Procedure for Purification of Recombinant preMsk1p from E. coli Determines Its Properties as a Factor of tRNA Import into Yeast Mitochondria
E. V. Smirnova1,2,3, I. V. Chicherin1,2, M. V. Baleva1,2, N. S. Entelis2, I. A. Tarassov2, and P. A. Kamenski1*
1Lomonosov Moscow State University, Faculty of Biology, 119991 Moscow, Russia; E-mail: kamenski_pa@mail.bio.msu.ru2Strasbourg University, UMR No. 7156 GMGM, 21 rue Rene Descartes, 67084 Strasbourg, France; E-mail: i.tarassov@unistra.fr
3Present address: Strasbourg University, Institute of Plant Molecular Biology, 4 rue General Zimmer, 67084 Strasbourg, France; E-mail: e.renyxa@gmail.com
* To whom correspondence should be addressed.
Received April 27, 2016; Revision received May 23, 2016
Mitochondrial genomes of many eukaryotic organisms do not code for the full tRNA set necessary for organellar translation. Missing tRNA species are imported from the cytosol. In particular, one out of two cytosolic lysine tRNAs of the yeast Saccharomyces cerevisiae is partially internalized by mitochondria. The key protein factor of this process is the precursor of mitochondrial lysyl-tRNA synthetase, preMsk1p. In this work, we show that recombinant preMsk1p purified from E. coli in native conditions, when used in an in vitro tRNA import system, demonstrates some properties different from those shown by the renatured protein purified from E. coli in the denatured state. We also discuss the possible mechanistic reasons for this phenomenon.
KEY WORDS: mitochondria, tRNA, tRNA import into mitochondria, preMsk1p, Eno2p, import mechanisms, protein conformationDOI: 10.1134/S0006297916100060
In eukaryotic cells, mitochondria play an important role in many important processes such as ATP and iron-sulfur (Fe-S) cluster synthesis, apoptosis, etc. [1]. According to the endosymbiotic theory [2], mitochondria evolved from ancient free-living bacteria. Throughout evolution, mitochondrial genomes have undergone considerable reduction, so nowadays they contain no more than a few genes, the rest having been transferred to the nucleus, and products of their expression are imported to the organelle from the cytosol [3]. Ribonucleic acids are also imported into mitochondria. Several types of RNA (mostly tRNAs) were shown to be imported [4]. This process has been described in different eukaryotic organisms. The set of tRNAs imported from the cytosol is not conserved and generally depends on which tRNA genes needed for mitochondrial translation are missing in the mitochondrial genome. Mechanisms of tRNA mitochondrial import also vary from one species to another [5].
Baker’s yeast Saccharomyces cerevisiae is a convenient model organism for studying tRNA mitochondrial import, since in this organism only three tRNAs are imported: lysine tRNA (with CUU anticodon) [6] (further referred to as tRK1) and two glutamine tRNAs (with UUG and CUG anticodons) [7]. It should be mentioned that import of the two latter tRNAs is questioned by some researchers [8], but even if such a process really takes place, its mechanism differs radically from the mechanism of tRK1 import. Yeast cells bear three different lysine tRNAs – the above-mentioned tRK1, encoded in the nucleus and partially imported into mitochondria [9], tRK2 (with UUU anticodon), also encoded in the nucleus but localized exclusively in the cytosol, and tRK3 (with UUU anticodon), encoded in the mitochondrial genome, which assures the incorporation of lysine residues into newly synthesized polypeptide chains in the organelles.
The role of tRK1 in the mitochondrial matrix remained for a long time a mystery, since tRK3 was supposed to be able to decode both lysine codons AAA and AAG in mitochondrial mRNAs. In fact, under normal growth conditions tRK1 is not essential for the mitochondrial translation. However, at elevated temperature the first nucleotide of the tRK3 anticodon becomes hypomodified and loses its ability to recognize efficiently the AAG codon [10], so tRK1 becomes indispensable for mitochondrial protein synthesis [11]. Since under normal growth conditions in the absence of tRK1 yeast mitochondria function normally, the main role of tRK1 in the mitochondrial matrix is to assure mitochondrial protein biosynthesis at elevated temperature.
Some nucleotides have been shown to be important for tRK1 mitochondrial import, namely the first nucleotide of the anticodon, the discriminator nucleotide, and the first base pair of the amino acceptor stem [12, 13]. Moreover, it was shown that before being imported, tRK1 adopts an alternative conformation different from the canonical L-shape form of tRNA, and that the ability to adopt such conformation is the major factor defining the ability of tRNA to be imported into yeast mitochondria [14].
The mechanisms of tRK1 translocation through mitochondrial membranes are still unknown; the translocation probably occurs through the channels of protein mitochondrial import [15]. However, the cascade of events in the cytosol preceding the translocation of tRK1 through the membranes is well defined. First, tRK1 has to be aminoacylated by cytosolic lysyl-tRNA-synthetase [16]. Then, aminoacylated tRK1 interacts with a glycolytic enzyme enolase (Eno2p) [17] adopting the above-mentioned alternative conformation [14]. The complex of tRK1 with Eno2p is transported towards the surface of mitochondria, where tRK1 is released from the complex and binds another protein import factor – the precursor of mitochondrial lysyl-tRNA synthetase (preMsk1p), being synthesized by ribosomes associated with the outer mitochondrial membrane [18]. Perhaps tRK1 is imported into the mitochondria in a complex with preMsk1p. In any case, binding to this protein is one of the most important steps in the mechanism of import.
All experiments of tRNA import into isolated mitochondria in our early works were carried out with recombinant preMsk1p purified from E. coli under denaturing conditions, with subsequent renaturation. In this report, we describe a method for purification of a fully soluble native preMsk1p and show that its properties as a factor of tRNA import into isolated yeast mitochondria differ to some extent from those of the renatured recombinant protein.
MATERIALS AND METHODS
Strains of microorganisms and genetic constructs. Escherichia coli strain Rosetta (F– ompT hsdSB(RB– mB–) gal dcm λ(DE3 [lacI lacUV5-T7 gene 1 ind1 sam7 nin5]) pLysSRARE (CamR)) was used for expression of heterologous proteins; Saccharomyces cerevisiae strain BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) was used for purification of mitochondria.
MSK1 and ENO2 genes (with addition of 6HIS coding sequences just before the stop codon) were cloned in expression vector pET3a.
Purification of recombinant preMsk1p from E. coli under native conditions. The protocol for expression and purification of recombinant preMsk1p in E. coli described previously [19] was used, with some modifications. Escherichia coli cells were grown in LB medium (containing 0.5 M NaCl, 0.2% glucose and 1 mM betaine) at 37°C to the logarithmic phase (OD600 = 0.4). The cells were cooled to 25°C, biosynthesis of the recombinant protein was induced by addition of IPTG to 0.2 mM, and the cells were incubated at 25°C for 4 h. The cells were lysed in buffer containing 10 mM Na2HPO4, 0.5 M NaCl, 75 mM imidazole, pH 8.0, and incubated for 1 h at 4°C. To break the cell wall, lysozyme was added to the final concentration 1 mg/ml; DNA was further sheared by sonication. Cell debris was pelleted by centrifugation at 16,400g for 30 min at 4°C. The recombinant protein was further purified by metal-affinity chromatography on Ni-agarose column. Cleared lysate was loaded on the column, the column was washed, and the recombinant protein was eluted with a gradient of imidazole (from 75 to 500 mM). Washing and elution buffers contained 0.2% CHAPSO detergent.
RNAs. tRK1 in its aminoacylated form was purified from yeast cells using oligonucleotide complementary to a fragment of tRK1 (nucleotides 1-34) (GAGTCATACGCGCTACCGATTGCGCCAACAAGGC) fused to streptavidin-Sepharose beads. The protocol described elsewhere [20] was slightly modified: all buffers used had pH 5.0 to stabilize tRNA in its aminoacylated form.
tRK2 was kindly provided by Dr. Robert Martin (Strasbourg University, France). tRK2 was aminoacylated with lysine using yeast recombinant cytosolic lysyl-tRNA synthetase Krs1p (protein expressed and purified by Estelle Pfister, Strasbourg University, France).
Electrophoretic Mobility Shift Assay (EMSA). 5ʹ-end 32P-labeled tRNAs were mixed with recombinant proteins in buffer containing 20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 10 mM MgCl2, 5 mM DTT, 10% glycerol, and 0.1 mg/ml BSA. The samples were incubated for 10 min at 30°C; the complexes formed were separated on 6% polyacrylamide gel containing 5% glycerol [21]. Bands corresponding to free tRNAs and tRNAs bound to proteins were visualized using Typhoon Trio (GE Healthcare, USA).
Northwestern. Proteins were separated by SDS-PAGE and transferred to the nitrocellulose membrane Hybond N+ (GE Healthcare). Protein bands were visualized with Ponceau Red staining. Proteins were renatured on the membrane by incubation of the membrane for 5 h at 4°C in buffer containing 100 mM Tris-HCl, pH 7.5, 0.1% NP-40, 20 mM KCl, and 3 mM MgCl2. The membrane was washed with buffer containing 10 mM Tris-HCl, pH 7.5, 20 mM KCl, 5 mM MgCl2, and 1% BSA. Then the BSA was removed by washing the membrane with the same buffer lacking BSA. Renatured proteins were hybridized with 32P-labeled tRNAs for at least 12 h at 4°C by incubation of the membrane with tRNAs in buffer containing 10 mM Tris-HCl, pH 7.5, 2 mM EDTA, 1 M NaCl, and 1 mM ATP. Radioactive signal was visualized using Typhoon Trio (GE Healthcare).
Import of tRNAs into isolated yeast mitochondria [22]. Labeled tRNAs were incubated with mitochondria purified from yeast cells in the presence of different combinations of recombinant proteins. Then the mitochondria were treated with RNase A to digest tRNAs that were not imported into the mitochondria. The mitochondria were washed several times to remove RNase A and then lysed. Small RNAs were extracted from the lysate by treatment with hot phenol and were separated by SDS-PAGE. Radioactive signals were visualized with the Typhoon Trio device (GE Healthcare).
RESULTS
According to our previous results, the organization of the tRK1·preMsk1p complex differs from that of canonical tRNA complexes with class IIb aminoacyl-tRNA synthetases (P. A. Kamenski, unpublished data). This might be explained by the fact that the formation of canonical complexes (such as tRK1·Krs1p) leads to aminoacylation of tRNA, while the formation of tRK1·preMsk1p complex results in tRNA import into mitochondria. Moreover, preMsk1p interacts poorly with deacylated tRK1 [18, 23] and is not able to aminoacylate it [16]. Thus, a high-resolution structure of the tRK1·preMsk1p complex would be extremely interesting to obtain. However, when standard protocols for heterologous protein expression in E. coli were used, preMsk1p was totally accumulated in inclusion bodies, and we therefore had to renature it using dialysis. The preMsk1p obtained according to this procedure efficiently and specifically bound tRK1 in vitro and efficiently directed its import into isolated yeast mitochondria. On the other hand, one cannot exclude that the structure of the renatured protein would not be identical to the structure of the protein functioning in a yeast cell. This consideration stimulated us to develop a technique for purification of the recombinant protein from E. coli under native conditions in order to avoid possible artifacts caused by structural changes during renaturation.
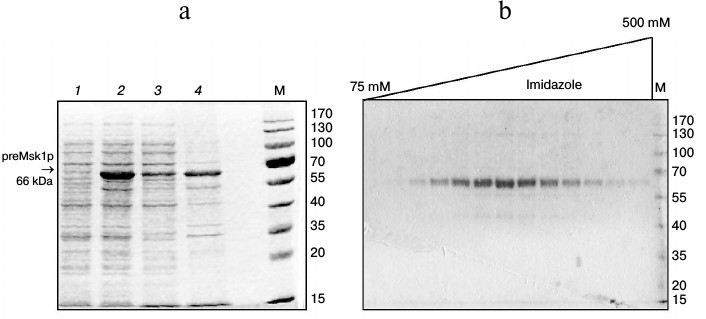
One of the most efficient ways to increase the solubility of recombinant proteins in E. coli is to use osmolytes as growth media additives with simultaneous stimulation of synthesis of cellular chaperones by means of a heat shock step [19]. When using this approach, a significant part of the preMsk1p was found in the soluble fraction (Fig. 1a). Further we will refer to such preMsk1p as “native”. Moreover, native preMsk1p bound efficiently to the Ni-NTA column and was eluted with ~90% purity (Fig. 1b).
Fig. 1. Purification of recombinant preMsk1p under native conditions. a) Synthesis of preMsk1p in E. coli after a heat shock step. SDS-PAGE separation of cell lysates and fractions. Lanes: 1) total cell protein fraction before induction of preMsk1p synthesis; 2) total cell protein fraction 12 h after induction; 3) soluble proteins fraction; 4) inclusion bodies. preMsk1p (MW ≈ 66 kDa) is shown by the arrow. b) Purification of the recombinant preMsk1p on the Ni-NTA column. SDS-PAGE separation of fractions after elution of the bound protein by the gradient of imidazole (indicated above). M, molecular weight standards (size of each band in kDa is designated on the right).
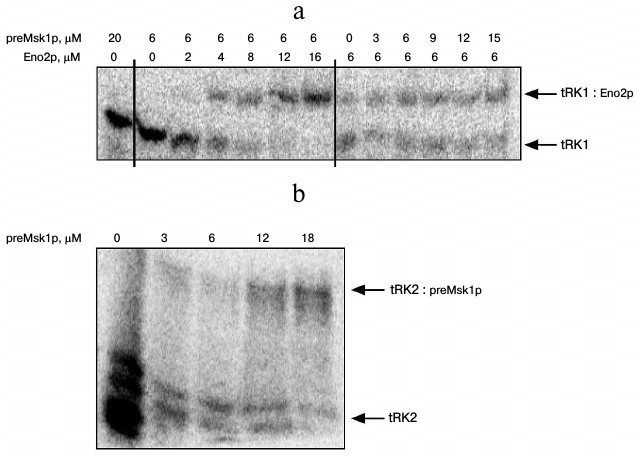
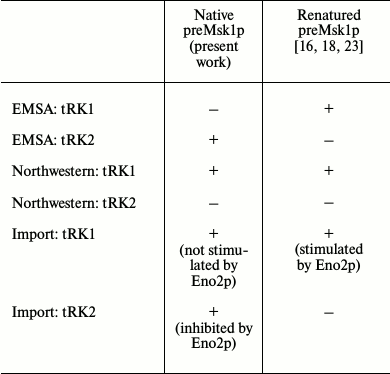
Since native preMsk1p has never been studied as a factor of tRNA import into isolated yeast mitochondria, we analyzed its ability to direct this process. The easiest way to study import-directing properties of preMsk1p is to analyze its ability to bind tRK1 using EMSA by monitoring changes in the migration of tRNA in a polyacrylamide gel upon its binding to the protein [24]. Using this approach, we showed earlier that renatured preMsk1p binds tRK1, but not tRK2, the latter being not imported into mitochondria [23]. Moreover, it was shown that another protein import factor (Eno2p) stimulates the binding of preMsk1p with tRK1 [18]. We carried out similar experiments with native preMsk1p and, surprisingly, found that this protein was not able to bind aminoacylated tRK1 (Fig. 2a). Binding was not observed even when the concentration of the protein was two orders of magnitude higher than the dissociation constant calculated for the complex of renatured preMsk1p with tRK1 (0.18 µM) [18]. Moreover, addition of Eno2p to the reaction mix did not stimulate the formation of tRK1·preMsk1p complex. Increasing the concentration of Eno2p led to the formation of larger amounts of tRK1·Eno2p complex, but not tRK1·preMsk1p complex. The dissociation constant for tRK1·Eno2p complex was approximately 3 µM, being similar to the values obtained earlier [18].
Fig. 2. Native preMsk1p does not bind tRK1, but binds tRK2 in EMSA experiments. An autoradiograph of the gel after electrophoretic separation of complexes of radioactively labeled tRNAs with proteins is presented. Bands corresponding to free tRNAs and their complexes are shown with arrows. Protein concentratons are indicated at the top. a) Analysis of tRK1 complexes with preMsk1p and Eno2p; b) analysis of tRK2 complexes with preMsk1p. Free tRK2 migrates as a double band, which is the result of partial degradation of this molecule always detected in these experiments.
Interestingly, under similar conditions native preMsk1p bound tRK2 (Fig. 2b) with a dissociation constant of approximately 10 µM. Therefore, this complex is much less stable than the complex of renatured preMsk1p with tRK1 (dissociation constant of the latter being 0.18 µM [18]). Nevertheless, this complex was systematically detected in EMSA experiments. Thus, by EMSA we have shown that native preMsk1p as a tRNA mitochondrial import factor shows the properties strictly opposite to that of the renatured protein.
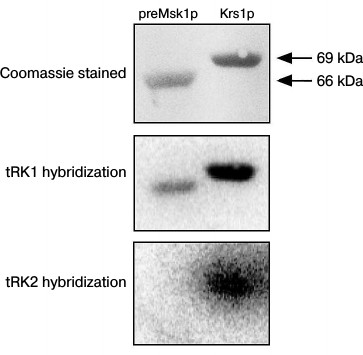
Following these results, we decided to analyze the tRNA binding properties of the native preMsk1p in a different way, namely using the Northwestern blot approach [25]. The rational of this method consists of electrophoretic separation of proteins under denaturing conditions, their transfer to a membrane, with their subsequent in-membrane renaturation and hybridization of a membrane with radioactively labeled tRNA. This method was used to demonstrate for the first time the interaction of Eno2p with tRK1 [18]. Moreover, this method was used to demonstrate the specific binding of imported tRNAs by renatured preMsk1p and its human ortholog [26]. The results of this experiment are presented in Fig. 3. We can see that native preMsk1p binds tRK1, but not tRK2 (in other words, behaves the same way as the renatured protein). Cytosolic lysyl-tRNA synthetase Krs1p used in this experiment as a positive control bound both tRK1 and tRK2, in full accordance with the fact that this enzyme aminoacylates both tRNAs in yeast cells.
Fig. 3. Native preMsk1p binds with tRK1, but not with tRK2, in Northwestern blot assays. SDS-PAGE separation of recombinant proteines (upper panel) and autoradiograph after hybridisation of labeled tRNAs with proteins transferred to the membrane (lower and middle pannels) are presented.
EMSA and Northwestern blot experiments were repeated three times each, leading to the same results: according to the Northwestern blot analysis, native preMsk1p showed exactly the same properties as the renatured protein (Fig. 3), while EMSA showed absolutely opposite results (Fig. 2).
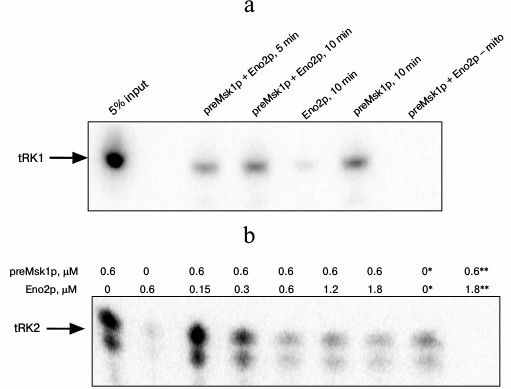
The properties of preMsk1p as a factor of tRNA import can be tested directly in an in vitro tRNA import system [9]. In this experiment, labeled tRNA is mixed with different combinations of the import factors and is incubated with mitochondria isolated from yeast cells. After treatment with RNases aimed at degradation of tRNA molecules that have been not imported, the mitochondria are lysed, and small mitochondrial RNAs are separated by SDS-PAGE. Import into isolated mitochondria of tRK1 and tRK2 in the presence of native preMsk1p and/or Eno2p was analyzed (Fig. 4). In the case of tRK1 import, native preMsk1p showed the same properties as the renatured protein (Fig. 4a); namely, its presence in the reaction led to the detection of a radioactive signal from tRK1 in mitochondria. On the other hand, it was shown earlier that addition of Eno2p to the renatured preMsk1p increased the efficiency of tRK1 import [18]. For native preMsk1p, such effect was not observed: addition of Eno2p to the native preMsk1p did not significantly change the amount of tRK1 in mitochondria.
Fig. 4. Native preMsk1p directs import of both tRK1 and tRK2 into isolated mitochondria, although in the latter case import is inhibited by Eno2p. Autoradiograph of SDS-polyacrylamide gel with fractions of small RNAs extracted from mitochondria after in vitro import reactions are presented. a) Import of tRK1. “5% input”: 5% of the total amount of tRK1 used for each reaction is loaded on the lane. Time of incubation of tRK1 with mitochondria and proteins added are indicated above. The concentrations of preMsk1p and Eno2p – 0.6 µM; “– mito” is reaction without mitochondria. b) Import of tRK2. Proteins added and their concentrations are indicated above. * 1% of the total ammount of tRK2 used for each reaction is loaded on the lane. ** Reaction without mitochondria. Free tRK2 migrates as a double band, which is the result of the partial degradation of this molecule always detected in these experiments.
Results that are even more interesting were obtained when analyzing import of tRK2 (Fig. 4b). Native preMsk1p, unlike renatured protein, was able to direct import of tRK2 into isolated mitochondria. It should be noted that in yeast cells tRK2 is localized exclusively in the cytosol. However, efficiency of tRK2 import decreased in a dose-dependent manner upon addition of Eno2p into the reaction mix.
Properties of native preMsk1p as a tRNA import factor compared to those of renatured protein are summarized in the table.
preMsk1p as a factor of tRNA import into isolated yeast mitochondria:
comparison of properties of recombinant proteins purified from E.
coli under native or denaturing conditions
DISCUSSION
We showed earlier that renatured preMsk1p purified from E. coli inclusion bodies simulate completely the situation in yeast cells when used in experiments of tRNA mitochondrial import in vitro. This protein was shown to bind efficiently in vitro only those tRNAs that can be imported into isolated mitochondria in vivo, and only these tRNAs were targeted to the isolated mitochondria upon incubation with renatured preMsk1p. According to the results presented in this report, preMsk1p purified from E. coli under native conditions shows slightly different properties in spite of the fact that the absence of denaturation/renaturation step during purification should better imitate the natural conditions within yeast cells. According to EMSA experiments, native preMsk1p does not form complexes with tRK1, but does form them with non-imported tRNA, tRK2 (Fig. 2). Moreover, native preMsk1p was able to direct tRK2 import into isolated yeast mitochondria (Fig. 4), although the efficiency of this process decreases upon addition of Eno2p into the reaction. At the same time, the only difference between native and renatured preMsk1p is the presence of a renaturation step in the case of purification of the recombinant protein from inclusion bodies. Thus, one can assume that the structure of native preMsk1p differs slightly from that of the renatured protein. Structural changes in proteins during renaturation were described for the first time several decades ago [27] and since then were many times confirmed. In general, proteins purified under native conditions are considered to have the “correct” structure, while renaturation may cause some structural changes. In the case of preMsk1p, on the contrary, properties of renatured protein are much closer to the properties of the yeast protein in a living cell, while native preMsk1p behaves slightly different. Thus, we assume that the structure of the renatured protein is more “correct” than the structure of the “native” protein.
We believe that this apparent contradiction can be explained by the particularities of preMsk1p biogenesis in a yeast cell. This protein is synthesized by cytosolic ribosomes as a precursor containing an N-terminal signal sequence. After being synthetized, preMsk1p is imported into mitochondria, where the signal sequence is cleaved and the mature protein acquires its enzymatic activity as aminoacyl-tRNA synthetase (for the details of mechanisms of protein import into mitochondria, we recommend the review [28]). On the other hand, the binding of preMsk1p with tRK1 occurs in the cytosol before the step of protein import. The preMsk1p is synthesized by ribosomes localized in the proximity of a mitochondrial surface [18], suggesting that the import of this protein into mitochondria probably starts shortly after its synthesis. We assume that binding with tRK1 occurs already at the moment when the process of preMsk1p import into mitochondria has started, i.e. when the N-terminal signal sequence of the protein is bound to the receptors on the outer mitochondrial membrane and the protein has started to cross the membrane. The structure of preMsk1p anchored in the membrane can be not exactly the same as the structure of the protein in solution: it is known that interaction with biological membranes can lead to considerable changing of protein structure and even to the formation of new secondary structure elements [29]. We cannot exclude the possibility that anchoring of preMsk1p to the mitochondrial membrane allows it to acquire a certain conformation that enables binding of tRK1.
According to our data presented herein, the properties of native preMsk1p corresponded to the model of tRNA mitochondrial import in vivo only when the protein had the possibility to be anchored to the membrane (experiments with tRNA import into isolated mitochondria; Fig. 4). Perhaps the interaction with membranes of isolated mitochondria allows the native preMsk1p to function correctly. In EMSA experiments (Fig. 2), on the contrary, anchoring of the protein was not possible, so its structure could have been different compared to the previous experiments. As a result, its specificity of tRNA binding was changed. As for renatured preMsk1p, which (as shown earlier [23]) demonstrates the “correct” properties in vitro even without anchoring to the membranes, it is possible to assume that this protein acquires “import-competent” conformation during renaturation after a long dialysis.
Figure 4 shows that addition of Eno2p to the in vitro import reaction leads to decrease in tRK2 import efficiency directed to isolated mitochondria by native preMsk1p. The more Eno2p was added to the reaction, the weaker tRK2 was imported into mitochondria. In other words, Eno2p “corrects” the behavior of preMsk1p, playing the role of an import specificity factor, as tRK2 is not imported into yeast mitochondria in vivo. Eno2p itself is not able to bind tRK2 in vitro [18]; therefore, it cannot “take” tRK2 from the complex with preMsk1p and, thus, weaken import. It is possible that the presence of Eno2p in the import reaction of tRK2 physically prevents the formation of a complex of tRK2 with native preMsk1p. It was shown earlier that Eno2p, after being synthesized by cytosolic ribosomes, is transported to the mitochondrial surface where it is incorporated in a macromolecular complex of glycolytic enzymes associated with the outer mitochondrial membrane [30]. Thus, in a yeast cell Eno2p is at least partially associated with the surface of mitochondria. Perhaps in the case of tRK2 import into isolated mitochondria Eno2p associated with mitochondria masks to some extent native preMsk1p, which was already bound to the outer mitochondrial membrane and has acquired its “import-competent” conformation. This might impair binding of preMsk1p with tRK2 and decrease the efficiency of import, respectively. In the case of tRK1 import, the presence of Eno2p does not have the same effect, as far as Eno2p is able to bind tRK1 and deliver it to the mitochondrial surface, where tRK1 interacts with preMsk1p [18].
Thus, the results of the present work seem to indicate the importance of structural changes in preMsk1p for the process of tRNA import into yeast mitochondria. We believe that structural studies that will allow confirmation or ruling out this hypothesis need to be done in the future. These studies will provide a more detailed description of the molecular mechanisms of tRNA mitochondrial import. Besides the fundamental importance, our results can also have some practical interest, since derivatives of yeast tRNAs can be imported into human mitochondria and suppress mutations in mitochondrial tRNAs genes associated with severe hereditary diseases [31, 32]. Import of tRNAs into human mitochondria is carried out by an ortholog of preMsk1p [26], and deeper understanding of the role of this protein in the process of import will allow coming closer to the development of the gene therapy approaches for treatment of mitochondrial disorders.
Acknowledgements
This work was done with a financial support from the Ministry of Education and Science of the Russian Federation (the FTP “Research and Development in Priority Areas of Development of the Russian Scientific and Technological Complex for 2014-2020”, agreement 14.604.21.0113, the identity No. RFMEFI60414X0113) and the French National Centre for Scientific Research LIA “ARN-Mitocure”.
REFERENCES
1.Scheffler, I. E. (2008) Mitochondria, Wiley,
New Jersey.
2.Martin, W. F., Garg, S., and Zimorski, V. (2015)
Endosymbiotic theories for eukaryote origin, Philos. Trans. R Soc.
Lond. B Biol. Sci., 370, 20140330.
3.Huot, J. L., Enkler, L., Megel, C., Karim, L.,
Laporte, D., Becker, H. D., Duchene, A. M., Sissler, M., and
Marechal-Drouard, L. (2014) Idiosyncrasies in decoding mitochondrial
genomes, Biochimie, 100, 95-106.
4.Schneider, A. (2011) Mitochondrial tRNA import and
its consequences for mitochondrial translation, Annu. Rev.
Biochem., 80, 1033-1053.
5.Sieber, F., Duchene, A. M., and Marechal-Drouard,
L. (2011) Mitochondrial RNA import: from diversity of natural
mechanisms to potential applications, Int. Rev. Cell Mol. Biol.,
287, 145-190.
6.Martin, R. P., Schneller, J. M., Stahl, A. J., and
Dirheimer, G. (1979) Import of nuclear deoxyribonucleic acid coded
lysine-accepting transfer ribonucleic acid (anticodon C-U-U) into yeast
mitochondria, Biochemistry, 18, 4600-4605.
7.Rinehart, J., Krett, B., Rubio, M. A., Alfonzo, J.
D., and Soll, D. (2005) Saccharomyces cerevisiae imports
the cytosolic pathway for Gln-tRNA synthesis into the mitochondrion,
Genes Dev., 19, 583-592.
8.Frechin, M., Senger, B., Braye, M., Kern, D.,
Martin, R. P., and Becker, H. D. (2009) Yeast mitochondrial
Gln-tRNA(Gln) is generated by a GatFAB-mediated transamidation pathway
involving Arc1p-controlled subcellular sorting of cytosolic GluRS,
Genes Dev., 23, 1119-1130.
9.Tarassov, I. A., and Entelis, N. S. (1992)
Mitochondrially-imported cytoplasmic tRNA(Lys)(CUU) of
Saccharomyces cerevisiae: in vivo and in
vitro targeting systems, Nucleic Acids Res., 20,
1277-1281.
10.Kamenski, P., Kolesnikova, O., Jubenot, V.,
Entelis, N., Krasheninnikov, I. A., Martin, R. P., and Tarassov, I.
(2007) Evidence for an adaptation mechanism of mitochondrial
translation via tRNA import from the cytosol, Mol. Cell,
26, 625-637.
11.Tarassov, I., Kamenski, P., Kolesnikova, O.,
Karicheva, O., Martin, R. P., Krasheninnikov, I. A., and Entelis, N.
(2007) Import of nuclear DNA-encoded RNAs into mitochondria and
mitochondrial translation, Cell Cycle, 6, 2473-2477.
12.Entelis, N., Kieffer, S., Kolesnikova, O.,
Martin, R., and Tarassov, I. (1998) Structural requirement of
tRNALys for its import into yeast mitochondria, Proc.
Natl. Acad. Sci. USA, 95, 2838-2843.
13.Kolesnikova, O., Entelis, N., Kazakova, H.,
Brandina, I., Martin, R. P., and Tarassov, I. (2002) Targeting of tRNA
into yeast and human mitochondria: the role of anticodon nucleotides,
Mitochondrion, 2, 95-107.
14.Kolesnikova, O., Kazakova, H., Comte, C.,
Steinberg, S., Kamenski, P., Martin, R. P., Tarassov, I., and Entelis,
N. (2010) Selection of RNA aptamers imported into yeast and human
mitochondria, RNA, 16, 926-941.
15.Tarassov, I., Entelis, N., and Martin, R. P.
(1995) An intact protein translocating machinery is required for
mitochondrial import of a yeast cytoplasmic tRNA, J. Mol. Biol.,
245, 315-323.
16.Tarassov, I., Entelis, N., and Martin, R. P.
(1995) Mitochondrial import of a cytoplasmic lysine-tRNA in yeast is
mediated by cooperation of cytoplasmic and mitochondrial lysyl-tRNA
synthetases, EMBO J., 14, 3461-3471.
17.Entelis, N. S., Krasheninnikov, I. A., Martin, R.
P., and Tarassov, I. A. (1996) Mitochondrial import of a yeast
cytoplasmic tRNA (Lys): possible roles of aminoacylation and modified
nucleosides in subcellular partitioning, FEBS Lett., 384,
38-42.
18.Entelis, N., Brandina, I., Kamenski, P.,
Krasheninnikov, I. A., Martin, R. P., and Tarassov, I. (2006) A
glycolytic enzyme, enolase, is recruited as a cofactor of tRNA
targeting toward mitochondria in Saccharomyces cerevisiae,
Genes Dev., 20, 1609-1620.
19.Oganesyan, N., Ankoudinova, I., Kim, S. H., and
Kim, R. (2007) Effect of osmotic stress and heat shock in recombinant
protein overexpression and crystallization, Protein Express.
Purif., 52, 280-285.
20.Yokogawa, T., Kumazawa, Y., Miura, K., and
Watanabe, K. (1989) Purification and characterization of two serine
isoacceptor tRNAs from bovine mitochondria by using a hybridization
assay method, Nucleic Acids Res., 17, 2623-2638.
21.Kaminska, M., Deniziak, M., Kerjan, P.,
Barciszewski, J., and Mirande, M. (2000) A recurrent general RNA
binding domain appended to plant methionyl-tRNA synthetase acts as a
cis-acting cofactor for aminoacylation, EMBO J.,
19, 6908-6917.
22.Entelis, N., Kolesnikova, O., Kazakova, H.,
Brandina, I., Kamenski, P., Martin, R. P., and Tarassov, I. (2002)
Import of nuclear encoded RNAs into yeast and human mitochondria:
experimental approaches and possible biomedical applications, Genet.
Eng. (N.Y.), 24, 191-213.
23.Kamenski, P., Smirnova, E., Kolesnikova, O.,
Krasheninnikov, I. A., Martin, R. P., Entelis, N., and Tarassov, I.
(2010) tRNA mitochondrial import in yeast: mapping of the import
determinants in the carrier protein, the precursor of mitochondrial
lysyl-tRNA synthetase, Mitochondrion, 10, 284-293.
24.Molloy, P. L. (2000) Electrophoretic mobility
shift assays, Methods Mol. Biol., 130, 235-246.
25.Zang, S., and Lin, R. J. (2016) Northwestern blot
analysis: detecting RNA–protein interaction after gel separation
of protein mixture, Methods Mol. Biol., 1421,
111-125.
26.Gowher, A., Smirnov, A., Tarassov, I., and
Entelis, N. (2013) Induced tRNA import into human mitochondria:
implication of a host aminoacyl-tRNA-synthetase, PLoS One,
8, e66228.
27.Teipel, J. W., and Koshland, D. E., Jr. (1971)
Kinetic aspects of conformational changes in proteins. II. Structural
changes in renaturation of denatured proteins, Biochemistry,
10, 798-805.
28.Schulz, C., Schendzielorz, A., and Rehling, P.
(2015) Unlocking the presequence import pathway, Trends Cell
Biol., 25, 265-275.
29.Baumgart, T., Capraro, B. R., Zhu, C., and Das,
S. L. (2011) Thermodynamics and mechanics of membrane curvature
generation and sensing by proteins and lipids, Annu. Rev. Phys.
Chem., 62, 483-506.
30.Brandina, I., Graham, J., Lemaitre-Guillier, C.,
Entelis, N., Krasheninnikov, I., Sweetlove, L., Tarassov, I., and
Martin, R. P. (2006) Enolase takes part in a macromolecular complex
associated to mitochondria in yeast, Biochim. Biophys. Acta,
1757, 1217-1228.
31.Kolesnikova, O. A., Entelis, N. S., Mireau, H.,
Fox, T. D., Martin, R. P., and Tarassov, I. A. (2000) Suppression of
mutations in mitochondrial DNA by tRNAs imported from the cytoplasm,
Science, 289, 1931-1933.
32.Kolesnikova, O. A., Entelis, N. S.,
Jacquin-Becker, C., Goltzene, F., Chrzanowska-Lightowlers, Z. M.,
Lightowlers, R. N., Martin, R. P., and Tarassov, I. (2004) Nuclear
DNA-encoded tRNAs targeted into mitochondria can rescue a mitochondrial
DNA mutation associated with the MERRF syndrome in cultured human
cells, Hum. Mol. Genet., 13, 2519-2534.