binding domain (CBD). For example, it was shown that the Fusarium solani f. sp. phaseoli modified the chitinase from the plant cell, which facilitated colonization of the plant host [57], and the Fusarium solani f. sp. eumartii secreted a subtilisin-like proteinase for the same purpose, which modified the chitinase and β-1,3-glucanases [58].
It was shown that the corn pathogens Fusarium verticillioides, Bipolaris zeicola, and Stenocarpella maydis secrete two types of proteinases that change the structure of the CBD in the corn chitinase molecule [59, 60]. It was found that a metalloprotease from F. verticillioides cleaved the CBD between Gly and Cys residues, while the polyglycine hydrolase present in many fungi belonging to the Pleosporineae family cleave the polyglycine linker in the chitinase hinge domain [60, 61].
The tomato pathogens B. cinerea, V. dahlia, and F. oxysporum f. sp. lycopersici secrete proteinases that modify the CBD of the plant chitinase [62]. The cleavage and removal of the CBD from the tomato chitinase during F. oxysporum f. sp. lycopersici infection was performed only during synergetic action of the serine proteinase FoSep1 and metalloproteinase FoMep1 (an ortholog of fungalisin of the F. verticillioides fungus) [62]. Removal of the CBD from the plant chitinases by these two enzymes decreased their chitinase and anti-pathogenic activity. Moreover, F. oxysporum f. sp. lycopersici mutants not containing both the FoSep1 and FoMep1 demonstrated reduced virulence with respect to tomato plants. Hence, the secreted fungal proteinases comprise important virulence factors, while the chitinase – an important component of the basal plant defense [62].
It is likely that the secretion of proteinases and their inhibitors by pathogens plays a more important role during coevolution of the pathogens and their host plants [63]. The presented data allowed understanding the reason why the overexpression of plant chitinase in transgenic plants did not result in the generation of tomato species resistant to the action of pathogens. In this connection, overexpression of chitinase by the heterologous gene, for which there has been no mutual evolution stage, could present a more promising approach for generation of species resistant to the action of pathogenic microorganisms.
The genes of phytopathogens encode proteinases belonging to different subfamilies, and their number depends of the nature of the pathogen. As a rule, hemibiotrophs and saprotrophs contain a larger number of secreted proteinases than biotrophs [64]. Nevertheless, the number of proteinases secreted by the biotrophic fungus C. fulvum was comparable with the phylogenetically close hemibiotroph Dothistroma septosporum [55], which was likely due to their adaptation to different plant families [65, 66]. It is generally accepted that proteinases play an important role in adaptation of two species of Phytophthora – P. infestans and P. mirabilis – to their respective host plants [67]. This provides an example that diversification and adaptation of the proteinase–inhibitor complex can occur at the molecular level and corroborates a hypothesis on the role of proteinases and their inhibitors in the “arms race” between the plant and its pathogen. The targeted removal of one or even two proteinase genes did not affect the virulence of plant pathogenic fungi Glomerella cingulata and B. cinerea [68].
MECHANISM OF PATHOGEN INTERACTION WITH PLANT HOST
Physiological and molecular mechanisms underlying interactions of necrotrophic phytopathogenic fungi with respective to host plants remain poorly understood. Three stages of the pathogenic process are recognized. The first includes formation of appressoria – specialized structures facilitating the attachment of the mycelium to the plant. Formation of colonies on cells of the host plant occurs during the second (biotrophic) stage, and degradation of cell walls – during the third stage (necrotrophic) [69, 70]. The pH of surrounding tissues becomes more alkaline during the stage of appressoria formation and later during the necrotrophic stage due to secretion of ammonium. This initiates programmed cell death. It has been suggested that this process optimizes conditions for further colonization of the host plant, which is accompanied by the induction of pathogenic factors [71, 72]. For example, the secreted pectate lyase enzyme (PelB) that destroys plant cell wall is controlled by transcription factor pacC, which is subjected to proteolytic activation with alkaline pH environment [73, 74]. It was shown that metabolic activity of the enzymes destroying cell wall increased significantly during proteolysis [71, 72].
The transition from the stage of appressoria to the biotrophic stage is accompanied by the transcription of genes encoding secreted proteinases – serine proteinases (subtilisin- and trypsin-like). Considering that these enzymes operate outside the cell, their activity as well as mechanisms controlling their synthesis and secretion is under the effect of many environmental factors [75]. It was shown that pH of the cultivation medium changed from neutral to slightly alkaline during ascomycete culture growth, and it reached a constant value when the activity of exoproteinases was at its maximum [76]. Increase in the medium pH caused by formation of ammonium induces the expression of pathogenicity genes of the fungus, including genes encoding proteolytic enzymes [77]. The pathogen is likely under oxidative stress generated by the host plant during the infection process, and availability of an active detoxication system is required for successful colonization. It was found that the pH of the medium during the invasion of ripe avocado fruits by the pathogenic Colletotrichum gloeosporioides fungi increased due to secretion of high ammonium concentration by the fungus [78, 79]. It has been suggested that a certain amount of ammonium could form during nitrate reduction, because fungal mutants (nit) lacking the nitroreductase were not capable of ammonium accumulation, and their virulence was significantly reduced [78, 80]. During increase in pH due to ammonium accumulation, the expression of genes encoding proteolytic enzymes with high pH optimum is induced [78]. It was shown that the site-directed deletion of the major nitrogen regulator (AREA) and of the ammonium transporter (AMET) affected ammonium accumulation and virulence of the C. coccodes pathogen [80-82]. The host plant becomes more susceptible to infection with a decrease in nitrogen content [83]. For example, it was shown that the expression of pathogenicity genes such as of proteolytic enzymes was induced in the Magnaporthe grisea, C. gloeosporioides, and Cladosporium fulvum fungi under conditions of nitrogen deficit in plants, which resulted in accumulation of ammonium ions [78, 84-88].
A nonpathogenic strain of C. lindemuthianum that is incapable of colonization of plant cells and transition to the necrophilic stage was generated by mutations in the AreA genes, which is a gene responsible for nitrogen catabolic repression [89]. It was found that the nit mutants of C. coccodes secreted less ammonium and demonstrated lower rate of infection of tomato plants [80], and the nit mutants of C. gloeosporioides attacking avocado fruits and characterized by inability of ammonium accumulation exhibited significantly lower virulence [78].
It can be suggested that the change in the medium pH is, on one hand, an adaptation mechanism facilitating absorption of proteinaceous nutrients by the pathogen, and on the other hand – a physiological signal initiating synthesis and secretion of proteinases that comprise one of the virulence factors [58, 78].
It was shown that during the infection of avocado fruits with the C. gloeosporioides pathogen, the infection pegs of appressoria penetrated the epidermal cells of its exocarp, but remained inactive until the ripening of the fruit. The secretion and synthesis of serine proteinases as the fungus pathogenicity factors were absent during the rest stage, which were stimulated by medium alkalinization, and the unripe fruits induced the synthesis of antipathogenic compounds. In turn, accumulation of ammonium in the decaying fruit tissues ensured transition of the biotrophic rest stage into the necrotrophic stage [78].
Proteinases play an important role in physiology and metabolism of fungi, which are heterotrophic organisms using both organic and inorganic compounds as carbon and nitrogen sources. Saprotrophic and pathogenic fungi secrete proteolytic enzymes; however, the secretion dynamics are different for these two groups [90-92]. Serine proteinases belonging to two major subfamilies – subtilisin-like and trypsin-like – are predominant among the proteinases of bacterial and fungal origin. The same catalytic triad (Asp-His-Ser) is present in the active center of enzymes from both subfamilies, which as suggested has been formed during convergent evolution [93, 94]. Participation of the extracellular microbial proteinase in pathogenesis can be of different nature: from destruction of structural proteins of the plant cell wall and plant protective proteins to processing of their own extracellular microbial proteins essential for the disease development. In several cases, direct dependence of the disease intensity on the activity of extracellular microbial serine proteinases was observed, which indicated an important role of proteinases in pathogenesis [95-97]. This agrees with data that protein inhibitors of proteinases contained in plants not only inhibit the enzyme activity, but also suppress their growth and development [77].
The subtilisin-like proteinases from pathogenic fungi can disrupt physiological integrity of the host plant cell ensuring penetration of the pathogen into it and colonization by breaking down both the cell wall proteins and the plant protective proteins [58]. In this connection, they can be considered as a virulence factor of phytopathogens [98]. For example, it was shown that proteinases from the subtilisin family were secreted during the infection of rapeseed (Brassica napus L.) by the Pyrenopeziza brassicae fungus, and synthesis of serine proteinase inhibitors was observed in rice species resistant to infection by this pathogen [98]. The degree of degradation of storage proteins in kernels of wheat plants (Triticum) infected with the F. culmorum (Wm. G. Sm.) Sacc. fungus also depended on the activity level of the secreted alkaline proteinase [97]. Targeted deletion of the Spm1 gene encoding a subtilisin-like proteinase significantly reduced pathogenicity of the Conidiobolus coronatus fungus [99]. It was shown that the degree of conidial germination of C. coronatus depended on the activity of alkaline serine proteinase. The subtilisin-like proteinases (APN 2) that affected virulence of the A. fumigatus fungus were also required for sporulation of this type of fungus [99].
Considering that proteinases from pathogens not only facilitate penetration of the fungus into the tissues of the host plant, but also actively affect the plant defense barriers, they comprise the required component of the pathogenesis process [100]. Correlation between the activity of serine proteinases and fungal pathogenicity suggests that these enzymes can serve as markers of phytopathogenicity [98, 100].
The presence of subtilisin-like proteinases was reported in the cultural medium of both saprotrophic and phytopathogenic fungi on the 4-5th day of growth, while the presence of trypsin-like proteinases from phytopathogens was observed on the 7th day [76, 92, 101]. It can be suggested that the subtilisin-like enzymes with wide substrate specificity are the first to cleave cell wall proteins of the host plant, thus ensuring access for the action of trypsin-like proteinases. That is why the latter are observed later in the cultivation medium, likely indicating their specific role in phytopathogenesis.
It was found that the saprotrophic species Trichoderma harcianum, Penicillium terlikowskii, and P. chrysogenum were characterized by high activity of the secreted subtilisin-like proteinases and lack of secretion of trypsin-like proteinases, while the phytopathogenic species Alternaria alternata, B. cinerea and Ulocladium botrytis secreted trypsin-like proteinases along with subtilisin-like ones [101]. The high level of trypsin-like activity of extracellular proteinases is likely determined high pathogenicity of the mycelial fungal species [95]. Hence, formation and secretion of the trypsin-like proteinases is a characteristic feature of phytopathogens and of the subtilisin-like enzymes of saprotrophic and entomopathogenic fungi.
Analysis of the mechanism of interaction within the host plant–pathogen system including the defense response of the pathogen can help to develop more effective tools to protect plants from diseases caused by fungi and oomycetes, which are responsible for large crop losses.
REFERENCES
# Van Loon, L. C., Rep, M., and Pieterse, C. M. (2006) Significance of
inducible defense-related proteins in infected plants, Annu. Rev.
Phytopathol., 44, 135-162.
# El Hadrami, A., El-Bebany, A.
F., Yao, Z., Adam, L. R., El Hadrami, I., and Daayf, F. (2012) Plants
versus fungi and oomycetes: pathogenesis, defense and counter-defense
in the proteomics era, Int. J. Mol. Sci., 13,
7237-7259.
# Lee, S.-J., Kelley, B. S., Damasceno, C. M. B.,
Bonnie, St. J., Kim, B.-S., Kim, B.-D., and Rose, J. K. C. (2006) A
functional screen to characterize the secretomes of eukaryotic
pathogens and their hosts in planta, Mol. Plant–Microbe
Interact., 19, 1368-1377.
# Cook, D. E., Mesarich, C.
H., and Thomma Bart, P. H. J. (2015) Understanding plant immunity as a
surveillance system to detect invasion, Annu. Rev. Phytopathol.,
53, 541-563.
# Van der Hoorn, R. A., and Jones, J. D.
(2004) The plant proteolytic machinery and its role in defense, Curr.
Opin. Plant Biol., 7, 400-407.
# Valueva, T. A., and
Mosolov, V. V. (2002) The role of inhibitors of proteolytic enzymes in
plant defense, Uspekhi Biol. Khim., 42, 193-216.
# Rast, D. M., Baumgartner, D., Mayer, C., and Hollenstein, G.
O. (2003) Cell wall-associated enzymes in fungi, Phytochemistry,
64, 339-366.
# Huitema, E., Bos, J. I., Tian, M., Win, J.,
Waugh, M. E., and Kamoun, S. (2004) Linking sequence to phenotype in
Phytophthora–plant interactions, Trends Microbiol.,
12, 193-200.
# Vorwerk, S., Somerville, S., and
Somerville, C. (2004) The role of plant cell wall polysaccharide
composition in disease resistance, Trends Plant Sci., 9,
203-209.
# Abramovitch, R. B., and Martin, G. B. (2004)
Strategies used by bacterial pathogens to suppress plant defenses,
Curr. Opin. Plant Biol., 7, 356-364.
# Rose, J. K.
C., Ham, K. S., Darvill, A. G., and Albersheim, P. (2002) Molecular
cloning and characterization of glucanase inhibitor proteins:
coevolution of a counter defense mechanism by plant pathogens, Plant
Cell, 14, 1329-1345.
# Alfano, J. R., and Collmer, A.
(2004) Type III secretion system effector proteins: double agents in
bacterial disease and plant defense, Annu. Rev. Phytopathol.,
42, 385-414.
# Guttman, D. S., Vinatzer, B. A., Sarkar, S.
F., Ranall, M. V., Kettler, G., and Greenberg, J. T. (2002) A
functional screen for the type III (Hrp) secretome of the plant
pathogen Pseudomonas syringae, Science, 295,
1722-1726.
# Joo, H. S., and Chang, C. S. (2005) Production of
protease from a new alkalophilic Bacillus sp. I-312 grown on
soybean meal: optimization and some properties, Process
Biochem., 40, 1263-1270.
# Rep, M. (2005) Small
proteins of plant-pathogenic fungi secreted during host colonization,
FEMS Microbiol. Lett., 253, 19-27.
# Armstrong, M.
R., Whisson, S. C., Pritchard, L., Bos, J. I. B., Venter, E., Avrova,
A. O., Rehmany, A. P., Böhme, U., Brooks, K., Cherevach, I.,
Hamlin, N., White, B., Fraser, A., Lord, A., Quail, M. A., Churcher,
C., Hall, N., Berriman, M., Huang, S., Kamoun, S., Beynon, J. L., and
Birch, P. R. (2005) An ancestral oomycete locus contains late blight
avirulence gene Avr3a, encoding a protein that is recognized in the
host cytoplasm, Proc. Natl. Acad. Sci. USA, 102,
7766-7771.
# Bryant, M. K., Schardl, C. L., Hesse, U., and Scott,
B. (2009) Evolution of a subtilisin-like protease gene family in the
grass endophytic fungus Epichloe festucae, BMC Evol.
Biol., 9, 168-175.
# Rehmany, A. P., Gordon, A., Rose,
L. E., Allen, R. L., Armstrong, M. R., Whisson, S. C., Kamoun, S.,
Tyler, B. M., Birch, P. R. J., and Beynon, J. L. (2005) Differential
recognition of highly divergent downy mildew avirulence gene alleles by
RPP1 resistance genes from two Arabidopsis lines, Plant
Cell, 17, 1839-1850.
# Innes, R. (2003) New effects of
type III effectors, Mol. Microbiol., 50, 363-365.
#
Petnicki-Owieja, T., Schneider, D. J., Tam, V. C., Chancey, S. T.,
Shan, L., Jamir, Y., Schechter, L. M., Janes, M. D., Buell, C. R.,
Tang, X., Collmer, A., and Alfano, J. R. (2002) Genome wide
identification of proteins secreted by the Hrp type III protein
secretion system of Pseudomonas syringae pv. tomato
DC3000, Proc. Natl. Acad. Sci. USA, 99, 7652-7657.
# Deslandes, L., Olivier, J., Peeters, N., Feng, D. X., Khounlotham,
M., Boucher, C., Somssich, I., Genin, S., and Marco, Y. (2003) Physical
interaction between RRS1-R, a protein conferring resistance to
bacterial wilt, and PopP2, a type III effector targeted to the plant
nucleus, Proc. Natl. Acad. Sci. USA, 100, 8024-8029.
# Arnold, D. L., Jackson, R. W., Fillingham, A. J., Gross, S. C.,
Taylor, J. D., Mansfield, J. W., and Vivian, A. (2001) Highly conserved
sequences flank avirulence genes: isolation of novel avirulence genes
from Pseudomonas syringae pv. pisi, J. Microbiol.,
147, 1171-1182.
# Hotson, A., and Mudgett, M. B. (2004)
Cysteine proteases in phytopathogenic bacteria: identification of plant
targets and activation of innate immunity, Curr. Opin. Plant.
Biol., 7, 384-390.
# Hotson, A., Chosed, R., Shu, H.,
Orth, K., and Mudgett, M. B. (2003) Xanthomonas type III
effector XopD targets SUMO-conjugated proteins in planta, Mol.
Microbiol., 50, 377-389.
# Shao, F., Merritt, P. M.,
Bao, Z., Innes, R. W., and Dixon, J. E. (2002) A Yersinia
effector and a Pseudomonas avirulence protein define a family of
cysteine proteases functioning in bacterial pathogenesis, Cell,
109, 575-588.
# Shao, F., Golstein, C., Ade, J.,
Stoutemyer, M., Dixon, J. E., and Innes, R. W. (2003) Cleavage of
Arabidopsis PBS1 by a bacterial type III effector,
Science, 301, 1230-1233.
# Lopez-Solanilla, E.,
Bronstein, P. A., Schneider, A. R., and Collmer, A. (2004) HopPtoN is a
Pseudomonas syringae Hrp (type III secretion system) cysteine
protease effector that suppresses pathogen-induced necrosis associated
with both compatible and incompatible plant interactions, Mol.
Microbiol., 54, 353-365.
# Gottfert, M.,
Rothlisberger, S., Kundig, C., Beck, C., Marty, R., and Hennecke, H. J.
(2001) Potential symbiosis-specific genes uncovered by sequencing a
410-kilobase DNA region of the Bradyrhizobium japonicum
chromosome, Bacteriology, 183, 1405-1412.
# Kaneko,
T., Nakamura, Y., Sato, S., Minamisawa, K., Uchiumi, T., Sasamoto, S.,
Watanabe, A., Idesawa, K., Iriguchi, M., Kawashima, K., Kohara, M.,
Matsumoto, M., Shimpo, S., Tsuruoka, H., Wada, T., Yamada, M., and
Tabata, S. (2002) Complete genomic sequence of nitrogen-fixing
symbiotic bacterium Bradyrhizobium japonicum USDA110, DNA
Res., 9, 189-197.
# Axtell, M. J., Chisholm, S.,
Dahlbeck, D., and Staskawicz, B. J. (2003) Genetic and molecular
evidence that the Pseudomonas syringae type III effector protein
AvrRpt2 is a cysteine protease, Mol. Microbiol., 49,
1537-1546.
# Zhu, M., Shao, F., Innes, R. W., Dixon, J. E., and
Xu, Z. (2004) The crystal structure of Pseudomonas avirulence
protein AvrPphB: a papain-like fold with a distinct substrate-binding
site, Proc. Natl. Acad. Sci. USA, 101, 302-307.
#
Mudgett, M. B. (2005) New insights to the function of phytopathogenic
bacterial type III effectors in plants, Annu. Rev. Plant Biol.,
56, 509-531.
# Xia, Y. (2004) Proteases in pathogenesis
and plant defense, Cell Microbiol., 6, 905-913.
#
Mudgett, M. B., and Staskawicz, B. J. (1999) Characterization of the
Pseudomonas syringae pv. tomato AvrRpt2 protein:
demonstration of secretion and processing during bacterial
pathogenesis, Mol. Microbiol., 32, 927-941.
# Jin,
P., Wood, M. D., Wu, Y., Xie, Z., and Katagiri, F. (2003) Cleavage of
the Pseudomonas syringae type III effector AvrRpt2 requires a
host factor(s) common among eukaryotes and is important for AvrRpt2
localization in the host cell, Plant Physiol., 133,
1072-1082.
# Mackey, D., Belkhadir, Y., Alonso, J. M., Ecker, J.
R., and Dangl, J. L. (2003) Arabidopsis RIN4 is a target of the
type III virulence effector AvrRpt2 and modulates RPS2-mediated
resistance, Cell, 112, 379-389.
# Coake, G.,
Falick, A., and Staskawicz, B. J. (2005) Activation of a
phytopathogenic bacterial effector protein by a eukaryotic cyclophilin,
Science, 308, 548-550.
# Orth, K., Xu, Z., Mudgett,
M. B., Bao, Z. Q., Palmer, L. E., Bliska, J. B., Mangel, W. F.,
Staskawicz, B., and Dixon, J. E. (2000) Disruption of signaling by
Yersinia effector YopJ, a ubiquitin-like protein protease,
Science, 290, 1594-1597.
# Roden, J., Eardley, L.,
Hotson, A., Cao, Y., and Mudgett, M. B. (2004) Characterization of the
Xanthomonas AvrXv4 effector, a SUMO protease translocated into
plant cells, Mol. Plant–Microbe Interact., 17,
633-643.
# Noel, L., Thieme, F., Nennstiel, D., and Bonas, U.
(2002) Two novel type III-secreted proteins of Xanthomonas
campestris pv. vesicatoria are encoded within the hrp
pathogenicity island, J. Bacteriol., 184, 1340-1348.
# Kurepa, J., Walker, J. M., Smalle, J., Gosink, M. M., Davis, S.
J., Durham, T. L., Sung, D. Y., and Vierstra, R. D. (2003) The small
ubiquitin-like modifier (SUMO) protein modification system in
Arabidopsis. Accumulation of SUMO1 and -2 conjugates is
increased by stress, J. Biol. Chem., 278, 6862-6872.
# Deslandes, L., Olivier, J., Theulieres, F., Hirsh, J., Feng, D.
X., Bittner-Eddy, P., Beynon, J., and Marco, Y. (2002) Resistance to
Ralstonia solanacearum in Arabidopsis thaliana is
conferred by the recessive RRS1-R gene, a member of a novel family of
resistance genes, Proc. Natl. Acad. Sci. USA, 99,
2404-2409.
# Orbach, M. J., Farrall, L., Sveigard, J. A.,
Chumley, F. G., and Valent, B. (2000) A telomeric avirulence gene
determines efficacy for the rice blast resistance gene Pi-ta, Plant
Cell, 12, 2019-2032.
# Jia, Y., McAdams, S. A., Bryan,
G. T., Hershey, H. P., and Valent, B. (2000) Direct interaction of
resistance gene and avirulence gene products confers rice blast
resistance, EMBO J., 19, 4004-4014.
# Axtell, M.
J., and Staskawicz, B. J. (2003) Initiation of RPS2-specified disease
resistance in Arabidopsis is coupled to the AvrRpt2-directed
elimination of RIN4, Cell, 112, 369-377.
# El
Hadrami, A., El Hadrami, I., and Daayf, F. (2009) Suppression of
induced plant defense responses by fungal and oomycete pathogens, in:
Molecular Plant–Microbe Interactions (Bouarab, K.,
Brisson, N., and Daayf, F., eds.) CABI, pp. 231-268.
# Knogge,
W., and Scheel, D. (2006) LysM receptors recognize friend and foe,
Proc. Natl. Acad. Sci. USA, 103, 10829-10830.
#
Stergiopoulos, I., and de Wit, P. J. G. M. (2009) Fungal effector
proteins, Annu. Rev. Phytopathol., 47, 233-263.
#
Stergiopoulos, I., van den Burg, H. A., Ökmen, B., Beenen, H. G.,
van Liere, S., Kema, G. H. J., and de Wit, P. J. (2010) Tomato Cf
resistance proteins mediate recognition of cognate homologous effectors
from fungi pathogenic on dicots and monocots, Proc. Natl. Acad. Sci.
USA, 107, 7610-7615.
# Win, J., Morgan, W., Bos, J.,
Krasileva, K. V., Cano, L. M., Chaparro-Garcia, A., Ammar, R.,
Staskawicz, B. J., and Kamoun, S. (2007) Adaptive evolution has
targeted the C-terminal domain of the RXLR effectors of plant
pathogenic oomycetes, Plant Cell, 19, 2349-2369.
#
Thatcher, L. F., Manners, J. M., and Kazan, K. (2009) Fusarium
oxysporum hijacks COI1-mediated jasmonate signaling to promote
disease development in Arabidopsis, Plant J., 58,
927-939.
# El Oirdi, M., and Bouarab, K. (2007) Plant signaling
components EDS1 and SGT1 enhance disease caused by the necrotrophic
pathogen Botrytis cinerea, New Phytol., 175,
131-139.
# El Oirdi, M., Abd El Rahman, T., Rigano, L., El
Hadrami, A., Rodriguez, M., Daayf, F., Vojnov, A., and Bouarab, K.
(2011) Botrytis cinerea manipulates the antagonistic effects
between immune pathways to promote disease development in tomato,
Plant Cell, 23, 2405-2421.
# Van den Burg,
H. A., Harrison, S. J., Joosten, M. H., Vervoort, J., and de Wit, P. J.
(2006) Cladosporium fulvum Avr4 protects fungal cell walls
against hydrolysis by plant chitinases accumulating during infection,
Mol. Plant–Microbe Interact., 19, 1420-1430.
# De
Wit, P. J., van der Burg, A., Ökmen, B., Stergiopoulos, I.,
Abd-Elsalam, K. A., Aerts, A. L., Bahkali, A. H., Beenen, H. G.,
Chettri, P., Cox, M. P., Datema, E., de Vries, R. P., Dhillon, B.,
Ganley, A. R., Griffiths, S. A., Guo, Y., Hamelin, R. C., Henrissat,
B., Kabir, M. S., Jashni, M. K., Kema, G., Klaubauf, S., Lapidus, A.,
Levasseur, A., Lindquist, E., Mehrabi, R., Ohm, R. A., Owen, T. J.,
Salamov, A., Schwelm, A., Schijlen, E., Sun, H., van den Burg, H. A.,
van Ham, R. C., Zhang, S., Goodwin, S. B., Grigoriev, I. V., Collemare,
J., and Bradshaw, R. E. (2012) The genomes of the fungal plant
pathogens Cladosporium fulvum and Dothistroma septosporum
reveal adaptation to different hosts and lifestyles but also signatures
of common ancestry, PLoS Genet., 8, e1003088.
# Mesarich,
C. H., Stergiopoulos, I., Beenen, H. G., Cordovez, V., Guo, Y., Karimi
Jashni, M., Bradshaw, R. E., and de Wit, P. J. (2016) A conserved
proline residue in Dothideomycete Avr4 effector proteins is
required to trigger a Cf-4-dependent hypersensitive response, Mol.
Plant Pathol., 17, 84-95.
# Lange, J., Mohr U.,
Wiemken, A., Boller, T., and Vögeli-Lange, R. (1996) Proteolytic
processing of class IV chitinase in the compatible interaction of bean
roots with Fusarium solani, Plant Physiol., 111,
1135-1144.
# Olivieri, F., Zanetti, E. M., Oliva, C. R.,
Covarrubias, A. A., and Casalongué, C. A. (2002)
Characterization of an extracellular serine protease of Fusarium
eumartii and its action on pathogenesis related proteins, Eur.
J. Plant Pathol., 108, 63-72.
# Naumann, T. A. (2011)
Modification of recombinant maize Chit A chitinase by fungal
chitinase-modifying proteins, Mol. Plant Pathol., 12,
365-372.
# Naumann, T. A., Wicklow, D. T., and Price, N. P. J.
(2014) Polyglycine hydrolases secreted by Pleosporineae fungi
that target the linker region of plant class IV chitinases, Biochem.
J., 460, 187-198.
# Naumann, T. A., Naldrett, M. J.,
Ward, T. J., and Price, N. P. J. (2015) Polyglycine hydrolases: fungal
β-lactamase-like endoproteases that cleave polyglycine regions
within plant class IV chitinases, Protein Sci., 24,
1147-1157.
# Jashni, M. K., Mehrabi, R., Collemare, J.,Mesarich,
C. H., and de Wit, P. J. G. M. (2015) The battle in the apoplast:
further insights into the roles of proteases and their inhibitors in
plant–pathogen interactions, Front. Plant Sci., 6,
584.
# Hörger, A. C., and van der Hoorn, R. A. L. (2013) The
structural basis of specific protease–inhibitor interactions at
the plant–pathogen interface, Curr. Opin. Struct. Biol.,
23, 842-850.
# Ohm, R. A., Feau, N., Henrissat, B.,
Schoch, C. L., Horwitz, B. A., Barry, K. W. Condon, B. J., Copeland, A.
C., Dhillon, B., Glaser, F., Hesse, C. N., Kosti, I., LaButti, K.,
Lindquist, E. A., Lucas, S., Salamov, A. A., Bradshaw, R. E.,
Ciuffetti, L., Hamelin, R. C., Kema, G. H., Lawrence, C., Scott, J. A.,
Spatafora, J. W., Turgeon, B. G., de Wit, P. J., Zhong, S., Goodwin, S.
B., and Grigoriev, I. V. (2012) Diverse lifestyles and strategies of
plant pathogenesis encoded in the genomes of eighteen
Dothideomycetes fungi, PLoS Pathog., 8,
e1003037.
# Valueva, T. A., Kudryavtseva, N. N., Sof’in, A.
V., Zaitchik, B. T., Pobedinskaya, M. A., Kokaeva, L. Y., and Elansky,
S. N. (2015) Serine exoproteinases secreted by the pathogenic fungi of
Alternaria genus, J. Plant Pathol. Microb., 6,
272-279.
# Van der Burgt, A., Karimi, J. M., Bahkali, A. H., and
de Wit, P. J. (2014) Pseudogenization in pathogenic fungi with
different host plants and lifestyles might reflect their evolutionary
past, Mol. Plant Pathol., 15, 133-144.
# Dong, S.,
Stam, R., Cano, L. M., Song, J., Sklenar, J., Yoshida, K., Bozkurt, T.
O., Oliva, R., Liu, Z., Tian, M., Win, J., Banfield, M. J., Jones, A.
M., van der Hoorn, R. A., and Kamoun, S. (2014) Effector specialization
in a lineage of the Irish potato famine pathogen, Science,
343, 552-555.
# Plummer, K. M., Clark, S. J., Ellis, L.
M., Loganathan, A., Al-Samarrai, T. H., Rikkerink, E. H. A., Sullivan,
P. A., Templeton, M. D., and Farley, P. C. (2004) Analysis of a
secreted aspartic peptidase disruption mutant of Glomerella
cingulata, Eur. J. Plant Pathol., 110, 265-274.
# Alkan, N., Friedlander, G., Ment, D., Prusky, D., and Fluhr, R.
(2015) Simultaneous transcriptome analysis of Colletotrichum
gloeosporioides and tomato fruit pathosystem reveals novel fungal
pathogenicity and fruit defense strategies, New Phytol.,
205, 801-815.
# Lee, S.-J., and Rose, J. K. C. (2010)
Mediation of the transition from biotrophy to necrotrophy in
hemibiotrophic plant pathogens by secreted effector proteins, Plant
Signal. Behav., 5, 769-772.
# Alkan, N., Espeso, E.
A., and Prusky, D. (2013) Virulence regulation of phytopathogenic fungi
by pH, Antioxid. Redox Signal., 19, 1012-1025.
#
Alkan, N., Meng, X., Friedlander, G., Reuveni, E., Sukno, S., Sherman,
A., Thon, M., Fluhr, R., and Prusky, D. (2013) Global aspects of pacC
regulation of pathogenicity genes in Colletotrichum
gloeosporioides as revealed by transcriptome analysis, Mol.
Plant–Microbe Interact., 26, 1345-1358.
#
Miyara, I., Shafran, H., Haimovich, H. K., Rollins, J., Sherman, A.,
and Prusky, D. (2008) Multifactor regulation of pectate lyase secretion
by Colletotrichum gloeosporioides pathogenic on avocado fruits,
Mol. Plant Pathol., 9, 281-291.
# Miyara, I.,
Shafran, H., Davidzon, M., Sherman, A., and Prusky, D. (2010) pH
regulation of ammonia secretion by Colletotrichum
gloeosporioides and its effect on appressorium formation and
pathogenicity, Mol. Plant–Microbe Interact., 23,
304-316.
# Maccheroni, W., Jr., Araújo, W. L., and
Azevedo, J. L. (2004) Ambient pH-regulated enzyme secretion in
endophytic and pathogenic isolates of the fungal genus
Colletotrichum, Scientia Agricola, 61, 298-302.
# Kudryavtseva, N. N., Gvozdeva, E. L., Sof’in, A. V., and
Valueva, T. A. (2010) The influence of cultural medium composition on
the proteolytic enzyme secretion of fungus Rhizoctonia solani,
Appl. Biochem. Microbiol., 46, 324-330.
# Mosolov,
V. V., and Valueva, T. A. (2006) Participation of proteolytic enzymes
in the interaction of plants with phytopathogenic microorganisms,
Biochemistry (Moscow), 71, 838-845.
#
Kramer-Haimovich, H., Servi, E., Katan, T., Rollins, J., Okon, Y., and
Prusky, D. (2006) Effect of ammonia production by Colletotrichum
gloeosporioides on pelB activation, pectate lyase secretion, and
fruit pathogenicity, Appl. Environ. Microbiol., 72,
1034-1039.
# Miyara, I., Shnaiderman, C., Meng, X., Vargas, W.
A., Diaz-Minguez, J. M., Sherman, A., Thon, M., and Prusky, D. (2012)
Role of nitrogen-metabolism genes expressed during pathogenicity of the
alkalinizing Colletotrichum gloeosporioides and their
differential expression in acidifying pathogens, Mol.
Plant–Microbe Interact., 25, 1251-1263.
# Alkan,
N., Fluhr, R., Sherman, A., and Prusky, D. (2008) Role of ammonia
secretion and pH modulation on pathogenicity of Colletotrichum
coccodes on tomato fruit, Mol. Plant–Microbe
Interact., 21, 1058-1066.
# Alkan, N., Davydov, O.,
Sagi, M., Fluhr, R., and Prusky, D. (2009) Ammonium secretion by
Colletotrichum coccodes activates host NADPH oxidase activity
enhancing host cell death and fungal virulence in tomato fruits,
Mol. Plant–Microbe Interact., 22, 1484-1491.
# Shnaiderman, C., Miyara, I., Kobiler, I., Sherman, A., and Prusky, D.
(2013) Differential activation of ammonium transporters during the
accumulation of ammonia by Colletotrichum gloeosporioides and
its effect on appressoria formation and pathogenicity, Mol.
Plant–Microbe Interact., 26, 345-355.
#
Inguagiato, J. C., Murphy, J. A., and Clarke, B. B. (2008) Anthracnose
severity on annual bluegrass influenced by nitrogen fertilization,
growth regulators, and verticutting, Crop Sci., 48,
1595-1607.
# Donofrio, N. M., Oh, Y., Lundy, R., Pan, H., Brown,
D. E., Jeong, J. S., Coughlan, S., Mitchell, T. K., and Dean, R. A.
(2006) Global gene expression during nitrogen starvation in the rice
blast fungus, Magnaporthe grisea, Fungal Genet. Biol.,
43, 605-617.
# Perez-Garcia, A., Snoeijers, S. S.,
Joosten, M. H. A. J., Goosen, T., and De Wit, P. J. G. M. (2001)
Expression of the avirulence gene Avr9 of the fungal tomato pathogen
Cladosporium fulvum is regulated by the global nitrogen response
factor NRF1, Mol. Plant–Microbe Interact., 14,
316-325.
# Stephenson, S.-A., Hatfield, J., Rusu, A. G., Maclean,
D. J., and Manners, J. M. (2000) CgDN3: an essential pathogenicity gene
of Colletotrichum gloeosporioides necessary to avert a
hypersensitive-like response in the host Stylosanthes
guianensis, Mol. Plant–Microbe Interact., 13,
929-941.
# Talbot, N. J., Ebbole, D. J., and Hamer, J. E. (1993)
Identification and characterization of MPG1, a gene involved in
pathogenicity from the rice blast fungus Magnaporthe grisea,
Plant Cell, 5, 1575-1590.
# Talbot, N. J., and
Foster, A. J. (2001) Genetics and genomics of the rice blast fungus
Magnaporthe grisea: developing an experimental model for
understanding fungal diseases of cereals, Adv. Bot. Res.,
34, 263-287.
# Pellier, A.-L., Lauge, R.,
Veneault-Fourrey, C., and Langin, T. (2003) CLNR1, the AREA/NIT2-like
global nitrogen regulator of the plant fungal pathogen
Colletotrichum lindemuthianum is required for the infection
cycle, Mol. Microbiol., 48, 639-655.
# Dunaevskii,
Ya. E., Matveeva, A. R., Fatkhullina, G. N., Belyakova, G. A., and
Belozerskii, M. A. (2008) Extracellular proteases of mycelial fungi as
participants of pathogenic process, Russ. J. Bioorg. Chem.,
34, 286-289.
# Valueva, T. A., Kudryavtseva, N .N.,
Gvozdeva, E. L., Sof’in, A. V., Iľina, N. A., Pobedinskaya,
M. A., and Elansky, S. N. (2013) Serine proteinases secreted by two
isolates of the fungus Alternaria solani, J. Basic. App.
Sci., 9, 105-115.
# Valueva, T. A., Kudryavtseva, N.
N., Sof’in, A. V., Revina, T. A., Gvozdeva, E. L., and Ievleva,
E. V. (2011) Comparative analyses of exoproteinases produced by three
phytopathogenic microorganisms, J. Pathog., 2011,
947218.
# Bryan, G. T., Wu, K.-S., Farrall, L., Jia, Y., Hershey,
H. P., McAdams, S. A., Faulk, K. N., Donaldson, G. K., Tarchini, R.,
and Valent, B. (2000) A single amino acid difference distinguishes
resistant and susceptible alleles of the rice blast resistance gene
Pi-ta, Plant Cell, 12, 2033-2045.
# Yike, I. (2011)
Fungal proteases and their pathophysiological effects,
Mycopathology, 171, 299-323.
# Kokaeva, L. Yu.,
Kudryavtseva, N. N., Pobedinskaya, M. A., Zaichik, B. T., and Elanskii,
S. N. (2015) Virulence of Alternaria alternata strains to
different potato and tomato cultivars, Zashchita Kartofelya,
1, 14-18.
# Kudryavtseva, N. N., Sof’in, A. V.,
Revina, T. A., Gvozdeva, E. L., Ievleva, E. V., and Valueva, T. A.
(2013) Secretion of proteolytic enzymes by three phytopathogenic
microorganisms, Appl. Biochem. Microbiol., 49,
514-520.
# Chand, R., Kumar, M., Kushwaha, C., Shah, K., and
Joshi, A. K. (2014) Role of melanin in release of extracellular enzymes
and selection of aggressive isolates of Bipolaris sorokiniana in
barley, Curr. Microbiol., 69, 202-211.
#
Chandrasekaran, M., Thangavelu, B., Chun, S. Ch., and Sathiyabama, M.
J. (2016) Proteases from phytopathogenic fungi and their importance in
phytopathogenicity, J. Gen. Plant Pathol., 82,
233-239.
# Oh, Y., Donofrio, N., Pan, H., Coughlan, S., Brown,
D., Meng, S., Mitchell, T., and Dean, R. A. (2008) Transcriptome
analysis reveals new insight into appressorium formation and function
in the rice blast fungus Magnaporthe oryzae, Genome
Biol., 9, R85.
# Dubovenko, A. G., Dunaevsky, Y. E.,
Belozersky, M. A., Oppert, B., Lord, J. C., and Elpidina, E. N. (2010)
Trypsin-like proteins of the fungi as possible markers of
pathogenicity, Fungal Biol., 114, 151-159.
#
Dunaevskii, Ya. E., Gruban, T. N., Belyakova, G. A., and Belozerskii,
M. A. (2006) Extracellular proteinases of filamentous fungi as
potential markers of phytopathogenesis, Microbiology, 75,
649-652.