REVIEW: The Mitochondrial Genome. The Nucleoid
A. A. Kolesnikov
Lomonosov Moscow State University, Faculty of Biology, 119991 Moscow, Russia; E-mail: aak330@yandex.ru
Received May 30, 2016; Revision received July 1, 2016
Mitochondrial DNA (mtDNA) in cells is organized in nucleoids containing DNA and various proteins. This review discusses questions of organization and structural dynamics of nucleoids as well as their protein components. The structures of mt-nucleoid from different organisms are compared. The currently accepted model of nucleoid organization is described and questions needing answers for better understanding of the fine mechanisms of the mitochondrial genetic apparatus functioning are discussed.
KEY WORDS: mitochondrial nucleoid, mitochondrial genomeDOI: 10.1134/S0006297916100047
Abbreviations: mtDNA, mitochondrial DNA; mt genome, mitochondrial genome.
It is now clear that understanding of genome organization of subcellular
structures is necessary for understanding of cellular processes
occurring through the cell cycle, including questions of storage,
realization, and transmission of genetic information. A considerable
amount of information about forms and sizes of mitochondrial DNA
(mtDNA) throughout evolution has been accumulated [1]. Since the discovery of mtDNA [2, 3], it has been clear that
organization of mtDNA differs significantly from the organization of
nuclear chromatin. For a long time, mtDNA was not supposed to be
covered with proteins, and some authors even used the term
“naked” DNA, which is not true. Up to now, the list of
proteins that can be associated with mtDNA in most characterized
mt-nucleoids of human cells includes more than 50 items and continues
to grow [4]. Classical methods of cellular biology
allow analysis of nucleoid topology in a cell and the presence of
certain components (proteins, RNA, etc.) in a nucleoid, but they are
not able to provide detailed information about specific features of
nucleoid structure. The main approach to studying nucleoids is to
analyze natural DNA–protein complexes.
It was recently suggested that the mitochondrial chromosome and not the mtDNA is a unit of inheritance of a mitochondrial genome, which makes studies of nucleoid structure quite important. Moreover, deep interest in the nucleoid is stimulated by, so-called, “mitochondrial diseases” (diseases of multicellular organisms associated with accumulation of numerous mutations in mtDNA). In fact, proteins associated with a nucleoid can influence the speed of accumulation of mutations.
In the last decade, reviews devoted to this subject appeared regularly [5-9]. Generally, they cover the subject of nucleoid organization in metazoan mitochondria (mainly in human cells) and fungi (mainly yeast). Reviews touching plant cells are much less frequent [10, 11]. This can be explained by more complex organization of the plant mt-genome and, first of all, by the extremely big size of mtDNA (in some species, mtDNA is more than 1,000,000-bp-long); thus, more complicated experimental approaches are needed to analyze the organization of the mt-genome in this group of organisms.
NUCLEOID MORPHOLOGY
Several words must be said about terminology. The nucleoid is an area in a mitochondrion (similarly in chloroplasts) that contains DNA associated with proteins necessary for the maintenance of mtDNA integrity, complexes involved in transfer of genetic information to daughter cells and its realization. Although the nucleoid is not separated from the matrix by a classical membrane, one can expect that the nucleoid forms a separate compartment due to some substance. The nature of this substance remains unclear. The term nucleoid was proposed by analogy to prokaryotic cells. Some authors use this term to designate only the DNA–protein complex (DNP), which is not completely correct in our opinion. Sometimes the term mitochondrial chromosome is used instead of DNP.
Direct experimental studies of nucleoid structure are being done using several approaches: (i) by use of electron, fluorescence, and confocal microscopy [12, 13]; (ii) by analysis of DNP purified from isolated mitochondria; (iii) by genetic methods. Microscopy reveals the form and the size of a nucleoid and allows estimation of the colocalization of individual proteins with DNA. Nucleoid can be visualized using either fluorescent dyes (DAPI, PicoGreen, EtBr, etc.) [14, 15]; fluorescently labeled antibodies to DNA or protein components of a nucleoid [12, 16]; by means of expression of fluorescent fusion proteins (in human cells, this approach was used to localize TFAM or Twinkle proteins in DNP) [16, 17]; or by means of expression of fluorescent photoactivated proteins mEos2 or Dronpa [18]. The latter approach allows monitoring of the lifetime dynamics of a nucleoid. Mitochondrial nucleoids appear as spherical or ellipsoid structures of 70-100 nm diameter in human cells and of 400 nm in yeast [6, 19]. In mitochondria of plant cells, the spindle-shaped form of an mt-chromosome is observed more often [10, 11]. A limited number of species was used for these studies: mammalian cells (generally human cell cultures), Xenopus oocytes, yeast, protozoa, and higher plants (beans, tobacco cultured cells, etc.) [20-24]. It is important to mention that the number of nucleoids in human cells varies from 500 to 12,900 depending on the cell type and the method of nucleoid visualization (cited by [7]), in fibroblasts about 1000 nucleoids can be detected [7], in yeast – about 8-10, in trypanosomatids – one complex associate [25]. The number of nucleoids per cell and not per mitochondrion is calculated since in many cells mitochondria are not discrete small spherical bodies but have a form of “mitochondrial network”, constantly changing its form because of the continuous fusion/fission process [19]. The mitochondrial network is often seen in young, actively dividing cells. Constant progress in microscopy techniques allowed obtaining images of individual DNA–protein complexes with resolution to 10 nm. These techniques are, for example, STED microscopy, and PALM and STORM [19, 26-32]. Standard fluorescence microscopy allows obtaining resolution of 200-350 nm.
STED (stimulated emission depletion) microscopy – a modification of fluorescence microscopy enabling resolution 30-150 nm below the diffraction limit.
STORM (stochastic optical reconstruction microscopy) – resolution of individual molecules to 10-25 nm.
PALM (photo activated localization microscopy) allows distinguishing individual molecules.
Data collected with these methods suggest that the density of mt-chromosome packing excludes the diffusion of soluble proteins of mitochondrial matrix, at least for mammalian cells.
How many molecules of mtDNA does one nucleoid contain? Until recently, it was believed that individual yeast nucleoid contains 1-2 molecules of mtDNA, whereas the mt-nucleoid of vertebrates usually contains 5-7 molecules [20, 21]. In recent years, the development of high-resolution microscopy adjusted this parameter for mammalian cells. It is now accepted that a nucleoid contains 1-2 mtDNA molecules in mammals [19]. At the same time, it was shown that although the main portion of mtDNA in mammalian cells is represented by single circular molecules, in mouse cells from different tissues about 10% of mtDNA molecules form catenanes, and in human NEK cell culture about 30%. It is clear that in this case one nucleoid can contain more than 1-2 mtDNA molecules [33, 34]. A similar situation is observed in embryonic cells, where the number of mtDNA molecules is 3-4 orders of magnitude higher than in somatic cells. In trypanosomatids, several thousand individual DNA molecules are unified in a single complex associate that constitutes a basis of one (unique) nucleoid. Concerning the number of mtDNA molecules composing a nucleoid, it is probably more appropriate to consider not DNA molecules, but the number of full-size mitochondrial genomes.
How many nucleoids can one mitochondrion contain? Considering the dynamic nature of mitochondria, it is difficult to answer this question. During the major part of a cell cycle, mitochondria are multinucleoid structures (mt-reticulum). However, in synchronized culture of osteosarcoma (143B) cells, it was shown that mt-reticulum is fragmented, and one nucleoid can be seen in a discrete mitochondrion [14]. An interesting fact was observed: if mitochondria were studied after mechanical breakage of cells, a significant number of mitochondria (~40%) did not contain DNA (a nucleoid).
DNP COMPLEXES. PROTEINS ASSOCIATED WITH mtDNA
The second and, in fact, the main approach for studying the molecular organization of nucleoids is to analyze proteins, specifically bound to or interacting with mt-DNA (DNP). The strategy of studying nucleoid proteins consists of preparation of highly pure mitochondria with subsequent purification of DNP complexes. Sometimes, the size of a nucleoid is so big that it is possible to purify it using a combination of differential and gradient-density centrifugation [27, 35, 36].
Electron microscopy shows that the purified sample consists of spherical particles with uniform mean size. The main problem is to avoid reorganization (remodeling) of the complex, which can happen during purification of DNP. To exclude misinterpretation of the results, the composition of native DNP and DNP fixed with formaldehyde [22, 37] is often compared. Fixation with formaldehyde allows using more stringent conditions for DNP purification. Cross-linking is reversible; the reaction can be reversed by heating the sample at 70°C.
It must be kept in mind that formaldehyde forms DNA–protein, RNA–protein, and protein–protein bonds, but it does not interact with double-stranded DNA. However, formaldehyde is more efficient in fixation of protein–protein complexes compared to protein complexes with nucleic acids. Cross-linking can occur when the distance between interacting molecules is less than 10 Å. It was shown that Twinkle easily forms aggregates at lower ionic strength [38, 39]. Thus, a standard fixation procedure should be used, with precise pH, temperature, and concentration of reaction components, and the detergent. Another point should be mentioned: many studies were made in vitro; therefore, the result should be extrapolated to the in vivo situation with care.
The volume of purified mtDNA exceeds significantly the volume of the organelle itself. Therefore, it is logical to assume that there are factors assuring the compactization of DNA molecules. A chromosome in a prokaryotic cell has to be compressed 104 times to fit into the cell [40], but in the case of a metazoan mitochondria, DNA (15-20 kb) has to be compressed 20-30 times [19]. Proteins are the most probable in vivo component providing the mtDNA packaging.
To date more than 50 proteins specifically associated with mitochondrial nucleoid have been identified [4]. Among them are proteins interacting with DNA as well as proteins associated with DNP through protein–protein interactions. To analyze proteins associated with a nucleoid, researchers often purify in parallel native DNP (without fixing) and DNP fixed with formaldehyde. The subsequent comparison of the set of proteins coprecipitated with DNA defines what proteins are directly associated with DNA and what proteins are associated through interaction with other proteins.
Proteins identified in DNP can be divided into several functional groups: packaging, remodeling, functional, covering, and auxiliary.
A number of proteins were identified as packaging components: TFAM in human cells, Abf2 in Saccharomyces cerevisiae, KAP3-4 in trypanosomatids. Originally, TFAM was identified as a transcription initiation factor. Abf2 is the major protein purified with mtDNA. Both proteins bind DNA nonspecifically (with some preference to G–C pairs), and the amount of these proteins is sufficient to entirely cover the mtDNA molecule. Most likely, each species should have a limited number of variants of such proteins. According to Kukat and Larsson [28], a packaging protein has to fit several conditions: it has to interact with DNA; it has to possess properties allowing interaction with DNA; it has to be present in a large number of copies to cover DNA; it has to be colocalized with DNA under the high-resolution electron microscope; it has to be essential for maintenance of the nucleoid structure in vivo. In human cells, the most probable candidate for this role is TFAM. An interesting work was published by Kukat et al. [41] illustrating mtDNA condensation by means of mtTFAM in an in vitro system. Using electron microscopy, they analyzed mtDNA structure at various ratios of protein to DNA. Gradual increase in the amount of protein provided condensation of a circular DNA molecule. At a ratio one molecule TFAM to 6-bp DNA molecule, mtDNA formed a compact structure; further increase in the amount of protein did not influence the packaging. As for the number of TFAM molecules in a mitochondrion, different authors give numbers that differ significantly: from 50 to 2700 protein molecules per one mt-chromosome. The analysis was done for human cell culture, fibroblast, murine tissues, and Xenopus oocytes [6]. TFAM was also identified in mitochondria of sea urchin [42].
In other organisms, the role of the main packaging proteins can be played by: Gcf1 (Candida parapsilosis [43]), YlMhb1 (Yarrowia lipolytica [44]), Glom and Glom2 (Physarum polycephalum [23, 24]), KAP 3-4 (Trypanosomatidae [45]).
Besides the proteins mentioned above, many other less represented proteins can be associated with DNA. Unlike the main packaging proteins, their interaction with DNA is often limited to a specific sequence.
Another group of proteins is specific to linear mt-genomes. These are the proteins bound to telomeres [46].
The DNP complex is dynamic. To maintain this state, the cell needs factors that will regulate the density of genome packaging, i.e. DNP remodelers. This means that each packaging factor needs to have an antagonist. In human cells, two proteins can play this role: PGC-1a (a protein regulating mt-biogenesis and activating transcription of mt-genome) and a deacetylase SIRT1; in yeast cells – heat shock protein HSP60, aconitase Aco1 and Ilv5 [6]; in kinetoplast DNA associate of trypanosomatids – UMSBP [47]. In spinach Spinacia oleracea SWIB-1,-2,-3,-5 (12-kDa SWIB domain-containing proteins) can be considered as candidates for the role of nucleoid remodelers [48].
Generally, the number of these proteins in DNP is low and changes depending on the state of the cell [49]. However, the dynamics of the structure can be maintained through a reversible modification of packaging proteins (methylation, acetylation, phosphorylation, etc.), as happens in nuclear chromatin. Unfortunately, there are almost no publications on this subject.
Protein complexes involved in replication and transcription, i.e. DNA and RNA polymerases, as well as factors regulating these processes have to be localized in close proximity to DNA. For mitochondria of human cells, these are phage type RNA polymerase (POLRMT), mtDNA polymerase γA and γB (POLG and POLG2), transcription factor mtTFB2, DNA helicase Twinkle, and LONP1 protease [4]. One may also include in this list TEFM (mitochondrial transcription elongation factor), which binds POLRMT and prevents transcription termination in the CSB II region, resulting in a switch from transcription to replication, and the MTERF protein family, involved in transcription termination on HSP1 and LSP promoters. In yeast, these are RNA polymerase Rpo41, DNA polymerase γ Mip1, helicase Pif1, as well as a multifunctional protein Mgm101 [49]. In mitochondria of plant cells, these are DNA polymerase POLIA, POLIB (bacterial type), topoisomerase II GYRA, GYRB, recombinase RECA2, RECA3 (similar to bacterial ones), RNA polymerase RPROTm, RPROTmp [50], and transcription termination factor MOC1 in Chlamydomonas reinhardtii [51]. In plants, many identical proteins are found in both mitochondria and chloroplasts [50].
Another group of proteins can be regarded as “covering”, since the role of these proteins is to protect single-stranded DNA from nucleases. In the nucleoid of metazoan cells, this is mtssB protein, in yeast – Rim1, and in mitochondria of plant cells there is a whole set of proteins: SSB1, SSB2, OSB1, OSB3, WHY2(WHIRLY), ODB1 [50].
Auxiliary factors are proteins associated with DNP, but their role in the nucleoid remains unclear and is actively discussed in the literature. Here we can mention chaperons, proteases, etc. The possible role in the nucleoid of proteins OPA1 and mitofusin 1 and 2 (GTPase) involved in fusion/fission process and of non-muscular myosin and β-actin is actively discussed in the literature [7, 8].
Genetic analysis is generally used to understand the role of individual proteins purified with DNP. This approach is most actively used with fungi. The rational of this approach consists in switching off the synthesis of certain genes (through mutations) and subsequent analysis of growth and development of the mutant.
In summary we can say, that the “core” of a nucleoid always contains a certain number of proteins (core proteins):
– core proteins of mt-nucleoid of mammalian cells (circular mtDNA from 16 to 18 kb) include 6-7 proteins;
– core proteins of fungal mt-nucleoid (mainly yeast): (circular and linear mtDNA molecules from 19 to 108 kb long) included in Saccharomyces cerevisiae – 7-9 proteins [22] and in Pichia jadinii – 8 proteins [36];
– core proteins of mt-nucleoid in protozoa: Trypanosomatidae (1 associate per cell; 20-50 maxicircles and several thousand minicircles) – 8-9 proteins [47, 52-54];
– core proteins of plant mt-nucleoid (mt-genome from 208 to 2400 kb) [50] – more than 10 proteins;
– core proteins of plastid nucleoid: (circular chloroplast genome from 100 to 250 kb) – 8-10 proteins [48, 55, 56];
– core proteins of a bacterial nucleoid of Escherichia coli: (4.7·106 kb) [57] – 5-7 proteins.
The number of different proteins in a nucleoid can vary. DNA-packaging proteins are the most abundant. For example, the amount of TFAM protein in mitochondria of human or mouse cells, according to some authors, is sufficient to evenly bind mtDNA with an interval of about 20 bp. Considering that TFAM functions in a cell in the form of a homodimer, the distance between binding sites can be 35-40 nucleotides [8, 13, 58, 59]. Treatment of mt-nucleoid purified from BY-2 cultured tobacco cells with micrococcal nuclease gave a specific ladder-like pattern of DNA on the gel. The interval between bands was 75 nucleotides. This result suggested the existence of nucleosome-like structure in an mt-nucleoid of these cells [60].
Properties of proteins associated with DNA. Comparison of DNA-packaging proteins shows that they are basic proteins containing HMG box domains, or they are histone H1-like proteins containing lysine rich domains. Proteins TFAM, Abf2, and Glom belong to the first group (mitochondria of metazoan cells, yeast, and myxomycetes, respectively). The second group of proteins includes Glom2 (myxomycetes) and KAP 1-4 proteins (trypanosomatids) [45]. The second special feature of proteins associated with DNA, noted by the majority of researchers, is their bifunctionality.
Generally, DNA-packaging proteins have low molecular weight.
DNA-packaging and DNA-covering proteins function as multimers: TFAM and Abf2 are homodimers and mtSSB is a tetramer.
It seems that DNA-packaging proteins bind DNA mainly through their HMG domains. Other components (lysine rich fragments, zinc fingers, etc.) might be involved in binding in organisms staying at a lower stage of evolution, especially in protozoa. Exactly this was observed in monocellular eukaryotes. However, it must be kept in mind that the presence of a HMG domain may not assure the interaction of the protein with DNA. In particular, in S. cerevisiae it was shown that out of two HMG domains of Abf2 protein, one domain loses its ability to bind DNA (or, at least, this ability is significantly weakened) [61].
We have analyzed protein properties from the UniProt KB database.
NUCLEOID ORGANIZATION MODELS
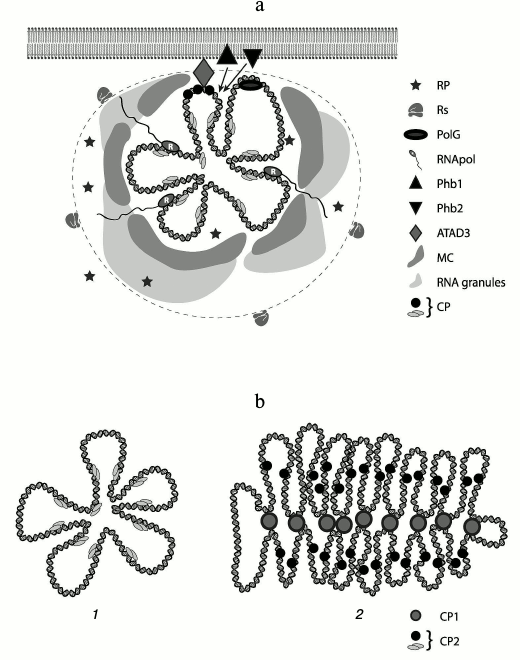
A “multi-layer” model of mt-nucleoid organization in human cells was proposed [37] according to which there is the central part – “core” – where DNA and proteins (DNA-packaging proteins and proteins involved in replication and transcription processes) are concentrated; the next layer contains proteins temporarily recruited to execute special functions. These proteins are generally retained in DNP through protein–protein interaction. On the periphery of a nucleoid, there is a zone in which translation and other processes take place. It is believed that the nucleoid is positioned on the internal mitochondrial membrane via ATAD3 protein, at least in human mitochondria [37]. This protein itself binds specifically only to the D-loop of mtDNA. Two other proteins of the inner mitochondrial membrane – prohibitin 1 and 2 [37] – might also be involved in keeping a nucleoid. Ribosomes are located next to the second layer. This model was proposed for mammalian cells, but it can be extended to all metazoan cells.
Recently, specific bodies called “mitochondrial RNA granules” were found in mitochondrial matrix. Granules are complexes of RNase P with newly synthesized mtRNA. RNA-binding protein GRSF1 (G-rich sequence factor 1) was also identified in the complex. RNase P has three subunits (MRPP1, MRPP2, and MRPP3) and assures the processing of the 5′-end of mitochondrial tRNAs in the in vitro system. Processing of the 3′-end of tRNAs is assured by ELAC2 enzyme (Z type RNase). GRSF1 protein directly participates in the processing of mitochondrial RNA [62-65].
After discovery of these granules, the model was modified slightly. Brown et al. [31] placed to the periphery of DNP a zone called “RNA cloud”. Moreover, outside of this cloud proteins providing remodeling of a nucleoid were placed. Formation of macromolecular “crowding” within the second layer is not excluded. The editosome in trypanosomatids, which assures editing of mitochondrial cryptogenes, is probably also localized at the periphery of a nucleoid. A scheme of possible nucleoid organization is shown in the figure.
Scheme of mitochondrial nucleoid organization. a) mtDNA with “core” proteins is in the middle. The dashed line defines the boundaries of a nucleoid. The shadowed regions at the periphery of a nucleoid represent “RNA granules”. Legend: CP, compacting (packaging) proteins; RNApol, RNA polymerase; PolG, DNA polymerase; PHB1, PHB2, prohibitin 1, 2; MC, macromolecular crowding (region, where functional proteins of a nucleoid are localized); Rs, ribosomes; RP, remodeling proteins [8, 19, 31, 37, 73]. b) A model of DNA organization in an mt-nucleoid: 1) metazoan, fungi; 2) plants and prokaryotes. Legend: CP1, first level packaging proteins; CP2, functional packaging proteins
IS THE NUCLEOID SEPARATED FROM THE MATRIX?
Absence of a membrane in a nucleoid does not mean the absence of any other substance separating the nucleoid compartment from the matrix. This hypothesis is favored by the fact that lipids were found in nuclei of eukaryotic cells and in nucleoid of Pseudomonas aurantiaca [66, 67]. Dispersed lipid droplets can a factor providing structured “crowding” [68]. On the other hand, long noncoding RNAs (longer than 150 nucleotides) with possible structural role have been extensively studied in recent years. Even the new term “architectural RNA” was introduced [69]. It is quite possible that these RNAs can actively participate in the formation of a “nucleoid backbone”. It is also possible that mitochondrial RNA granules can be such a backbone.
To define the general principles of nucleoid organization, one needs to take into account different types of mitochondrial genomes [1]. In this regard, several variants of organization of a central part of a nucleoid can be proposed for mitochondria of metazoan cells and fungi, plants, and trypanosomatids: spherical, spindle-shaped, and complex concatenated associate.
Absence of proteins with HMG domains and presence of high amount of histone-like proteins in mt-nucleoid of plants suggests the existence of two variants of basic mtDNA packaging. When an mt-genome exceeds 100 kb, histone-like proteins play a role of the major packaging factor (plants, some protozoa, and fungi), forming a “spindle” with large functional loops. They function as the main structural proteins. Specific packaging of the loops can be carried out by another set of proteins. It is a two-level system of mtDNA packaging. With the reduction of the genome size (for genomes smaller than 100 kb), the need for the first level of packaging disappears, and histone-like proteins are replaced by proteins with HMG domains.
In the bacterial nucleoid, the ratio protein to DNA changes from the logarithmic to the stationary growth stage [70]. In yeast, repression of aerobic respiration (growth in glucose-containing media) or amino acid starvation results in increase in the Abf2/DNA ratio. HSP60 in recruited to a nucleoid (in glucose-containing media) and Ilv5 (during amino acid starvation). In actively growing aerobic culture, the number of mtDNA copies increases [71]. These are only several examples of nucleoid dynamics. Considering this, there should be mechanisms providing remodeling of a condensed structure. At least, any model of nucleoid organization has to explain how initiation of replication and transcription occur in the compact structure.
It becomes increasingly obvious that replication, recombination, and segregation of mtDNA are closely related to the structure of a mitochondrial nucleoid [72].
Considering the large variety of forms of mitochondrial genome organization in nature, there is a significant lack of data for a full comparative analysis of organization of mitochondrial nucleoids in different groups of organisms.
According to Martin Kuchey and Ronald Butou [6], during evolution a considerable part of proteins was included in mtDNP taking into account the specificity of metabolism in different species.
UNSOLVED QUESTIONS
The general principles of nucleoid organization are still clear. However, many fine details demand further research. In our opinion, the most urgent questions are the following.
The mitochondrial genome of many organisms is represented not by monomorphic circular or linear molecules, but by a set of molecules. Do these genomes form a single nucleoid?
Is the nucleoid isolated from the mitochondrial matrix, and if so, what is the nature of the substance providing this separation?
Does a nucleoid have an internal structure?
What are the precise mechanisms assuring the dynamics of a nucleoid structure?
What are the dynamics of a nucleoid structure in a cell cycle?
Is a system of reversible modification of proteins used to regulate nucleoid dynamics?
Acknowledgements
The author is grateful to A. V. Fedyakov for his help in figure preparation.
REFERENCES
1.Kolesnikov, A. A., and Gerasimov, E. S. (2012)
Diversity of mitochondrial genome organization, Biochemistry
(Moscow), 77, 1424-1435.
2.Nass, S., Nass, M. M., and Hennix, U. (1965)
Deoxyribonucleic acid in isolated rat-liver mitochondria, Biochim.
Biophys. Acta, 95, 426-435.
3.Nass, S. (1969) The significance of the structural
and functional similarities of bacteria and mitochondria, Int. Rev.
Cytol., 25, 55-129.
4.Bogenhagen, D. F., Wang, Y., Shen, E. L., and
Kobayashi, R. (2003) Protein components of mitochondrial DNA nucleoids
in higher eukaryotes, Mol. Cell. Proteomics, 2,
1205-1216.
5.Holt, I. J., He, J., Mao, C.-C., Boyd-Kirkup, J.
D., Martinsson, P., Sembongi, H., Reyes, A., and Spelbrink, J. N.
(2007) Mammalian mitochondrial nucleoids: organizing an independently
minded genome, Mitochondrion, 7, 311-321.
6.Kucej, M., and Butow, R. A. (2007) Evolutionary
tinkering with mitochondrial nucleoids, Trends Cell Biol.,
17, 586-592.
7.Bogenhagen, D. F. (2012) Mitochondrial DNA nucleoid
structure, Biochim. Biophys. Acta, 1819, 914-920.
8.Spelbrink, J. N. (2010) Functional organization of
mammalian mitochondrial DNA in nucleoids: history, recent development,
and future challenges, IUBMB Life, 62, 19-32.
9.Gilkerson, R., Bravo, L., Garcia, I., Gaytan, N.,
Herrera, A., Maldonado, A., and Quintanilla, B. (2013) The
mitochondrial nucleoid: integrating mitochondrial DNA into cellular
homeostasis, Cold Spring Harb. Perspect. Biol., 5,
a011080.
10.Dai, H., Lo, Y.-S., Litvinchuk, A., Wang, Y.-T.,
Jane, W.-N., Hsiao, L.-J., and Chiang, K.-S. (2005) Structural and
functional characterizations of mung bean mitochondrial nucleoids,
Nucleic Acids Res., 33, 4725-4739.
11.Gualberto, J. M., Mileshina, D., Wallet, C.,
Niazi, A. K., Weber-Lotfi, F., and Dietrich, A. (2014) The plant
mitochondrial genome: dynamics and maintenance, Biochimie,
100, 107-120.
12.Satoh, M., and Kuroiwa, T. (1991) Organization of
multiple nucleoids and DNA molecules in mitochondria of a human cell,
Exp. Cell Res., 196, 137-140.
13.Alam, T. I., Kanki, T., Muta, T., Ukaji, K., Abe,
Y., Nakayama, H., Takio, K., Hamasaki, N., and Kang, D. (2003) Human
mitochondrial DNA is packaged with TFAM, Nucleic Acids Res.,
31, 1640-1645.
14.Hayashi, J., Takemitsu, M., Goto, Y., and Nonaka,
I. (1994) Human mitochondria and mitochondrial genome function as a
single dynamic cellular unit, J. Cell Biol., 125,
43-50.
15.Margineantu, D. H., Cox, W. G., Sundell, L.,
Sherwood, S. W., Beechem, J. M., and Capaldi, R. A. (2002) Cell cycle
dependent morphology changes and associated mitochondrial DNA
redistribution in mitochondria of human cell lines,
Mitochondrion, 1, 425-435.
16.Garrido, N., Griparic, L., Jokitalo, E.,
Wartiovaara, J., Van der Bliek, A. M., and Spelbrink, J. N. (2003)
Composition and dynamics of human mitochondrial nucleoids, Mol.
Biol. Cell, 14, 1583-1596.
17.Spelbrink, J. N., Li, F.-Y., Tiranti, V., Nikali,
K., Yuan, Q.-P., Tariq, M., Wanrooij, S., Garrido, N., Comi, G.,
Morandi, L., Santoro, L., Toscano, A., Fabrizi, G.-M., Somer, H.,
Croxen, R., Beeson, D., Poulton, J., Suomalainen, A., Jacobs, H. T.,
Zeviani, M., and Larsson, C. (2001) Human mitochondrial DNA deletions
associated with mutations in the gene encoding Twinkle, a phage T7 gene
4-like protein localized in mitochondria, Nat. Genet.,
28, 223-231.
18.Gerald, D., Keller, J., Medd, R., Andrei, A. M.,
Rizzoli, S. O., Luhrmann, R., Jahn, R., Eggeling, C., and Stefan, W. H.
(2006) Macromolecular-scale resolution in biological fluorescence
microscopy, Proc. Natl. Acad. Sci. USA, 103,
11440-11445.
19.Kukat, C., Wurm, C. A., Spahr, H., Falkenberg,
M., Larsson, N.-G., and Jacobs, S. (2011) Super-resolution microscopy
reveals that mammalian mitochondrial nucleoids have a uniform size and
frequently contain a single copy of mtDNA, Proc. Natl. Acad. Sci.
USA, 108, 13534-13539.
20.Legros, F., Malka, F., Frochon, P., Lombes, A.,
and Rojo, M. (2004) Organization and dynamics of human mitochondrial
DNA, J. Cell Sci., 117, 2653-2662.
21.Iborra, F. J., Kimura, H., and Cook, P. R. (2004) The
functional organization of mitochondrial genomes in human cells, BMC
Biol., 2, 9.
22.Kaufman, B. A., Newman, S. M., Hallberg, R. L.,
Slaugter, C. A., Perlman, P. S., and Butow, R. A. (2000) In organelle
formaldehyde crosslinking of proteins to mtDNA: identification of
bifunctional proteins, Proc. Natl. Acad. Sci. USA, 97,
7772-7777.
23.Itoh, K., Izumi, A., Mori, T., Dohmae, N., Yui,
R., Maeda-Sano, K., Shirai, Y., Kanaoka, M. M., Kuroiwa, T.,
Higashiyama, T., Sugita, M., Murakami-Murofushi, K., Kawano, S., and
Sasaki, N. (2011) DNA packaging proteins Glom and Glom2 coordinately
organize the mitochondrial nucleoid of Physarum polycephalum,
Mitochondrion, 11, 575-586.
24.Sasaki, N., Kuroiva, H., Nishitani, C., Takano,
H., Higashiyama, T., Kobayashi, T., Shirai, Y., Sakai, A., Kawano, S.,
Murakami-Murofushi, K., and Kuroiwa, T. (2003) Glom is a novel
mitochondrial DNA packaging protein in Physarum polycephalum and
cause intense chromatin condensation without suppressing DNA functions,
Mol. Biol. Cell, 14, 4758-4769.
25.Simpson, L. (1986) Kinetoplast DNA in
trypanosomatids flagellates, Int. Rev Cytol., 99,
119-179.
26.Kukat, C., Wurm, C. A., Spahr, H., Falkenberg, M., Larsson,
N.-G., and Jacobs, S. (2011) Super-resolution microscopy reveals that
mammalian mitochondrial nucleoids have a uniform size and frequently
contain a single copy of mtDNA, Proc. Natl. Acad. Sci. USA,
108, 13534-13539.
27.Shiiba, D., Fumoto, S.-I., Miyakawa, I., and
Sando, N. (1997) Isolation of giant nucleoids from the yeast
Saccharomyces cerevisiae, Protoplasma, 198
177-185.
28.Kukat, C., and Larsson, N.-G. (2013) mtDNA makes a U-turn for
the mitochondrial nucleoid, Trends Cell Biol., 23,
457-463.
29.Westphal, V., Rizzoli, S. O., Lauterbach, M. A.,
Kamin, D., Jahn, R., and Hell, S. W. (2008) Video-rate far-field
optical nanoscopy dissects synaptic vesicle movement, Science,
320, 246-249.
30.Singh, H., Lu, R., Rodriguez, P. F. G., Wu, Y.,
Bopassa, J. C., Stefani, E., and Toro, L. (2012) Visualization and
quantification of cardiac mitochondrial protein clusters with STED
microscopy, Mitochondrion, 12, 230-236.
31.Brown, T. A., Tkachuk, A. N., Shtengel, G.,
Kopek, B. G., G., Kopek, B. G., Bogenhagen, D. F., Hess, H. F., and
Clayton, D. A. (2011) Super-resolution fluorescence imaging of
mitochondrial nucleoids reveals their spatial range, limits, and
membrane interaction, Mol. Cell. Biol., 31,
4994-5010.
32.Kwiatek, J. M., Hinde, E., and Gaus, K. (2014)
Microscopy approaches to investigate protein dynamics and lipid
organization, Mol. Membr. Biol., 31, 141-151.
33.Pohjoismaki, J. L. O., and Goffart, S. (2011) Of
circles, forks and humanity: topological organization and replication
of mammalian mitochondrial DNA, BioEssays, 33,
290-299.
34.Pohjoismaki, J. L. O., Goffart, S., Tyynismaa,
H., Willcox, S., Ide, T., Kang, D., Suyomalainen, A., Karhunen, P. J.,
Griffith, J. D., Holt, I. J., and Jacobs, H. T. (2009) Human heart
mitochondrial DNA is organized in complex catenated networks containing
abundant four-way junctions and replication forks, J. Biol.
Chem., 284, 21446-21457.
35.Wang, Y., and Bogenhagen, D. F. (2006) Human
mitochondrial DNA nucleoids are linked to protein folding machinery and
metabolic enzymes at the mitochondrial inner membrane, J. Biol.
Chem., 281, 25791-25802.
36.Miyakawa, I., Okazaki-Higashi, C., Higashi, T.,
Furutani, Y., and Sando, N. (1996) Isolation and characterization of
mitochondrial nucleoids from the yeast Pichia jadinii, Plant
Cell Physiol., 37, 816-824.
37.Bogenhagen, D. F., Rousseau, D., and Burke, S.
(2008) The layered structure of human mitochondrial DNA nucleoids,
J. Biol. Chem., 283, 3665-3675.
38.Hensen, F., Cansiz, S., Gerhold, J. M., and
Spelbrink, J. N. (2014) To be or not to be a nucleoid protein: a
comparison of mass-spectrometry based approaches in the identification
of potential mtDNA-nucleoid associated proteins, Biochimie,
100, 219-226.
39.Korhonen, J. A., Gaspari, M., and Falkenberg, M.
(2003) TWINKLE has 5′-3′ DNA helicase activity and is
specifically stimulated by mitochondrial single-stranded DNA-binding
protein, J. Biol. Chem., 278, 48627-48632.
40.De Vries, R. (2010) DNA condensation in bacteria:
interplay between macromolecular crowding and nucleoid proteins,
Biochimie, 92, 1715-1721.
41.Kukat, C., Davies, K. M., Wurm, C. A., Spahr, H.,
Bonekamp, N. A., Kuhl, I., Joos, F., Polosa, P. L., Park, C. B., Posse,
V., Falkenberg, M., Jacobs, S., Kulbrandt, W., and Larsson, N.-G.
(2015) Cross-strand binding of TFAM to a single mtDNA molecule forms
the mitochondrial nucleoid, Proc. Natl. Acad. Sci. USA,
112, 11288-11293.
42.Malarkey, C. S., Lionetti, C., Deceglie, S.,
Roberti, M., Churchil, M. E. A., Cantatore, P., and Polosa, P. L.
(2016) The sea urchin mitochondrial transcription factor A binds and
bends DNA efficiently despite its unusually short C-terminal tail,
Mitochondrion, 29, 1-6.
43.Miyakawa, I., Okamuro, A., Kinski, S., Visacka,
K., Tomashka, L., and Nosek, J. (2009) Mitochondrial nucleoids from the
yeast Candida parapsilosis: expansion of the repertoire of
proteins associated with mitochondrial DNA, Microbiology,
155, 1558-1568.
44.Bakkalova, J., Arata, K., Matsunobu, M., Ono, B.,
Aoki, T., Lajdova, D., Nebohacova, M., Nosek, J., Miyakawa, I., and
Tomaska, L. (2014) The strictly aerobic yeast Yarrowia
lipolytica tolerates loss of a mitochondrial DNA-packaging protein,
Eukaryot. Cell, 13, 1143-1157.
45.Xu, C., and Ray, D. S. (1993) Isolation of
proteins associated with kinetoplast DNA networks in vivo,
Proc. Natl. Acad. Sci. USA, 90, 1786-1789.
46.Tomaska, L., Nosek, J., and Fukuhara, H. (1997)
Identification of a putative mitochondrial telomere-binding protein of
the yeast Candida parapsilosis, J. Biol. Chem.,
272, 3049-3056.
47.Kapeller, I., Milman, N., Yaffe, N., and Shlomai,
J. (2011) Interaction of a replication initiator with histone H1-like
proteins remodel the condensed mitochondrial genome, J. Biol.
Chem., 286, 40566-40574.
48.Melonek, J., Matros, A., Trosch, M., Mock, H. P.,
and Krupinska, K. (2012) The core of chloroplast nucleoids contains
architectural SWIB domain proteins, Plant Cell,
24, 3060-3073.
49.Kucej, M., Kucejova, B., Subramanian, R., Chen,
X. J., and Butow, R. A. (2008) Mitochondrial nucleoids undergo
remodeling in response to metabolic cues, J. Cell Sci.,
121, 1861-1868.
50.Gualberto, J. M., and Kuhn, K. (2014) DNA-binding
proteins in plant mitochondria: implications for transcription,
Mitochondrion, 19, 323-328.
51.Wobbe, L., and Nixon, P. J. (2013) The mTERF
protein MOC1 terminates mitochondrial DNA transcription in the
unicellular green alga Chlamydomonas reinhardtii, Nucleic
Acids Res., 41, 6553-6567.
52.Tzfati, Y., Abiliovich, H., Avrahami, D., and
Shlomai, J. (1995) Universal minicircle sequence binding protein, a
CCHC-type zinc finger protein that binds the universal minicircle
sequence of trypanosomatids. Purification and characterization, J.
Biol. Chem., 270, 21339-21345.
53.Hines, J. C., and Ray, D. S. (2010) A
mitochondrial DNA primase is essential for cell growth and kinetoplast
DNA replication in Trypanosoma brucei, Mol. Cell. Biol.,
30, 1319-1328.
54.Hines, J. C., and Ray, D. S. (2011) A second
mitochondrial DNA primase is essential for cell growth and kinetoplast
minicircle DNA replication in Trypanosoma brucei, Eukaryot.
Cell, 10, 445-454.
55.Krupinska, K., Oetke, S., Desel, C., Mulisch, M.,
Schafer, A., Hollmann, J., Kumlehn, J., and Hensel, G. (2014) WHIRLY1
is a major organizer of chloroplast nucleoids, Front. Plant
Sci., 5, doi: 10.3389/fpls.2014.00432.
56.Kobayashi, T., Takahara, M., Miyagishima, S. Y.,
Kuroiva, H., Sasaki, N., Ohta, N., Matsuzaki, M., and Kuroiva, T.
(2002) Detection and localization of a chloroplast-encoded HU-like
protein that organizes chloroplast nucleoids, Plant Cell,
14, 1579-1589.
57.Wang, W., Li, G.-W., Chen, C., Xie, X. S., and
Zhuang, X. (2011) Chromosome organization by a nucleoid-associated
protein in live bacteria, Science, 333, 1445-1449.
58.Takamatsu, C., Umeda, S., Ohsato, T., Ohno, T.,
Abe, Y., Fukuoh, A., Shinagawa, H., Hamasaki, N., and Kang, D. (2002)
Regulation of mitochondrial D-loops by transcription factor A and
single-stranded DNA-binding protein, EMBO Rep., 3,
451-456.
59.Kaufman, B. A., Durisic, N., Mativetsky, J. M.,
Costantino, S., Hancock, M. A., Grutter, P., and Shoubridge, E. A.
(2007) The mitochondrial transcription factor TFAM coordinates the
assembly of multiple DNA molecules into nucleoid-like structures,
Mol. Biol. Cell, 18, 3225-3236.
60.Takusagawa, M., Hayashi, T., Takano, H., and
Sakai, A. (2009) Organization of mitochondrial-nucleoids in BY-2
cultured tobacco cells, Cytologia, 74, 329-341.
61.Kurashenko, A. V., Samoylova, E. O., Baleva, M.
V., Chicherin, I. V., Petrov, D. Y., Kamenski, P. A., and Levitsky, S.
A. (2016) Two HMG-domains of yeast mitochondrial protein Abf2p has
different affinity to DNA, Vestnik RSMU, 1, 68-72.
62.Antonicka, H., Sasarman, F., Nishimura, T.,
Paupe, V., and Shoubridge, E. A. (2013) The mitochondrial RNA-binding
protein GRSF1 localizes to RNA granules and is required for
posttranscriptional mitochondrial gene expression, Cell Metab.,
17, 386-398.
63.Jourdain, A. A., Koppen, M., Wydro, M., Rodley,
C. D., Lightowlers, R. N., Chrzanovska-Lightowlers, Z. M., and
Martinou, J.-C. (2013) GRSF1 regulates RNA processing in mitochondrial
RNA granules, Cell Metab., 17, 399-410.
64.Brzezniak, L. K., Bijata, M., Szczesny, R. J.,
and Stepien, P. P. (2011) Involvement of human ELAC2 gene product in
3′-end processing of mitochondrial tRNAs, RNA Biol.,
8, 616-626.
65.Jourdain, A. A., Boehm, E., Maundrell, K., and
Martinou, J.-C. (2016) Mitochondrial RNA granules: compartmentalizing
mitochondrial gene expression, J. Cell Biol., 212,
611-614.
66.Albi, E., and Magni, M. P. V. (2004) The role of
intranuclear lipids, Biol. Cell, 96, 657-667.
67.Zhdanov, R. I., Kern, D., Lorenc, V., and
Ibragimova, M. Y. (2015) Lipid and fatty acid profiles of
Pseudomonas aurantiaca DNA-bound lipids determined by mass
spectrometry, Mikrobiologiya, 84, 50-57.
68.Kory, N., Thiam, A.-R., Farese, R. V., and
Walther, T. C. (2015) Protein crowding is a determinant of lipid
droplet protein composition, Dev. Cell, 34, 1-13.
69.Chujo, T., Yamazaki, T., and Hirose, T. (2016)
Architectural RNAs (arcRNAs): a class of long noncoding RNAs that
function as the scaffold of nuclear bodies, Biochim. Biophys.
Acta, 1859, 139-146.
70.Azam, T. A., Iwata, A., Nishimura, A., Ueda, S.,
and Ishihama, A. (1999) Growth phase-dependent variation in protein
composition of the Escherichia coli nucleoid, J.
Bacteriol., 181, 6361-6370.
71.Cavelier, L., Johannisson, A., and Gyllensten, U.
(2000) Analysis of mtDNA copy number and composition of single
mitochondrial particles using flow cytometry and PCR, Exp. Cell
Res., 259, 79-85.
72.Gilkerson, R. W. (2009) Mitochondrial DNA
nucleoids determine mitochondrial genetics and dysfunction, Intern.
J. Biochem. Cell Biol., 41, 1899-1906.
73.Wang, W., Li, G., Chen, C., Xie, S., and Zhuang,
X. (2011) Chromosome organization by a nucleoid-associated protein in
live bacteria, Science, 333, 1445-1449.