Priming of Human Neutrophils Is Necessary for Their Activation by Extracellular DNA
A. S. Prikhodko1, M. V. Vitushkina2, L. A. Zinovkina1, E. N. Popova3, and R. A. Zinovkin2,3*
1Lomonosov Moscow State University, Faculty of Bioengineering and Bioinformatics, 119991 Moscow, Russia2Lomonosov Moscow State University, Faculty of Biology, 119991 Moscow, Russia; E-mail: roman.zinovkin@gmail.com
3Lomonosov Moscow State University, Belozersky Institute of Physico-Chemical Biology, 119991 Moscow, Russia
* To whom correspondence should be addressed.
Received February 19, 2016; Revision received March 13, 2016
Extracellular plasma DNA is thought to act as a damage-associated molecular pattern causing activation of immune cells. However, purified preparations of mitochondrial and nuclear DNA were unable to induce neutrophil activation in vitro. Thus, we examined whether granulocyte-macrophage colony-stimulating factor (GM-CSF) acting as a neutrophil priming agent can promote the activation of neutrophils by different types of extracellular DNA. GM-CSF pretreatment greatly increased p38 MAPK phosphorylation and promoted CD11b/CD66b expression in human neutrophils treated with mitochondrial and, to a lesser extent, with nuclear DNA. Our experiments clearly indicate that GM-CSF-induced priming of human neutrophils is necessary for their subsequent activation by extracellular DNA.
KEY WORDS: neutrophil activation, GM-CSF, extracellular DNA, nuclear DNA, mitochondrial DNADOI: 10.1134/S0006297916060079
Abbreviations: GM-CSF, granulocyte-macrophage colony-stimulating factor; mtDNA, mitochondrial DNA; nDNA, nuclear DNA.
Sterile inflammation can be mediated by damage-associated molecular
patterns (DAMPs) released from dying cells: proteins, lipids, ATP, and
DNA [1]. The extracellular plasma DNA includes
nuclear DNA (nDNA) and mitochondrial DNA (mtDNA). Increased
concentration of extracellular nDNA is typical for a number of diseases
including sepsis [2], ischemic shock [3], pulmonary embolism [4], and
cancer [5]. Likewise, the level of extracellular
mtDNA correlates with disease severity and prognosis in patients with
sepsis [2], pulmonary embolism [4], myocardial infarction [6],
cancer [7], and severe traumatic injuries [8, 9].
Neutrophils are among the first immune cells reacting to danger signals (DAMPs). These phagocytic cells play a key role in the development of both infectious and noninfectious inflammatory response [10]. Mitochondrial DNA released into the bloodstream is thought to act as a DAMP, resulting in neutrophil activation [11]. We previously showed that human neutrophils are not activated directly by purified preparations of mtDNA or nDNA in vitro [12]. Nevertheless, proinflammatory action of mtDNA was clearly established by in vivo experiments, since its injection results in inflammatory reactions in lungs and cartilages [13, 14]. Thus, there should be some additional factor(s) affecting the activation of neutrophils by mtDNA in vivo. Besides direct activation of neutrophils, priming is a well-known phenomenon leading to an increased response to other activating factors [15]. Granulocyte-macrophage colony-stimulating factor (GM-CSF) induces neutrophil priming and enhances neutrophil response to bacterial DNA in vitro [16].
In this work, we examined whether GM-CSF can influence the activation of neutrophils using purified preparations of mtDNA and nDNA. In addition to the conventional protocol for neutrophil activation in vitro, we used whole blood assay to induce neutrophil activation without sample manipulation.
MATERIALS AND METHODS
Isolation of human neutrophils. The study conformed to the code of ethics of the World Medical Association (Declaration of Helsinki) and was approved by the local ethics committee. Peripheral venous blood from healthy donors was collected in heparin-coated tubes. Polymorphonuclear leukocytes (neutrophils) were isolated by dextran sedimentation and subsequent Ficoll-Hypaque (1.077 g/ml) gradient centrifugation as described previously [12]. The cells were resuspended in RPMI-1640 (Paneco, Russia) supplemented with 10% low endotoxin fetal bovine serum (FBS) (PAA Laboratories, Germany). Neutrophil viability (>97%) was assessed by trypan blue dye exclusion.
Neutrophil activation. Neutrophil activation was detected using phycoerythrin-conjugated CD11b and AlexaFluor647-conjugated CD66b antibodies (BD Biosciences, USA) according to the manufacturer’s protocol: neutrophils were incubated with the activators in RPMI-1640 with 10% low-endotoxin FBS for 30 min at 37°C. After incubation, the antibodies were added and the sample kept on ice for 30 min. Recombinant human GM-CSF (R&D Systems, USA) was used for neutrophil priming. Fluorescence intensity was analyzed using a Beckman-Coulter FC 500 flow cytometer.
Whole blood experiments. Venous blood from healthy donors was collected in heparin-coated tubes. Incubation with the activators was performed in whole blood (0.5 ml) prior to isolation of leukocytes. Then the neutrophils were isolated from each individual sample and analyzed by flow cytometry as described above.
Western blotting. After incubation with the activators, neutrophils were immediately lysed in buffer (62.5 mM Tris-HCl, pH 6.8, 2% SDS, 10% glycerol, 50 mM dithiothreitol (DTT), 0.01% bromophenol blue) for 3 min at 95°C. Proteins were separated by SDS-PAGE and transferred onto a PVDF membrane (Amersham, USA). Primary antibodies against human MAPK p38, phospho-p38, and tubulin (Cell Signaling, USA) were used. Horseradish peroxidase-conjugated secondary antibodies (Sigma-Aldrich, USA) were developed by ECL kit (Amersham). ImageJ 1.44p software was used for densitometric analysis.
Mitochondrial and nuclear DNA isolation. Human endothelial cells (EA.hy926) were grown in DMEM supplemented with 10% FBS (HyClone, USA) in a humidified atmosphere of 5% CO2 at 37°C. Confluent cells were resuspended in DMEM and centrifuged at 600g for 10 min at 4°C. The pellet was resuspended in hypotonic buffer (10 mM Tris-MOPS, pH 7.8, 50 mM NaCl, 5% (w/v) sucrose, 10 mM EDTA), homogenized, and centrifuged at 1000g (10 min, 4°C) to pellet nuclei and cell debris. The supernatant was centrifuged at 10,000g (20 min, 4°C) to pellet mitochondria. Standard mtDNA samples were obtained from the mitochondrial pellet using a DNeasy Blood & Tissue kit (Qiagen, USA) according to the manufacturer’s protocol. Additional purification was performed by ethanol precipitation with sodium acetate.
For nDNA purification, EA.hy926 cells were treated with 50 ng/ml of ethidium bromide for 3 weeks to produce Rho0 cells lacking mtDNA [17]. nDNA was purified using a DNeasy Blood & Tissue kit (Qiagen).
DNA concentration and quality was determined by spectrophotometry and agarose gel electrophoresis. The purity of mtDNA and nDNA was confirmed by real-time PCR analysis [12].
Statistical analysis. The data are presented as mean ± SEM. The difference between means was assessed by the Mann–Whitney nonparametric test.
RESULTS
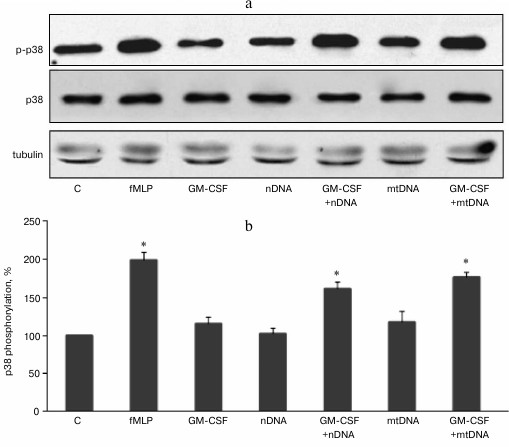
GM-CSF pretreatment enhances p38 MAPK phosphorylation in human neutrophils treated with mitochondrial and nuclear DNA. We previously showed that purified mtDNA does not activate unprimed human neutrophils in vitro and does not affect p38 MAPK phosphorylation [12]. Nuclear DNA also has no effect on p38 phosphorylation and MMP-9 metalloproteinase release [18]. Our goal was to test whether GM-CSF, a key regulator of hematopoietic neutrophil activation, can prime human neutrophils for further activation by extracellular DNA. Purified human neutrophils were stimulated with 2 ng/ml GM-CSF for 10 min at 37°C. The purified mitochondrial or nuclear DNA (10 µg/ml) was added to the neutrophils, and the cells were incubated for 20 min at 37°C and processed for Western blot analysis (Fig. 1, a and b). Tripeptide formylmethionine-leucine-phenylalanine (fMLP) was used as a positive control leading to a rapid activation (phosphorylation) of p38 MAPK. GM-CSF, mtDNA, or nuclear DNA alone did not cause increase in p38 phosphorylation. However, after GM-CSF pretreatment, exposure to mtDNA resulted in a significant increase in p38 MAPK phosphorylation (Fig. 1). Similarly, nDNA in combination with GM-CSF caused p38 MAPK activation (Fig. 1).
Fig. 1. GM-CSF promotes p38 MAPK activation in response to mtDNA and nDNA in human neutrophils. Neutrophils (106 cells/ml) were incubated in RPMI containing 10% FBS without activators (control, C) and with fMLP (10 nM) or GM-CSF (2 ng/ml) for 20 min. For GM-CSF priming experiments, neutrophils were preincubated for 10 min with GM-CSF (2 ng/ml) and treated for 10 min with mtDNA and nDNA (10 µg/ml). a) Representative Western blots for p-p38 MAPK, p38 MAPK, and tubulin as a loading control. b) Densitometric analysis of Western blots (n = 4, * p < 0.05 versus non-stimulated control neutrophils).
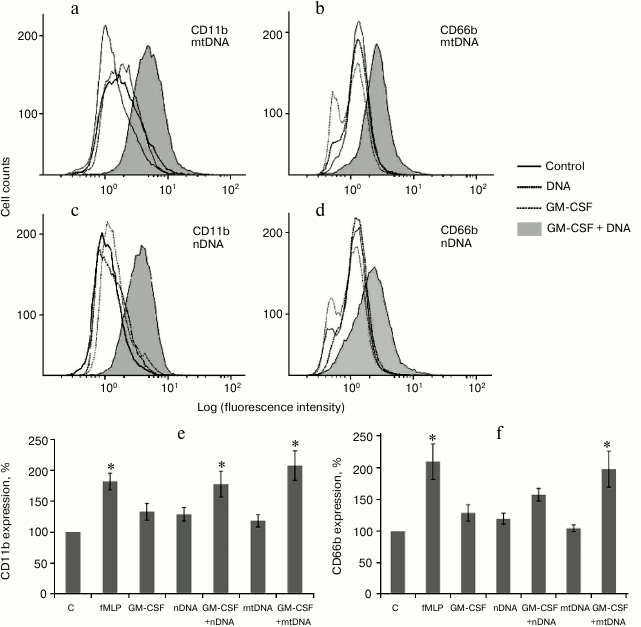
GM-CSF pretreatment promotes CD11b and CD66b expression in human neutrophils treated with mitochondrial and nuclear DNA. Human neutrophils were preincubated with GM-CSF (2 ng/ml) for 10 min at 37°C and then stimulated by nuclear or mitochondrial DNA (10 µg/ml) for 20 min at 37°C. GM-CSF, nDNA, and mtDNA alone did not increase expression of the degranulation markers CD11b/CD66b. However, GM-CSF pretreatment significantly increased CD11b/CD66b expression in the samples treated with mtDNA (Fig. 2). It is noteworthy that nDNA in combination with GM-CSF also increased expression of CD11b/CD66b, but to a lesser extent compared to mtDNA (Fig. 2, e and f).
Fig. 2. GM-CSF triggers CD11b and CD66b upregulation on human neutrophils in response to mtDNA and nDNA. Neutrophils (106 cells/ml) were incubated in RPMI containing 10% FBS without activators (control, C) and with fMLP (10 nM) or GM-CSF (2 ng/ml) for 30 min. For GM-CSF priming experiments, neutrophils were preincubated for 10 min with GM-CSF (2 ng/ml) and treated for 20 min with mtDNA and nDNA (10 µg/ml). a-d) Representative flow cytometry plots of CD11b (a, c) and CD66b (b, d) expression on human neutrophils. Mean channel fluorescence of neutrophils stained for CD11b (e) and CD66b (f) ± SEM (n = 4, * p < 0.05 versus non-stimulated control neutrophils).
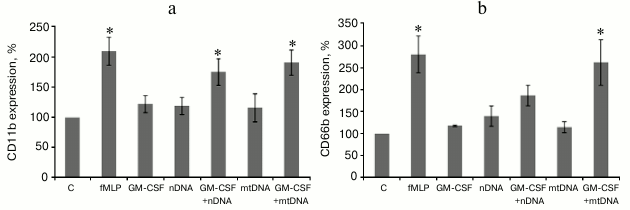
mtDNA and nDNA in combination with GM-CSF activate human neutrophils in whole blood. In a separate set of experiments, whole blood from healthy donors was used to confirm the results obtained for the isolated human neutrophils. Heparinized blood samples were sequentially treated with GM-CSF and then mtDNA or nDNA. The incubation was followed by isolation of the neutrophil fraction and CD11b/CD66b flow cytometry analysis (Fig. 3). GM-CSF, nDNA, and mtDNA alone did not cause a statistically significant increase in CD66b and CD11b expression level. However, GM-CSF pretreatment resulted in enhanced CD11b/CD66b expression in the mtDNA and nDNA samples, though nDNA was less effective. These results are in perfect agreement with the data obtained for the isolated neutrophils (Fig. 2).
Fig. 3. GM-CSF triggers CD11b (a) and CD66b (b) upregulation on human neutrophil induced by mtDNA and nDNA in whole blood. Whole blood was treated as indicated in Fig. 2. The neutrophils were subsequently isolated from individual samples and analyzed by flow cytometry for CD11b and CD66b expression. Mean channel fluorescence of neutrophils stained for CD11b and CD66b ± SEM (n = 4, * p < 0.05 versus non-stimulated control neutrophils).
DISCUSSION
Despite the fact that extracellular DNA level correlates well with severity and adverse outcome of numerous diseases [2-4], the mechanisms of its direct action on immune cells have not been studied in detail. Now there is no doubt that extracellular mtDNA is involved in the inflammatory response in vivo. mtDNA induces inflammation and increases TLR9/NF-κB expression in rat lung tissue and macrophages [14]. mtDNA also induces arthritis and infiltration of mononuclear cells when injected intraarticularly in mice [13]. Direct activation of neutrophils by mtDNA in vitro was described previously [18]. Later, we showed that purified mtDNA does not activate human neutrophils in vitro [12]. The results obtained in the current study confirmed our previous data: pure mtDNA was unable to activate neutrophils (Figs. 1-3). Indirectly, a recent report also confirms these data: incubation of neutrophils with mtDNA did not increase IL-6 expression [19].
As established previously, neutrophil priming by GM-CSF is required for their activation by bacterial DNA [16]. Similarly, human DAMP S100A9 protein primed IL-8 production only in GM-CSF-stimulated neutrophils [20]. We hypothesized that GM-CSF-induced neutrophil priming can also promote neutrophil activation by mtDNA. Our experiments clearly indicate that GM-CSF-induced priming of human neutrophils is required for their subsequent activation by mtDNA and, to a lesser extent, by nDNA. The activation was assessed by p38 MAPK phosphorylation (Fig. 1) and expression of cell surface markers CD11b and CD66b (Fig. 2). Since whole blood may contain additional factor(s) influencing neutrophil activation, we additionally used whole blood assay that confirmed these results (Fig. 3). Thus, only primed neutrophils are activated by mtDNA either in whole blood or after their isolation procedure. Though the activating action of mtDNA was rather anticipated, nDNA unexpectedly also showed this activity.
Immune cells activation by both bacterial and mtDNA is supposed to be TLR9-dependent due to the presence of CpG unmethylated dinucleotides TLR9 [14, 21]. This is in good agreement with the fact that TLR9 expression is enhanced in GM-CSF-treated PMNs [22]. So far, the role of nDNA in the development of proinflammatory response and various diseases is unclear. On one hand, nDNA as well as mtDNA level is elevated in many diseases [23]. This can be explained by necrotic death of damaged cells and subsequent release of unprocessed DNA [24]. On the other hand, nDNA has typical methylation pattern and there is no evidence about activation of immune cells by nDNA, which is often used as a negative control [18]. However, in our experiments, nDNA showed moderate ability to activate GM-CSF-pretreated PMNs. This fact raises further questions concerning proinflammatory action of extracellular DNA. According to a recently published report, bacterial, mtDNA, and nDNA all possess different proinflammatory properties [19]. Bacterial DNA and mtDNA, but not nDNA, delayed neutrophil apoptosis; bacterial DNA increased neutrophil IL-6 secretion, but mitochondrial and nuclear DNA did not; all three types of DNA similarly increased thrombin generation in platelet-poor plasma [19]. The impact and the mechanisms of the action of the extracellular DNA on the immune system require further investigation.
Besides theoretical importance, the current work also has some practical issues. GM-CSF is now undergoing clinical trials for immunomodulation in trauma patients (https://clinicaltrials.gov/ct2/show/results/NCT01495637), since sepsis is one of the most common dangerous complications after traumatic injury. In another clinical trial, GM-CSF treatment produced negative results in a randomized trial of acute lung injury and acute respiratory distress syndrome [25]. According to our data, GM-CSF pretreatment makes neutrophils sensitive to extracellular DNA. The level of extracellular DNA is highly elevated in trauma patients [26], thus GM-CSF treatment may potentially lead to the life-threatening excessive inflammatory response.
This work was supported by the Russian Science Foundation (project No. 14-24-00107).
REFERENCES
1.Piccinini, A. M., and Midwood, K. S. (2010)
DAMPening inflammation by modulating TLR signalling, Mediators
Inflamm., doi: 10.1155/2010/672395.
2.Kung, C. T., Hsiao, S. Y., Tsai, T. C., Su, C. M.,
Chang, W. N., Huang, C. R., Wang, H. C., Lin, W. C., Chang, H. W., Lin,
Y. J., Cheng, B. C., Su, B. Y., Tsai, N. W., and Lu, C. H. (2012)
Plasma nuclear and mitochondrial DNA levels as predictors of outcome in
severe sepsis patients in the emergency room, J. Transl. Med.,
10, 130.
3.Tsai, N. W., Lin, T. K., Chen, S. D., Chang, W. N.,
Wang, H. C., Yang, T. M., Lin, Y. J., Jan, C. R., Huang, C. R., Liou,
C. W., and Lu, C. H. (2011) The value of serial plasma nuclear and
mitochondrial DNA levels in patients with acute ischemic stroke,
Clin. Chim. Acta, 412, 476-479.
4.Arnalich, F., Maldifassi, M. C., Ciria, E.,
Codoceo, R., Renart, J., Fernandez-Capitan, C., Herruzo, R.,
Garcia-Rio, F., Lopez-Collazo, E., and Montiel, C. (2013) Plasma levels
of mitochondrial and nuclear DNA in patients with massive pulmonary
embolism in the emergency department: a prospective cohort study,
Crit. Care, 17, R90.
5.Jahr, S., Hentze, H., Englisch, S., Hardt, D.,
Fackelmayer, F. O., Hesch, R.-D., and Knippers, R. (2001) DNA fragments
in the blood plasma of cancer patients: quantitations and evidence for
their origin from apoptotic and necrotic cells, Cancer Res.,
61, 1659-1665.
6.Gonzalez-Masia, J. A., Garcia-Olmo, D., and
Garcia-Olmo, D. C. (2013) Circulating nucleic acids in plasma and serum
(CNAPS): applications in oncology, Onco Targets Ther., 6,
819-832.
7.Wang, L., Xie, L., Zhang, Q., Cai, X., Tang, Y.,
Hang, T., Liu, J., and Gong, J. (2015) Plasma nuclear and mitochondrial
DNA levels in acute myocardial infarction patients, Coron. Artery
Dis., 26, 296-300.
8.Yamanouchi, S., Kudo, D., Yamada, M., Miyagawa, N.,
Furukawa, H., and Kushimoto, S. (2013) Plasma mitochondrial DNA levels
in patients with trauma and severe sepsis: time course and the
association with clinical status, J. Crit. Care, 28,
1027-1031.
9.Nakahira, K., Hisata, S., and Choi, A. M. (2015)
The roles of mitochondrial damage-associated molecular patterns in
diseases, Antioxid. Redox Signal., 23, 1329-1350.
10.Mantovani, A., Cassatella, M. A., Costantini, C.,
and Jaillon, S. (2011) Neutrophils in the activation and regulation of
innate and adaptive immunity, Nat. Rev. Immunol., 11,
519-531.
11.Zhang, Q., Raoof, M., Chen, Y., Sumi, Y., Sursal,
T., Junger, W., Brohi, K., Itagaki, K., and Hauser, C. J. (2010)
Circulating mitochondrial DAMPs cause inflammatory responses to injury,
Nature, 464, 104-107.
12.Prikhodko, A. S., Shabanov, A. K., Zinovkina, L.
A., Popova, E. N., Aznauryan, M. A., Lanina, N. O., Vitushkina, M. V.,
and Zinovkin, R. A. (2015) Pure mitochondrial DNA does not activate
human neutrophils in vitro, Biochemistry (Moscow),
80, 629-635.
13.Collins, L. V., Hajizadeh, S., Holme, E.,
Jonsson, I. M., and Tarkowski, A. (2004) Endogenously oxidized
mitochondrial DNA induces in vivo and in vitro
inflammatory responses, J. Leukoc. Biol., 75,
995-1000.
14.Zhang, J. Z., Liu, Z., Liu, J., Ren, J. X., and
Sun, T. S. (2014) Mitochondrial DNA induces inflammation and increases
TLR9/NF-κB expression in lung tissue, Int. J. Mol. Med.,
33, 817-824.
15.Condliffe, A. M., Kitchen, E., and Chilvers, E.
R. (1998) Neutrophil priming: pathophysiological consequences and
underlying mechanisms, Clin. Sci. (London), 94,
461-471.
16.Fuxman Bass, J. I., Alvarez, M. E., Gabelloni, M.
L., Vermeulen, M. E., Amaral, M. M., Geffner, J. R., and Trevani, A. S.
(2008) GM-CSF enhances a CpG-independent pathway of neutrophil
activation triggered by bacterial DNA, Mol. Immunol., 46,
37-44.
17.King, M. P., and Attardi, G. (1996) Isolation of
human cell lines lacking mitochondrial DNA, Methods Enzymol.,
264, 304.
18.Zhang, Q., Itagaki, K., and Hauser, C. J. (2010)
Mitochondrial DNA is released by shock and activates neutrophils via
p38 map kinase, Shock, 34, 55-59.
19.Bhagirath, V. C., Dwivedi, D. J., and Liaw, P. C.
(2015) Comparison of the proinflammatory and procoagulant properties of
nuclear, mitochondrial, and bacterial DNA, Shock, 44,
265-271.
20.Simard, J. C., Noel, C., Tessier, P. A., and
Girard, D. (2014) Human S100A9 potentiates IL-8 production in response
to GM-CSF or fMLP via activation of a different set of transcription
factors in neutrophils, FEBS Lett., 588, 2141-2146.
21.Itagaki, K., Kaczmarek, E., Lee, Y. T., Tang, I.
T., Isal, B., Adibnia, Y., Sandler, N., Grimm, M. J., Segal, B. H.,
Otterbein, L. E., and Hauser, C. J. (2015) Mitochondrial DNA released
by trauma induces neutrophil extracellular traps, PLoS One,
10, e0120549.
22.Hayashi, F., Means, T. K., and Luster, A. D.
(2003) Toll-like receptors stimulate human neutrophil function,
Blood, 102, 2660-2669.
23.Tsang, J. C., and Dennis Lo, Y. (2007)
Circulating nucleic acids in plasma/serum, Pathology, 39,
197-207.
24.Pisetsky, D. S., and Fairhurst, A.-M. (2007) The
origin of extracellular DNA during the clearance of dead and dying
cells: review, Autoimmunity, 40, 281-284.
25.Paine, R., 3rd, Standiford, T. J., Dechert, R.
E., Moss, M., Martin, G. S., Rosenberg, A. L., Thannickal, V. J.,
Burnham, E. L., Brown, M. B., and Hyzy, R. C. (2012) A randomized trial
of recombinant human granulocyte-macrophage colony stimulating factor
for patients with acute lung injury, Crit. Care Med., 40,
90-97.
26.Lo, Y. D., Rainer, T. H., Chan, L. Y., Hjelm, N.
M., and Cocks, R. A. (2000) Plasma DNA as a prognostic marker in trauma
patients, Clin. Chem., 46, 319-323.