New Fluorescent Macrolide Derivatives for Studying Interactions of Antibiotics and Their Analogs with the Ribosomal Exit Tunnel
A. G. Tereshchenkov1, A. V. Shishkina2, V. V. Karpenko1, V. A. Chertkov1, A. L. Konevega3,4, P. S. Kasatsky3,4, A. A. Bogdanov1,2 and N. V. Sumbatyan1*
1Lomonosov Moscow State University, Faculty of Chemistry, 119991 Moscow, Russia; E-mail: sumbtyan@belozersky.msu.ru2Lomonosov Moscow State University, Belozersky Institute of Physico-Chemical Biology, 119991 Moscow, Russia
3Molecular and Radiation Biophysics Division, Konstantinov Petersburg Nuclear Physics Institute, National Research Center “Kurchatov Institute”, 188300 Gatchina, Russia
4Peter the Great Saint-Petersburg Polytechnic University, 195251 St. Petersburg, Russia
* To whom correspondence should be addressed.
Received June 21, 2016; Revision received July 18, 2016
Novel fluorescent derivatives of macrolide antibiotics related to tylosin bearing rhodamine, fluorescein, Alexa Fluor 488, BODIPY FL, and nitrobenzoxadiazole (NBD) residues were synthesized. The formation of complexes of these compounds with 70S E. coli ribosomes was studied by measuring the fluorescence polarization depending on the ribosome amount at constant concentration of the fluorescent substance. With the synthesized fluorescent tylosin derivatives, the dissociation constants for ribosome complexes with several known antibiotics and macrolide analogs previously obtained were determined. It was found that the fluorescent tylosin derivatives containing BODIPY FL and NBD groups could be used to screen the binding of novel antibiotics to bacterial ribosomes in the macrolide-binding site.
KEY WORDS: macrolides, tylosin, fluorescent derivatives, nascent peptide exit tunnel, fluorescent polarizationDOI: 10.1134/S0006297916100138
Abbreviations: Aoc, (aminooxy)acetic acid; Boc, tert-butyloxycarbonyl; BODIPY, 4,4-difluoro-5,7-dimethyl-4-bora-3a,4a-diaza-s-indacene-3-pentanoic acid (BODIPY FL C5); DCC, 1,3-dicyclohexylcarbodiimide; Des, desmycosin; DIPEA, diisopropylethylamine; Ery, erythromycin; LC-MS, chromato-mass-spectrometry; NBD, 7-nitro-2,1,3-benzoxadiazole-4-yl; OMT, 5-O-mycaminosyltylonolide; RT, ribosomal tunnel; TFA, trifluoroacetic acid; Tyl, tylosin.
The field of study of antibiotics whose bioactivity is mediated by
interaction with bacterial ribosomes expands in terms of both research
and development of new compounds, including those capable of
suppressing resistant bacterial strains, and investigation of ribosome
functions and regulation mechanisms using known antibiotics and their
analogs [1-3]. One of the first
steps in applied and fundamental research is to determine the ribosome
binding ability of a potential antibiotic (ligand).
Various experimental techniques are employed for studying ribosome interactions with small ligands, in particular antibiotics. Such methods as equilibrium dialysis [4-7], gel-exclusion chromatography [8, 9], filter-binding assay [4, 7, 10], and sedimentation [7] are generally based on the use of radiolabeled compounds. Other methods like footprinting [11-13], cross-linking [14-17], peptidyl transferase activity inhibition, and puromycin reaction [18-20] are highly informative but laborious, requiring considerable time, frequently including radiolabeled compounds, and not applicable for screening.
A separate group comprises techniques using fluorescent ligands. Initially, the binding of antibiotics to ribosomes was determined by a change in fluorescence intensity that could be enhanced or quenched upon complexing with a fluorescent ligand and return when the ligand was displaced by antibiotics [21-24]. Recently, an original method was suggested to study binding of potential antibiotics and other ligands to ribosomes based on the inclusion of a site-specific fluorescent label directly into the ribosome protein structure near the intended ligand-binding site [25]. With the development of the measurement technique for quantitative evaluation of antibiotic–ribosome complexes, a method using fluorescence polarization (anisotropy) has been proposed, which is widely applied for determination of the binding constants and studying the kinetics of reactions accompanied by the change in molecule rotation rate when fluorescently labeled compounds react with macromolecules [26]. The approach is based on the fact that the measured fluorescence anisotropy level is proportional to the share of the fluorescent derivative bound to a macromolecule. Since the bacterial ribosome has a huge molecular mass (2.5 MDa), its binding to a small fluorescently labeled compound results in a significant change in fluorescence anisotropy values, thus defining the high sensitivity of the method. Moreover, binding or displacement curves can be obtained without separation of bound and free fluorescent ligand. In addition, since fluorescence anisotropy despite its intensity is a ratiometric measurement, it is less sensitive to such interference factors as light scattering, fluorescence quenching, excitation light shielding by other molecules, energy migration, etc. This assay was applied for screening in the search of compounds capable of displacing a fluorescently labeled antibiotic from its complexes with 70S E. coli ribosomes [27]. Later, erythromycin-based fluorescent compounds bearing a BODIPY FL fluorescent label in different positions of the macrolide were synthesized [28, 29]. One of these substances, BODIPY-9-aminoerythromycin, has been applied for the detection of macrolide and chloramphenicol derivatives binding with E. coli ribosomes by displacement of the fluorescent erythromycin from its ribosomal complexes [28-32].
In the present work, new fluorescent derivatives based on tylosin and related 16-membered macrolides were proposed and synthesized; the dissociation constants of complexes of these compounds with E. coli ribosomes were measured using fluorescence polarization assay; the binding constants of known ribosomal antibiotics and previously synthesized macrolide analogs with bacterial ribosomes were determined with the new fluorescent compounds.
Rhodamine, fluorescein, Alexa Fluor 488, BODIPY FL, and nitrobenzoxadiazole (NBD) were chosen as fluorescent labels (Fig. 1), these varying in structure, fluorescent properties (excitation and emission wavelengths, fluorescence intensity, quantum yield), and size.
MATERIALS AND METHODS
Chemicals used in this work were as follows: (Boc-aminooxy)acetic acid and NBD-Cl from Aldrich (Germany), rhodamine B from Sigma (USA), fluorescein thiosemicarbazide from Fluka (Switzerland), Alexa Fluor 488 hydrazide from Molecular Probes (USA), BODIPY-OSu from Invitrogen (USA), and tylosin from Mosagrogen (Russia). Desmycosin [33] and erythromycin fluorescent derivative BODIPY-Ery [30] were synthesized earlier. The 50S ribosome subunits were isolated from E. coli strain BW25113 in accordance with a standard procedure [34]; 70S ribosomes from E. coli strain MRE-600 were prepared as previously described [35].
TLC was performed on Kieselgel 60 F254 plates from Merck (Germany), and column chromatography on Silica gel 60 (0.063-0.200 and 0.040-0.063 mm) from Merck. The compounds containing UV-absorbing groups were visualized on a UV cabinet Camag (England); the substances with free or Boc-protected amino groups were visualized by ninhydrin reagent; aldehyde groups were detected by 2,4-dinitrophenylhydrazine.
UV absorption spectra were registered using a Varian Cary 50 Bio spectrophotometer (Australia).
Analytical and preparative reverse phase HPLC was performed on a Knauer semipreparative chromatograph (Germany) with a Beckman Coulter Ultrasphere ODS (10 × 250 mm, 5 µm) column in a gradient of MeCN in aqueous solution of TFA (0.01%) with elution rate 5 ml/min.
MALDI-TOF mass spectra were recorded on an UltrafleXtreme MALDI-time-of-flight mass spectrometer (Bruker Daltonics, Germany) equipped with a UV laser (Nd) in the positive ion mode with a reflectron.
LC-MS (chromato-mass-spectrometry) was carried out with a UPLC/MS/MS system containing an Acquity UPLC System chromatograph (Waters, USA), Acquity BEH C18 (2.1 × 50 mm, 1.7 µm) column (3.55 ml/min, gradient 0-95% MeCN for 3 min), and TQD quadrupole mass spectrometer (Waters) (ESI ionization mode).
1H and 13C NMR spectra were registered on Bruker AM-300, Bruker DRX-500, and Bruker AV-600 NMR spectrometers (Bruker, USA) with 300, 500, and 600 MHz operating frequencies for 1H nuclei, correspondingly. The signal multiplicity in 13C spectra was determined with INEPT experiments. Coupled COSY NMR spectra were recorded in magnitude mode; coupled HSQC and HMBC NMR spectra were generated in phase-sensitive mode. The J-filter, optimal for 13C-1H correlation over one bond (about 135 Hz), was used for HSQC spectra registration. The low pass J-filter for long-range 13C-1H coupling constants (about 7 Hz) and refocusing 13C-1H constants over one bond were applied for HMBC experiments. For signal assignments, the following data were used: 13C NMR and coupled experiments COSY, HSQC, HMBC.
Fluorescence polarization was measured with a Varian Cary Eclipse fluorescence spectrophotometer (Australia) and VICTOR™ X5 Multilabel Plate Reader from Perkin Elmer (USA). The wavelengths were as follows: for BODIPY-Tyl and NBD-Tyl: λex = 485 nm, λem = 535 nm; for Flu-Tyl: λex = 492 nm, λem = 516 nm; for Alexa-Tyl: λex = 488 nm, λem = 512 nm; for Rho-Tyl and Rho-Des: λex = 535 nm, λem = 580 nm. The correcting G-factor was measured relative to fluorescein standard.
Synthesis of tylosin and desmycosin fluorescent derivatives.
For synthesis of Rho-Tyl (I), Rho-Des (II), Flu-Tyl (III), BODIPY-Tyl (IV), NBD-Tyl (V), Alexa-Tyl (VI), as well as rhodamine B hydrazide [36], N-(7-nitro-2,1,3-benzoxadiazol-4-yl)ethane-1,2-diamine (Va) [37], tert-butyloxycarbonyl-2-(aminooxy)-N-{2-[(7-nitro-2,1,3-benzoxadiazol-4-yl)amino]ethyl}acetamide (Vb), and 2-(aminooxy)-N-{2-[(7-nitro-2,1,3- benzoxadiazol-4-yl)amino]ethyl}acetamide trifluoroacetate (Vc), see the Supplement to this paper on the site of the journal (http://protein.bio.msu.ru/biokhimiya) and Springer site (Link.springer.com).
Study of ligand binding to bacterial E. coli ribosomes [28]. Ribosome concentration was determined by optical density at 260 nm (A260 = 1 a.u. at C(Rs) = 24 nM for 70S ribosomes and C(Rs) = 36 nM for 50S subunits). The 50S subunits and 70S ribosomes were thawed at 0°C, followed by incubation for 15 min at 37°C and dilution in BIND buffer with the following composition: 20 mM HEPES, pH 7.5, 50 mM NH4Cl, 10 mM MgCl2, 4 mM β-ME, 0.05% Tween 20. To obtain the binding curves, the fluorescently labeled compound (2.5-32 nM) was mixed with the ribosomes at different concentrations from 1 to 2000 nM and incubated at 25°C for 30 min to reach equilibrium. In ligand displacement experiments, the fluorescently labeled tylosin solutions (4-16 nM) were mixed with the ribosome solution (15-30 nM) in a 384-well plate and incubated at 25°C for 30 min. The solution of rival ligand at concentrations from 1 nM to 1 mM was added to the formed ribosomal complex and incubated for 3 h. Then the values of fluorescence polarization were measured and converted to anisotropy according to the formula [38]:
where A – fluorescence anisotropy, P – fluorescence polarization.
Data analysis. The binding assay data of fluorescent derivatives with ribosomes were approximated by the quadratic equation describing equilibrium binding of labeled ligand [39]:
where A – measured anisotropy, A0 – anisotropy level with non-bound ligand, Amax – anisotropy corresponding to total binding, [R]0 – ribosome concentration, [B]0 – labeled ligand concentration, KDRB – the dissociation constant of the fluorescent derivative.
In the experiments of labeled compound displacement, the data were approximated by a function that is the solution of the cubic equation corresponding to the equilibrium competitive binding of two ligands in one binding site [40]:
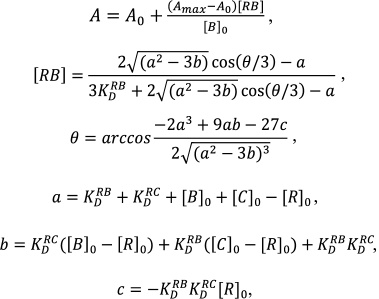 (3)
(3)where [C]0 – non-labeled compound concentration, KDRC – the dissociation constant of its complex with the ribosome, [RB] – the concentration of complex of labeled compound with the ribosome. The data were processed using GraphPad Prizm 6 software.
RESULTS
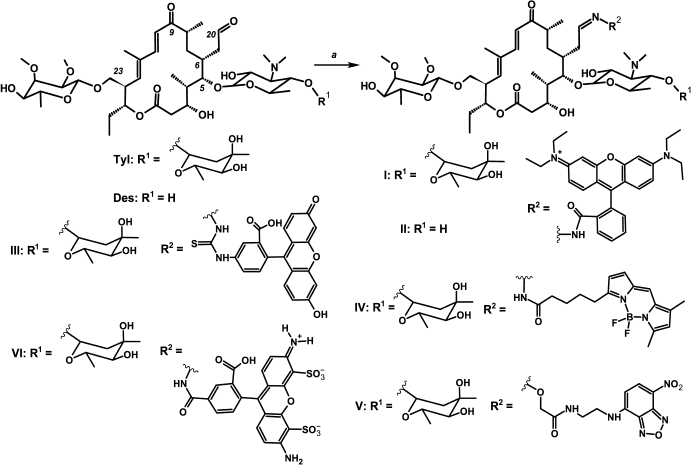
Synthesis. The C20 aldehyde group was chosen for introduction of fluorescent labels into the tylosin-related antibiotics (Fig. 1 and Fig. S1 in Supplement), since it can be modified in one step under mild conditions with good yield without any preliminary protection of functional groups of the antibiotic [33, 41]. The fluorescent dyes were modified by appropriate reagents so that they could interact with the tylosin or desmycosin aldehyde group, giving hydrazones, oximes, or thiosemicarbazones. Thus, Rho-tylosin (I) and Rho-desmycosin (II) were obtained by a reaction of previously synthesized rhodamine hydrazide [36] with the corresponding antibiotic. Fluorescein thiosemicarbazide was taken as a starting reagent for Flu-Tyl (III) synthesis. BODIPY FL C5 hydrazide was obtained from commercial BODIPY FL C5 succinimide ester for the synthesis of BODIPY-Tyl (IV). For tylosin modification resulting in NBD-Tyl (V), the NBD-derivative of (aminooxy)acetic acid (Vc) was used. Its structure was chosen so that compound V should be similar in spatial structure to the phenylalanine derivative of tylosin Phe-Tyl (VII; Fig. 2), which exhibited excellent inhibitory activity and good binding to bacterial ribosomes as we have shown previously [10]. Compound Vc was obtained via three stages from NBD-Cl and ethylenediamine with conjugation of the resulting NBD-ethylenediamine (Va) with (Boc-aminooxy)acetic acid giving Vb and subsequent amino group deblocking. The tylosin derivative bearing Alexa Fluor 488 residue (compound VI) was obtained as the substituted hydrazone. All reactions giving fluorescent macrolide derivatives were accomplished in acetate buffer at 25°C or heating to 50°C. Because of the low solubility in buffer of the starting fluorescent compounds, such solvents as DMSO, DMF, and THF were added to the reaction mixture if needed.
Fluorescent derivatives of macrolide antibiotics were purified by column chromatography and characterized by fluorescence and UV-spectrometry, LC-MS, and NMR spectroscopy. The stability of the compounds in the buffer used for ribosome binding assay was confirmed by HPLC analysis. The substances were stable at least for a day at 25°C.
The proof of the introduction of the fluorescent residue at the C20 position was based on structural analysis of compounds I, IV, V (1H and 13C NMR, COSY, HSQC, HMBC): in proton spectra the absence of a signal characteristic for the aldehyde proton in the 9.6-9.7 ppm region was observed, and the appearance of a broadened multiplet signal in 7.4-8.2 ppm characteristic for imine compounds –CH=N– appeared. As well as for other C20-derivatives of tylosin related antibiotics [33], one set of signals was observed in NMR spectra, and we therefore concluded that only one of the possible isomers of the –CH=N-bond (anti-isomer [33]) was synthesized.
Fig. 1. Scheme of synthesis of fluorescent derivatives of macrolide antibiotics. Rho-Tyl (I) a – rhodamine B hydrazide, 0.4 M acetate buffer (pH 4.7), DMSO, 25°C, 12 h; Rho-Des (II) a – rhodamine B hydrazide, 0.4 M acetate buffer (pH 4.7), DMSO, 25°C, 12 h; Flu-Tyl (III) a – fluorescein-5-thiosemicarbazide, 0.4 M acetate buffer (pH 4.7), DMSO, 50°C, 12 h; BODIPY-Tyl (IV) a – BODIPY FL C5 hydrazide, 0.4 M acetate buffer (pH 5.7), DMF, 25°C, 0.5 h; NBD-Tyl (V) a – Vc, 0.4 M acetate buffer (pH 5.7), DMSO, 40°C, 17 h; Alexa-Tyl (VI) a – Alexa Fluor 488, 0.4 M acetate buffer (pH 4.7), 50°C, 24 h.
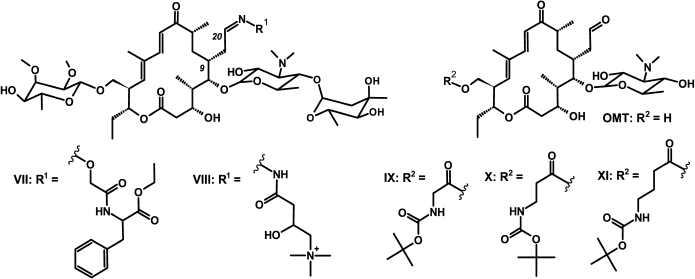
Fig. 2. Structures of amino acid derivatives of tylosin [10, 33] and 5-O-mycaminosyltylonolide (OMT) [30].
Binding assay of fluorescent derivatives of tylosin and desmycosin with E. coli ribosomes. Primarily for quantitative characterization of the binding of the synthesized fluorescent macrolide derivatives with E. coli ribosomes, the time required to reach equilibrium of complexes was determined. For this purpose, during one hour the fluorescence polarization level of a mixture containing ribosomes and fluorescently labeled antibiotics at concentrations equivalent about to 50% ligand binding was measured. It was found that equilibrium was established in 5-10 min and that the resulting complex was stable for several hours.
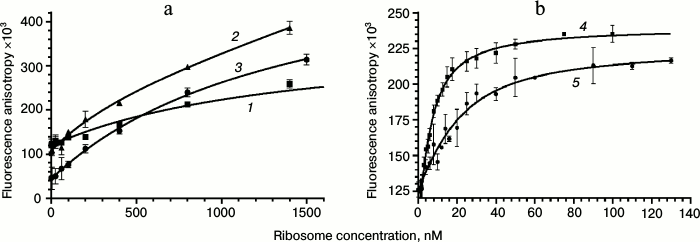
For obtaining binding isotherms, the solutions of fluorescent derivatives were titrated by 50S ribosome subunits (for Rho-Tyl (I), Rho-Des (II), Flu-Tyl (III), NBD-Tyl (V), and Alexa-Tyl (VI)) or 70S ribosomes (for BODIPY-Tyl (IV) and NBD-Tyl (V)) in the concentration range from 1 to 2000 nM. The mixture was incubated for 30 min at 25°C to reach equilibrium, and the values of fluorescence polarization were measured and converted to anisotropy. The binding isotherms were approximated by quadratic Eq. (2) corresponding to equilibrium binding of labeled ligand to the receptor without any assumptions (Fig. 3). This model was chosen because the concentrations of fluorescent compounds used were close to the putative KD values of their complexes with ribosomes. Fluorescent derivative Alexa-Tyl (VI) did not show any affinity to the bacterial ribosomes. The affinity of Rho-Tyl (I), Rho-Des (II), and Flu-Tyl (III) to 50S ribosomal subunits appeared to be quite low, the KD values of their complexes being in the micromolar range: 1.3 ± 0.5, 0.8 ± 0.3, and 1.5 ± 0.3 µM, correspondingly – three orders of magnitude greater than the KD of the complex of tylosin with ribosomes [10]. For the compounds that revealed the highest affinity, BODIPY-Tyl (IV) and NBD-Tyl (V), binding isotherms were measured at various concentrations of fluorescent derivatives in the ranges 2.5-20 and 4-32 nM, correspondingly. The resulting KD values turned to be similar, so their average values were taken: 3.1 ± 0.9 nM for BODIPY-Tyl (IV) and 6 ± 3 nM for NBD-Tyl (V). The affinity of these fluorescent compounds to ribosomes appeared to be close to that of tylosin, allowing their use in further experiments on the competitive displacement of macrolides. It is worth mentioning that unlike BODIPY-Tyl (IV), while NBD-Tyl (V) binding to ribosomes we observed an increase in the fluorescence level. For the used concentrations of NBD-Tyl (V), the enhancement comprised 20-25% (data not shown), which can be attributed to a change in fluorophore environment and its interaction with the nucleotide residues of the ribosomal tunnel.
Fig. 3. Binding curves of fluorescently labeled macrolides with E. coli ribosomes. a: 1) Rho-Tyl (I); 2) Rho-Des (II); 3) Flu-Tyl (III); [B]0 = 10 nM. b: 4) BODIPY-Tyl (IV); [B]0 = 4 nM; 5) NBD-Tyl (V); [B]0 = 16 nM. For each point, the mean value with standard deviation is shown.
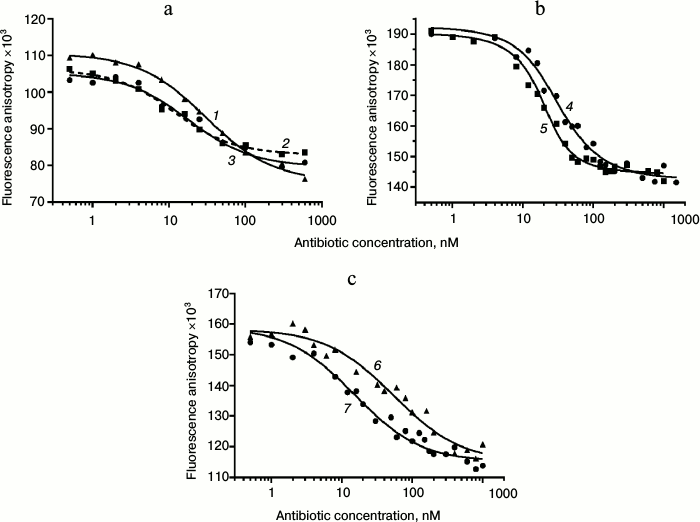
Displacement of BODIPY-Tyl and NBD-Tyl from their complexes with ribosomes by known antibiotics. Initially, the applicability of BODIPY-Tyl (IV) and NBD-Tyl (V) for ribosome binding assay was checked by competitive displacement of fluorescent ligands from their complexes with ribosomes. With this goal, the mentioned macrolide derivatives were used for determination of KD values of complexes of some well-known antibiotics, shown in Table 1, with E. coli ribosomes. Since tylosin, which serves a base for compounds IV and V, interacts with a rather vast ribosomal tunnel (RT) area, for this purpose we selected both macrolides (tylosin, erythromycin, clarithromycin, azithromycin, desmycosin and OMT) binding in the RT, and antibiotics (chloramphenicol, puromycin, and lincomycin) binding in the A site of the peptidyl transferase center. The time needed to reach equilibrium in the mixture of fluorescently labeled derivatives of tylosin, ribosomes and non-labeled antibiotic was in the range of 1 to 2 h. Thereafter, the fluorescence polarization values stay constant for at least 6 h. In displacement experiments, the concentration of macrolides was varied from 1 nM to 1 µM (Fig. 4 (a and b) and Fig. S2a in Supplement), whereas the concentration of poorly binding antibiotics, such as chloramphenicol, puromycin, and lincomycin were in the range 1-1000 µM (Fig. 4c and Fig. S2b in Supplement). Because the KD values of the chosen antibiotics and their concentrations in the experiments were close, the function corresponding to the exact solution (3) of the cubic equation, describing equilibrium binding of two ligands in a single site, was applied for data approximation. The calculated KD values are presented in Table 1. In the case of puromycin, partial displacement of fluorescent derivative was observed (Fig. S2b in Supplement), which may be due to the weak overlap of binding sites of puromycin and tylosin (KD value not determined). The KD values for tylosin, as well as for erythromycin, that were obtained using compounds IV and V, proved quite close. For comparison, Table 1 shows the literature binding data for antibiotics with bacterial ribosomes. For almost all antibiotics except azithromycin and desmycosin, the constants are in the value ranges given for respective constants in the literature.
Fig. 4. Displacement curves of BODIPY-Tyl (IV) and NBD-Tyl (V) from 70S ribosomes by different antibiotics. a) Displacement of BODIPY-Tyl: 1) OMT; 2) erythromycin; 3) tylosin. b) Displacement of NBD-Tyl: 4) erythromycin; 5) tylosin. c) Displacement of NBD-Tyl: 6) lincomycin; 7) chloramphenicol.
Table 1. KD values of
complexes of well-known antibiotics with E. coli ribosomes
determined by NBD-Tyl (V) and BODIPY-Tyl (IV)
displacement. The average KD values are presented
with confidence interval (α = 0.05). The literature data
obtained by 14C- and BODIPY-labeled erythromycin
displacement from their complexes with bacterial ribosomes or by other
methods are shown for comparison
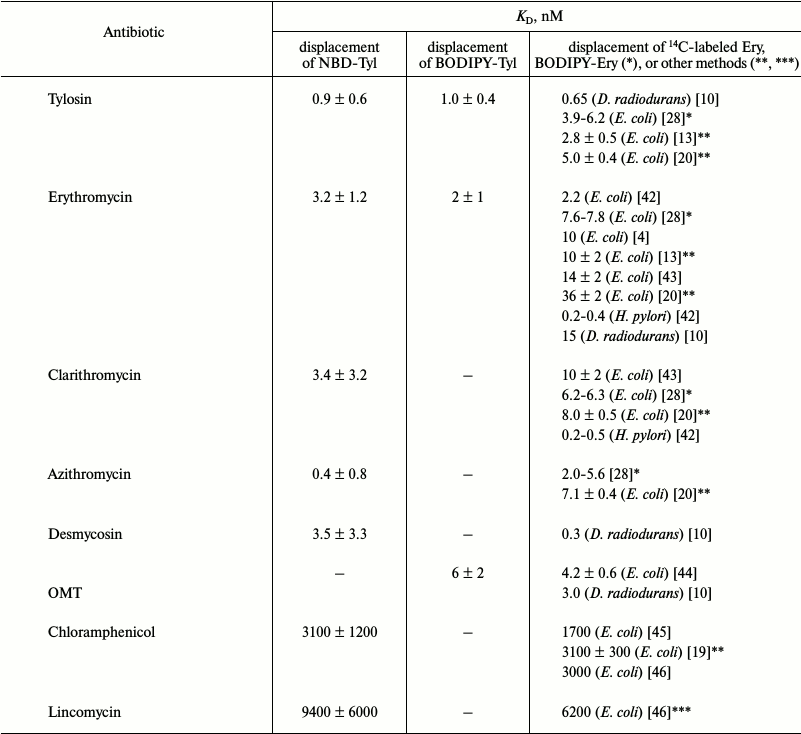
* Data from BODIPY-erythromycin displacement.
** Data from the puromycin reaction.
*** Data from [N-methyl-14C]lincomycin
displacement.
Study of binding of macrolide derivatives with ribosomes by displacement of fluorescently labeled tylosin. BODIPY-Tyl (IV) and NBD-Tyl (V) were used for determination of the quantitative characteristics of the binding with ribosome of previously synthesized tylosin-related macrolide derivatives (Fig. 2) [30, 33]. Binding with 70S D. radiodurans ribosomes of amino acid derivatives of tylosin, Phe-Tyl (VII), and Car-Tyl (VIII), bearing at C20 phenylalanine and carnitine residues, correspondingly, was earlier characterized by 14C-labeled erythromycin displacement assay [10]. The affinity of OMT derivatives, modified by Boc-protected amino acids at the C23 hydroxy group (compounds IX-XI), also was previously determined by BODIPY-erythromycin displacement from its complex with E. coli ribosomes [30].
The displacement curves of BODIPY-Tyl (IV) and NBD-Tyl (V) from their complexes with E. coli ribosomes by macrolide derivatives VII-XI are presented in Fig. S3 in Supplement, and the calculated dissociation constants are given in Table 2. According to the data, the binding with ribosome of compounds VII and VIII is inferior to tylosin binding. In contrast, the affinity to ribosomes of compounds IX-XI was shown to be greater than that of the starting antibiotic OMT, and compound IX possesses the highest affinity, which is consistent with previously reported data [30].
Table 2. KD values of
complexes of macrolide derivatives with ribosomes determined by
displacement of NBD-Tyl (V) and BODIPY-Tyl (IV). The
literature data obtained by displacement of 14C- or
BODIPY-labeled erythromycin from their complexes with bacterial
ribosomes are shown for comparison. The average KD
values are presented with confidence interval (α = 0.05)
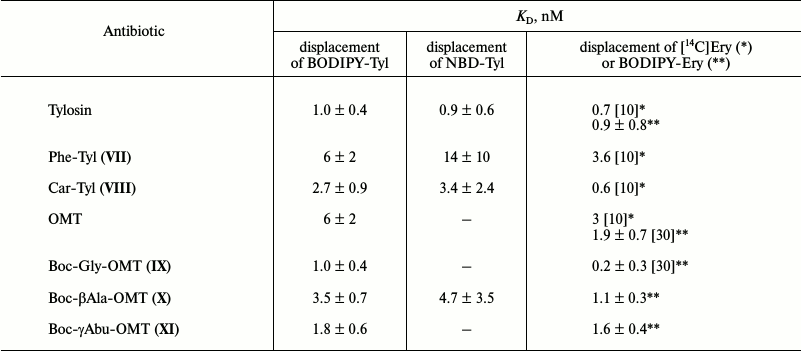
* Data for D. radiodurans ribosomes.
** Data for E. coli ribosomes.
DISCUSSION
Macrolides interact with bacterial ribosomes in the specific area of the ribosomal tunnel [47-49]. The binding sites of macrolides are not identical, but they are greatly overlapping [11, 47, 49]. When tylosin-related antibiotics bind to the ribosome, the substituent in the C5 position of the lactone ring (mycaminose or mycaminosylmycarose residue) is oriented towards the peptidyl transferase center, and the mycinose residue at C14 position of the lactone ring extends along the RT walls in the opposite direction [3, 47]. When tylosin-related antibiotics bind to ribosomes, the substituent at C20 position (or C6 position of the macrolide ring) orients into the cavity in the RT formed by nucleotide residue A2062 reorientation during the process of binding with the antibiotic, and it can interact with elements of 23S rRNA in this cavity [3, 10]. From the analysis of binding assay data of synthesized fluorescent derivatives of antibiotics I-VI (Table 1 and Fig. 1), it can be assumed that the low affinity of compounds I-III and VI is caused by significant volume of xanthene substituents (rhodamine, fluorescein, and Alexa Fluor 488 residues) in C20 position of desmycosin and tylosin, which apparently cannot occupy the cavity, thereby preventing the arrangement of the entire molecule in the macrolide binding site. In the case of compound VI, an additional factor destabilizing the complex with ribosomes may be the presence of negative charge at the fluorescent label, which may result in an electrostatic repulsion between the Alexa Fluor 488 residue and phosphate groups of the rRNA nucleotide backbone. Conversely, the smaller neutral substituents (BODIPY and NBD) in compounds IV and V insignificantly affect binding of the macrolide in the RT.
Moreover, compounds IV and V can be displaced under equilibrium conditions from their complexes with ribosomes by antibiotics or their analogs. And the dissociation constants calculated from curves of BODIPY- and NBD-derivatives of tylosin are very similar, as well as comparable with constants obtained by different methods for corresponding antibiotics and given in the literature or defined previously (Fig. 2 and Tables 1 and 2). However, it should be noted that the dissociation constant values of complexes of the same antibiotics with bacterial ribosomes often vary considerably depending on the experimental conditions, the method for binding determination, and the bacterial type and strain. For example, for such a well-established antibiotic like erythromycin, according to literature sources, the KD values of its complexes with E. coli ribosomes are from 2 to 36 nM [4, 42, 43] and with H. pylori bacteria the values are an order of magnitude lower, as in the case of clarithromycin [42]. Apparently, these same factors can be attributed to a slight difference between the values of constants defined by us for azithromycin and desmycosin from previously measured ones [10, 20].
It is worth mentioning that as fluorescent derivatives are based on tylosin, which while binding occupies a significant part of the RT [3, 47], they may be used to determine the binding of a broader range of compounds depending on their binding site than hitherto described macrolide analogs. Furthermore, as is known, there are some limitations of the fluorescence polarization method relating to the determination of binding constants of ligands with significantly higher affinity than the fluorescently labeled compound [50]. The dissociation constants of complexes of E. coli ribosomes with novel fluorescent tylosin derivatives IV and V are in the nanomolar range, which is an essential factor for more precise determination of binding constants of new substances with higher affinity to bacterial ribosomes.
Thus, it was shown that the synthesized fluorescent tylosin derivatives containing BODIPY FL and NBD groups at C20 position of the macrolide ring can be relatively easily obtained by modification of the aldehyde function of the original antibiotic and applied for determination of dissociation constants of complexes of bacterial ribosomes with ligands whose binding sites overlap with the characteristic for macrolides RT area, as well as for screening of the affinity of novel antibiotic derivatives and other ligands to bacterial ribosomes in the macrolide-binding site.
Acknowledgements
The authors are grateful to M. V. Serebryakova for mass-spectrometry analyses and V. N. Tashlitsky for LC-MS analyses.
This research was supported by the Russian Science Foundation (project No. 14-24-00061, study of antibiotics binding to bacterial ribosomes) and by the Russian Foundation for Basic Research (projects Nos. 16-04-00709-a, synthesis of tylosin analogs; 13-04-40211-H and 13-04-40212-H, isolation of ribosomes). The work was carried out using equipment purchased with funds of the Lomonosov Moscow State University Program of Development.
REFERENCES
1.Kannan, K., and Mankin, A. S. (2011) Macrolide
antibiotics in the ribosome exit tunnel: species-specific binding and
action, Ann. N. Y. Acad. Sci., 1241, 33-47.
2.Wilson, D. N. (2014) Ribosome-targeting antibiotics
and mechanisms of bacterial resistance, Nat. Rev. Microbiol.,
12, 35-48.
3.Bogdanov, A. A., Sumbatyan, N. V., Shishkina, A.
V., Karpenko, V. V., and Korshunova, G. A. (2010) Ribosomal tunnel and
translation regulation, Biochemistry (Moscow), 75,
1501-1516.
4.Pestka, S. (1974) Binding of
[14C]erythromycin to Escherichia coli ribosomes,
Antimicrob. Agents Chemother., 6, 474-478.
5.Teraoka, H., and Nierhaus, K. H. (1979) Measurement
of the binding of antibiotics to ribosomal particles by means of
equilibrium dialysis, Methods Enzymol., 59, 862-866.
6.Abelian, A., Walsh, A. P., Lentzen, G., Aboul-Ela,
F., and Gait, M. J. (2004) Targeting the A site RNA of the
Escherichia coli ribosomal 30S subunit by
2′-O-methyl oligoribonucleotides: a quantitative equilibrium
dialysis binding assay and differential effects of aminoglycoside
antibiotics, Biochem. J., 383, 201-208.
7.Fernandez-Munoz, R., Monro, R. E., Torres-Pinedo,
R., and Vazquez, D. (1971) Substrate- and antibiotic-binding sites at
the peptidyl-transferase center of Escherichia coli ribosomes.
Studies on the chloramphenicol, lincomycin and erythromycin sites,
Eur. J. Biochem., 23, 185-193.
8.Siegrist, S., Lagouardat, J., Moreau, N., and
LeGoffic, F. (1981) Mechanism of action of a 16-membered macrolide.
Binding of rosaramicin to the Escherichia coli ribosome and its
subunits, Eur. J. Biochem., 115, 323-327.
9.Llano-Sotelo, B., Dunkle, J., Klepacki, D., Zhang,
W., Fernandes, P., Cate, J. H., and Mankin, A. S. (2010) Binding and
action of CEM-101, a new fluoroketolide antibiotic that inhibits
protein synthesis, Antimicrob. Agents Chemother., 54,
4961-4970.
10.Starosta, A. L., Karpenko, V. V., Shishkina, A.
V., Mikolajka, A., Sumbatyan, N. V., Schluenzen, F., Korshunova, G. A.,
Bogdanov, A. A., and Wilson, D. N. (2010) Interplay between the
ribosomal tunnel, nascent chain, and macrolides influences drug
inhibition, Chem. Biol., 17, 504-514.
11.Moazed, D., and Noller, H. F. (1987)
Chloramphenicol, erythromycin, carbomycin and vernamycin B protect
overlapping sites in the peptidyl transferase region of 23S ribosomal
RNA, Biochimie, 69, 879-884.
12.Xiong, L., Korkhin, Y., and Mankin, A. S. (2005)
Binding site of the bridged macrolides in the Escherichia coli
ribosome, Antimicrob. Agents Chemother., 49, 281-288.
13.Petropoulos, A. D., Kouvela, E. C., Dinos, G. P.,
and Kalpaxis, D. L. (2008) Stepwise binding of tylosin and erythromycin
to Escherichia coli ribosomes, characterized by kinetic and
footprinting analysis, J. Biol. Chem., 283,
4756-4765.
14.Tejedor, F., and Ballesta, J. P. (1985) Ribosome
structure: binding site of macrolides studied by photoaffinity
labeling, Biochemistry, 24, 467-472.
15.Melançon, P., and Brakier-Gingras, L.
(1985) Cross-linking of streptomycin to the 50S subunit of
Escherichia coli with phenyldiglyoxal, Biochemistry,
24, 6089-6095.
16.Oehler, R., Polacek, N., Steiner, G., and Barta,
A. (1997) Interaction of tetracycline with RNA: photoincorporation into
ribosomal RNA of Escherichia coli, Nucleic Acids Res.,
25, 1219-1224.
17.Leach, K. L., Swaney, S. M., Colca, J. R.,
McDonald, W. G., Blinn, J. R., Thomasco, L. M., Gadwood, R. C.,
Shinabarger, D., Xiong, L., and Mankin, A. S. (2007) The site of action
of oxazolidinone antibiotics in living bacteria and in human
mitochondria, Mol. Cell., 26, 393-402.
18.McFarlan, S. C., and Vince, R. (1984) Inhibition
of peptidyltransferase and possible mode of action of a dipeptidyl
chloramphenicol analog, Biochem. Biophys. Res. Commun.,
122, 748-754.
19.Xaplanteri, M. A., Andreou, A., Dinos, G. P., and
Kalpaxis, D. L. (2003) Effect of polyamines on the inhibition of
peptidyltransferase by antibiotics: revisiting the mechanism of
chloramphenicol action, Nucleic Acids Res., 31,
5074-5083.
20.Dinos, G. P., Connell, S. R., Nierhaus, K. H.,
and Kalpaxis, D. L. (2003) Erythromycin, roxithromycin, and
clarithromycin: use of slow-binding kinetics to compare their in
vitro interaction with a bacterial ribosomal complex active in
peptide bond formation, Mol. Pharmacol., 63, 617-623.
21.Vince, R., Weiss, D., and Pestka, S. (1976)
Binding of N-substituted erythromycylamines to ribosomes,
Antimicrob. Agents Chemother., 9, 131-136.
22.Langlois, R., Cantor, C. R., Vince, R., and
Pestka, S. (1977) Interaction between the erythromycin and
chloramphenicol binding sites on the Escherichia coli ribosome,
Biochemistry, 16, 2349-2356.
23.Brandt-Rauf, P., Vince, R., LeMahieu, R., and
Pestka, S. (1978) Fluorescent assay for estimating the binding of
erythromycin derivatives to ribosomes, Antimicrob. Agents
Chemother., 14, 88-94.
24.Watkins, D., Norris, F. A., Kumar, S., and Arya,
D. P. (2013) A fluorescence-based screen for ribosome binding
antibiotics, Anal. Biochem., 434, 300-307.
25.Llano-Sotelo, B., Hickerson, R. P., Lancaster,
L., Noller, H. F., and Mankin, A. S. (2009) Fluorescently labeled
ribosomes as a tool for analyzing antibiotic binding, RNA,
15, 1597-1604.
26.Jameson, D. M., and Ross, J. A. (2010)
Fluorescence polarization/anisotropy in diagnostics and imaging,
Chem. Rev., 110, 2685-2708.
27.Turconi, S., Shea, K., Ashman, S., Fantom, K.,
Earnshaw, D. L., Bingham, R. P., Haupts, U. M., Brown, M. J. B., and
Pope, A. J. (2001) Real experience of uHTS: a prototypic 1536-well
fluorescence anisotropy-based uHTS screen and application of well-level
quality control procedures, J. Biomol. Screen., 6,
275-290.
28.Yan, K., Hunt, E., Berge, J., May, E., Copeland,
R. A., and Gontarek, R. R. (2005) Fluorescence polarization method to
characterize macrolide–ribosome interactions, Antimicrob.
Agents Chemother., 49, 3367-3372.
29.Li, J., Kim, I. H., Roche, E. D., Beeman, D.,
Lynch, A. S., Ding, C. Z., and Ma, Z. (2006) Design, synthesis, and
biological evaluation of BODIPY-erythromycin probes for bacterial
ribosomes, Bioorg. Med. Chem. Lett., 16, 794-797.
30.Shishkina, A., Makarov, G., Tereshchenkov, A.,
Korshunova, G., Sumbatyan, N., Golovin, A., Svetlov, M., and Bogdanov,
A. (2013) Conjugates of amino acids and peptides with
5-O-mycaminosyltylonolide and their interaction with the ribosomal exit
tunnel, Bioconjug. Chem., 24, 1861-1869.
31.Glassford, I., Lee, M., Wagh, B., Velvadapu, V.,
Paul, T., Sandelin, G., DeBrosse, C., Klepacki, D., Small, M. C.,
MacKerell, A. D., Jr., and Andrade, R. B. (2014) Desmethyl macrolides:
synthesis and evaluation of 4-desmethyl telithromycin, ACS Med.
Chem. Lett., 5, 1021-1026.
32.Tereshchenkov, A. G., Shishkina, A. V.,
Tashlitsky, V. N., Korshunova, G. A., Bogdanov, A. A., and Sumbatyan,
N. V. (2016) Interaction of chloramphenicol tripeptide analogs with
ribosomes, Biochemistry (Moscow), 81, 392-400.
33.Sumbatyan, N. V., Kuznetsova, I. V., Karpenko, V.
V., Fedorova, N. V., Chertkov, V. A., Korshunova, G. A., and Bogdanov,
A. A. (2010) Amino acid and peptide derivatives of the tylosin family
of macrolide antibiotics modified by aldehyde function, Russ. J.
Bioorg. Chem., 36, 245-256.
34.Noll, M., Hapke, B., and Noll, H. (1973)
Structural dynamics of bacterial ribosomes, II. Preparation and
characterization of ribosomes and subunits in the translation of
natural messenger RNA, J. Mol. Biol., 80, 519-529.
35.Milon, P., Konevega, A. L., Peske, F., Fabbretti,
A., Gualerzi, C. O., and Rodnina, M. V. (2007) Transient kinetics,
fluorescence, and FRET in studies of initiation of translation in
bacteria, Methods Enzymol., 43, 1-30.
36.Yang, X. F., Guo, X. Q., and Zhao, Y. B. (2002)
Development of a novel rhodamine-type fluorescent probe to determine
peroxynitrite, Talanta, 57, 883-890.
37.Onoda, M., Uchiyama, S., Santa, T., and Imai, K.
(2002) A photoinduced electron-transfer reagent for peroxyacetic acid,
4-ethylthioacetylamino-7-phenylsulfonyl-2,1,3-benzoxadiazole, based on
the method for predicting the fluorescence quantum yields, Anal.
Chem., 74, 4089-4096.
38.Jablonski, A. (1960) On the notion of emission
anisotropy, Bull. Acad. Pol. Sci., 8, 259-264.
39.Copeland, R. A. (2000) Enzymes: A Practical
Introduction to Structure, Mechanism and Data Analysis, 2nd Edn.,
Wiley-VCH, New York, N. Y.
40.Wang, Z. X. (1995) An exact mathematical
expression for describing competitive binding of two different ligands
to a protein molecule, FEBS Lett., 360, 111-114.
41.Sumbatyan, N. V., Korshunova, G. A., and
Bogdanov, A. A. (2003) Peptide derivatives of antibiotics tylosin and
desmycosin, protein synthesis inhibitors, Biochemistry (Moscow),
68, 1156-1158.
42.Goldman, R. C., Zakula, D., Flamm, R., Beyer, J.,
and Capobianco, J. (1994) Tight binding of clarithromycin, its
14-(R)-hydroxy metabolite, and erythromycin to Helicobacter
pylori ribosomes, Antimicrob. Agents Chemother., 38,
1496-1500.
43.Douthwaite, S., Hansen, L. H., and Mauvais, P.
(2000) Macrolide-ketolide inhibition of MLS-resistant ribosomes is
improved by alternative drug interaction with domain II of 23S rRNA,
Mol. Microbiol., 36, 183-193.
44.Karahalios, P., Kalpaxis, D. L., Fu, H., Katz,
L., Wilson, D. N., and Dinos, G. P. (2006) On the mechanism of action
of 9-O-arylalkyloxime derivatives of 6-O-mycaminosyltylonolide, a new
class of 16-membered macrolide antibiotics, Mol. Pharmacol.,
70, 1271-1280.
45.Mamos, P., Krokidis, M. G., Papadas, A.,
Karahalios, P., Starosta, A. L., Wilson, D. N., Kalpaxis, D. L., and
Dinos, G. P. (2013) On the use of the antibiotic chloramphenicol to
target polypeptide chain mimics to the ribosomal exit tunnel,
Biochimie, 95, 1765-1772.
46.Contreras, A., and Vazquez, D. (1977) Cooperative
and antagonistic interactions of peptidyl-tRNA and antibiotics with
bacterial ribosomes, Eur. J. Biochem., 74, 539-547.
47.Hansen, J., Ippolito, J., Ban, N., Nissen, P.,
Moore, P., and Steitz, T. (2002) The structures of four macrolide
antibiotics bound to the large ribosomal subunit, Mol. Cell,
10, 117-128.
48.Schlunzen, F., Zarivach, R., Harms, J., Bashan,
A., Tocilj, A., Albrecht, R., Yonath, A., and Franceschi, F. (2001)
Structural basis for the interaction of antibiotics with the peptidyl
transferase center in eubacteria, Nature, 413,
814-821.
49.Hansen, J. L., Moore, P. B., and Steitz, T. A.
(2003) Structures of five antibiotics bound at the peptidyl transferase
center of the large ribosomal subunit, J. Mol. Biol.,
330, 1061-1075.
50.Huang, X. (2003) Fluorescence polarization
competition assay: the range of resolvable inhibitor potency is limited
by the affinity of the fluorescent ligand, J. Biomol. Screen.,
8, 34-38.
Supplementary Figures S1, S2 and S3 (PDF)