REVIEW: Plant Proteases Involved in Regulated Cell Death
A. A. Zamyatnin, Jr.1,2
1Sechenov First Moscow State Medical University, Institute of Molecular Medicine, 119991 Moscow, Russia2Lomonosov Moscow State University, Belozersky Institute of Physico-Chemical Biology, 119991 Moscow, Russia; E-mail: zamyat@belozersky.msu.ru
Received August 28, 2015
Each plant genome encodes hundreds of proteolytic enzymes. These enzymes can be divided into five distinct classes: cysteine-, serine-, aspartic-, threonine-, and metalloproteinases. Despite the differences in their structural properties and activities, members of all of these classes in plants are involved in the processes of regulated cell death – a basic feature of eukaryotic organisms. Regulated cell death in plants is an indispensable mechanism supporting plant development, survival, stress responses, and defense against pathogens. This review summarizes recent advances in studies of plant proteolytic enzymes functioning in the initiation and execution of distinct types of regulated cell death.
KEY WORDS: programmed cell death, PCD, apoptosis, autophagy, vacuolar processing enzyme, metacaspase, phytaspase, papain-like protease, proteasomeDOI: 10.1134/S0006297915130064
Abbreviations: ER, endoplasmic reticulum; NCCD, Nomenclature Committee on Cell Death; PCD, programmed cell death; PS-SCL, positional scanning substrate combinatorial library; RCD, regulated cell death; VPE, vacuolar processing enzyme.
The influence of external stimuli, both mechanical and physicochemical,
often leads to uncontrolled death of living cells. This type of cell
death is generally referred to as “accidental cell death”.
However, in many cases cell death is genetically programmed, although
this program can be cancelled by means of specific genetic or
pharmacological factors. Such “regulated cell death” (RCD)
can occur as a part of physiological programs or can be activated at
the level of the whole organism by intracellular events or some
extracellular factors. Mechanisms of regulated cell death can also be
activated in response to external stimuli. According to the latest
recommendations formulated by the Nomenclature Committee on Cell Death
(NCCD), cellular death occurring as a part of physiologic program in
ontogenesis (i.e. a particular case of RCD) should be referred to as
programmed cell death (PCD) [1].
The term “programmed cell death” was first proposed by Richard Lockshin in the mid-1960s [2]. Intensive studies of PCD phenomenon during last 50 years has led to the discovery of different types of cell death and different mechanisms involved. Such a number of mechanisms necessitated the development of a special classification of cell death types. A classification based on morphological features distinguishing main types of cell death was proposed ten years ago [3]. However, it soon became clear that morphological features alone were not sufficient for exhaustive classification of all types of cell death. Therefore, it was suggested to complement morphological features with the unique molecular and biochemical features specific for certain types of RCD [4]. One of the major molecular markers of cell death is the activity of proteolytic enzymes, which control the initiation and play a role in execution of RCD [4, 5].
Studies of RCD phenomenon in plants have not been as intensive as studies of this process in human and animal cells. Since information about molecular mechanisms of RCD in plants was lacking, the classification of various types of RCD in plants was based only on morphological features [6]. Studies of RCD in plants revealed some similarities with these processes in animals, but also pointed out some differences [6]. One of the main differences was the absence of caspases in plants, while in animals these enzymes are key players in initiation and mediation of apoptosis [5]. At the same time, in plants there are many other proteolytic enzymes involved in various stages of RCD, including enzymes with caspase-like activities and caspase-like functions.
In plants, as in animals, RCD can be induced by both biotic and abiotic factors. Different pathogens (viruses, bacteria, or fungi) can trigger RCD, in a form of hypersensitive response, for example, which is a protective reaction of a plant. In this case, cell death can be induced either by elicitors – compounds specific to a pathogen, recognized by the immune system of a plant – or by pathogen-specific protein products, whose expression in a plant cell causes ER stress leading to the development of RCD [7-10]. Among the abiotic factors triggering the development of RCD in plants are metal ions, oxidative stress, salt stress, UV radiation, and heat shock [11-15]. At the same time, some cells undergo RCD to ensure normal development of a plant. For example, such phenomenon can be observed during the development of xylem [16, 17].
In this review, we summarize our knowledge about proteolytic enzymes involved in regulation and execution of regulated cell death, caused by biotic and abiotic factors, or necessary for normal plant development (table). Proteolytic enzymes involved in particular types of RCD associated with senescence of plant organs will be discussed less extensively.
Plant proteolytic enzymes involved in RCD
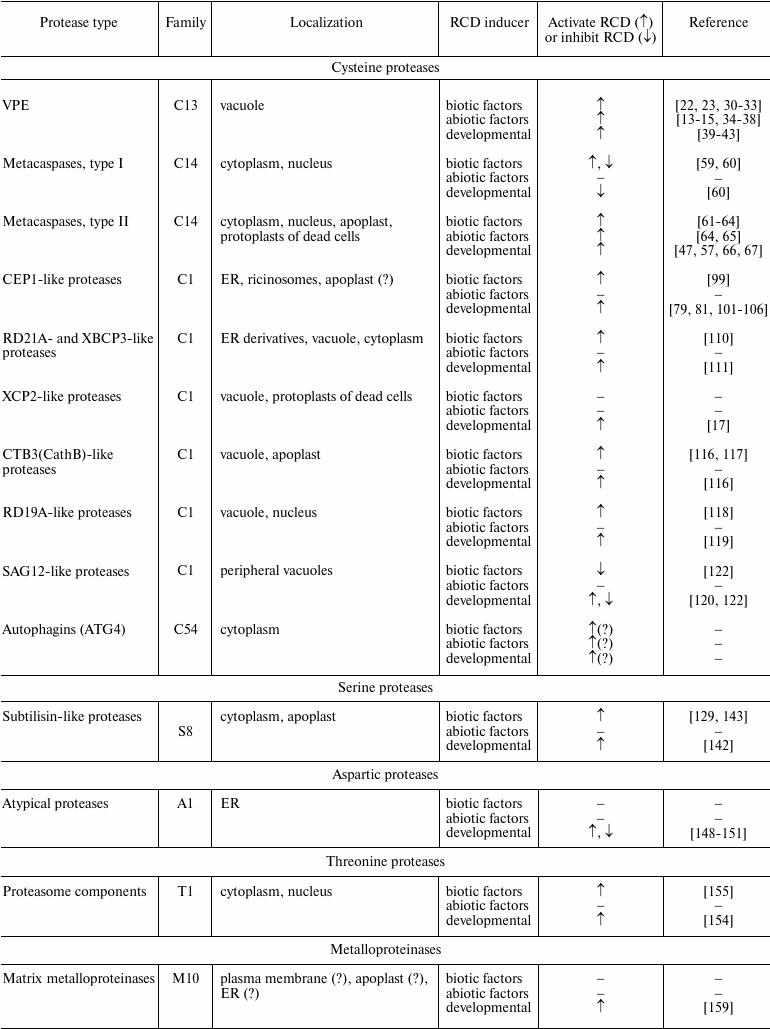
CYSTEINE PROTEASES
Vacuolar processing enzymes (VPE). The detection of caspase-1-like proteolytic activity in dying cells during the development of the hypersensitive response of tobacco to the infection induced by tobacco mosaic virus [18] stimulated active search for a plant enzyme with such properties. Meanwhile, other examples of caspase-like activities in plants were revealed by studies of different types of RCD [19]. However, sequencing of the whole genomes of Arabidopsis thaliana L. and rice Oryza sativa L. [20, 21] showed in plants there are no caspase genes that can be identified by means of simple homology search. Therefore, some groups focused their attention on plant proteolytic enzymes that possess caspase-like activities without being caspase orthologs.
The first identified protease with caspase-1 like activity that is involved in RCD is a vacuolar processing enzyme (VPE) [22, 23]. A VPE is a legumain-like cysteine protease that belongs to the C13 family of the CD clan according to the classification of proteolytic enzymes represented in the MEROPS peptidase database [24]. Like many proteases, VPE is translated as an inactive zymogene containing N- and C-terminal propeptides that are autocatalytically cleaved during activation of the enzyme. The N-terminal fragment contains a signal peptide that guides VPE into the vacuole, where the autocatalytic processing takes place [25, 26].
Homologs of VPEs are widespread throughout the plant kingdom; they are found both in mosses and ferns, as well as in higher plants. A VPE homolog known as asparagine endopeptidase (AEP) [27, 28] is also found in animals. There are four VPE genes in the genome of A. thaliana: αVPE, βVPE, γVPE, and σVPE. Expression of αVPE and γVPE is detected in vegetative organs of the plants, while βVPE is expressed in embryos, and σVPE is expressed during the formation of the seed coat [27, 28].
Collapse of the vacuole induced by a VPE is considered one of the key factors in RCD in plants. A plant VPE was shown to be involved in different types of RCD, including RCD induced by biotic and abiotic factors. Moreover, VPEs play a role in RCD indispensable for the normal development of a plant [28]. A VPE is necessary for the development of a hypersensitive response induced by tobacco mosaic virus in tobacco plants bearing a resistance gene N, as it stimulates degradation of a vacuole, DNA fragmentation, and the development of a necrotic reaction [22, 29]. A VPE is also involved in morphologically similar mechanisms of RCD induced by other viruses, fungi, bacteria, and their toxins [23, 28, 30-32]. It should be noted that VPEs also take part in the development of a special type of RCD induced by ER stress, which is the result of interaction between cells of A. thaliana with the fungus Piriformospora indica [33]. VPEs also play a role in the development of RCD induced by a number of abiotic factors, such as thermal shock [15], salt stress [14, 34], oxidative stress [34, 35], UV radiation [13], and metals [36-38]. Moreover, they are players in the mechanisms of RCD supporting ontogenesis and aging, such as formation of seed coat [39], death of pericarp and nucellus cells in the ovule [40, 41], and senescence of leaves [42] and petals [43].
In spite of the fact that VPEs are key players in the development of some types of cell death in plants, substrates of these enzymes remain largely unknown. Therefore, the conclusion that VPEs in plants have the same functions as caspases in animal cells is premature.
Metacaspases. Only special bioinformatic tools allowed researchers to find in plant genomes the very distant relatives of caspases – metacaspases [44]. Publication of these data stimulated studies of plant metacaspases with the aim to show their functional similarities with caspases. However, soon it became clear that substrate specificity of metacaspases differs dramatically from that of caspases: metacaspases appear to be arginyl/lysyl-specific endopeptidases, while caspases are aspartate-specific proteolytic enzymes [45-47].
Metacaspases are cysteine proteases belonging to peptidase family C14 of the CD clan according to the classification of proteolytic enzymes in the MEROPS database [24]. Metacaspases, similarly to caspases, consist of a large (p20) and a small (p10) subunit. As in caspases, catalytic His and Cys residues in metacaspases are located in the p20 subunit, while the p10 subunit takes part in the formation of a substrate-binding pocket [48, 49]. Plant metacaspases are subdivided into two main types. Type I metacaspases often contain an additional N-terminal proline-rich prodomain with a zinc finger motif. Type II metacaspases never have such N-terminal prodomain and, unlike type I metacaspases, the p20 and p10 subunits of type II enzymes are separated by a linker sequence [48, 50]. Metacaspases found in some phytoplankton species were united into type III. Unlike type I and type II metacaspases, the p10 domain of type III metacaspases is located closer to the N-terminus of the protein, while the p20 domain is closer to the C-terminus [51].
Plant genomes generally contain about ten genes coding metacaspases, although in some species the number of genes can reach twenty [50]. There are nine genes in the A. thaliana genome: three genes coding type I metacaspases (AtMC1-AtMC3) and six genes coding type II metacaspases (AtMC4-AtMC9). All nine genes are expressed in various tissues of the plant [44, 48].
Maturation of metacaspases, similarly to maturation of many others cysteine endopeptidases, is a result of an autocatalytic cleavage of the zymogen. However, autoproteolytic hydrolysis of metacaspases is a Ca2+-dependent process, with rare exceptions [45, 47, 52]. Moreover, as shown for the metacaspase AtMC4, during Ca2+-induced autocatalytic proteolysis the cleavage sites can vary slightly depending on the conditions of the reaction. This probably reflects the existence of a special mechanism that can increase or decrease protease activity at the early stage of its activation [53, 54]. Nitric oxide (NO) can also regulate the activity of metacaspases. It was shown that the catalytic Cys residue of metacaspase AtMC9 could be S-nitrosylated, which can be the result of excess NO and can lead to inhibition of the autoprocessing of the enzyme [55].
Intracellular localization of metacaspases can slightly vary depending on the enzyme, but these proteases are generally localized in the cytoplasm and/or in the nucleus [56]. Moreover, the intracellular localization of an enzyme can change during the development of RCD, as shown, for example, for the Norway spruce mcII-Pa metacaspase: at initial stages of RCD, the enzyme was found in the cytoplasm, while at later stages – in the nucleus [47]. AtMC9 metacaspase of A. thaliana can be found not only in the nucleus and cytoplasm, but also in the apoplast [57, 58].
Plant metacaspases were shown to be key players and regulators of RCD processes induced by different biotic and abiotic factors and in RCD supporting normal plant development. It was shown that type I metacaspase AtMC1 of A. thaliana participates in the activation of hypersensitive response of the plant to infection by the oomycete Hyaloperonospora arabidopsidis or bacterium Pseudomonas syringae, while another type I metacaspase AtMC2 was acting as an antagonist of AtMC1 [59]. Although AtMC2 blocked the development of RCD, it should be noted that in this case this function of the enzyme was not related to its proteolytic activity [59, 60]. Later, it became clear that AtMC1 also can interfere with the development of RCD; a pro-survival role of AtMC1 metacaspase during aging was shown [60]. Expression of the metacaspase-9 gene (CaMC9) in Capsicum annuum L. is increased in response to infection with Xanthomonas campestris. Silencing of the CaMC9 gene inhibits the development of pathogen-triggered RCD, while the overexpression if this gene, in contrast, increases the probability of cell death [61]. Metacaspases TaMC4 in wheat and NbMCA1 in Nicotiana benthamiana L. also participate in the development of RCD as protective mechanisms against infection of wheat with the fungus Puccinia striiformis or infection of tobacco plants with the fungus Colletotrichum destructivum and bacterium P. syringae [62, 63]. Plants with knockout of the type II metacaspase AtMC4 gene showed reduced sensitivity to RCD-inducing mycotoxin fumonisin B1 and other oxidative stress inducers, while overexpression of the AtMC4 gene, in contrast, increased plant sensitivity towards these factors [64]. Overexpression of the metacaspase AtMC8 gene stimulated RCD induced by UV irradiation or H2O2 in protoplasts, while silencing of the AtMC8 gene inhibited it [65]. Norway spruce metacaspase mcII-Pa induces autophagy, which triggers RCD mechanisms during the terminal differentiation of embryonic suspensor cells, and, on the other hand, participates in further development of RCD [47, 66]. Being localized in extracellular space, AtMC9 metacaspase can be considered as an effector of RCD activation, as it cleaves a peptide of 11 amino acids from GRI protein, which, once bound to the membrane receptor PRK5, initiates RCD [67]. Moreover, it turned out that one of the functions of the secreted AtMC9 metacaspase is degradation of xylem cell content after the collapse of the central vacuole. Thus, we conclude that in this particular case the functional role of the enzyme is manifested after the fact of cell death [57]. Is should be mentioned that the genome of A. thaliana contains a gene coding for the protein inhibitor of AtMC9 metacaspase – AtSerpin1, which is at the same time a substrate of AtMC9, since before the formation of a covalent bond with an enzyme, an irreversible inhibitor from the serpin family has to be cleaved by a protease [68]. However, the physiological role of such inhibition of AtMC9 in RCD is not yet clear.
Besides AtSerpin1, several dozens of other substrates of plant metacaspase AtMC9 were identified [58]. However, the functional role of proteolytic cleavage catalyzed by AtMC9 in vivo was demonstrated only for one of them – phosphoenolpyruvate carboxykinase 1 (PEPCK1), which is one of the key enzymes involved in gluconeogenesis in plants. It was shown that processing of PEPCK1 catalyzed by AtMC9 led to increase in enzyme activity [58]. Unfortunately, analysis of identified substrates of metacaspase AtMC9 does not allow making any conclusions about RCD mechanisms that involve AtMC9.
Studies of RCD during the terminal differentiation of embryonic suspensor cells in Norway spruce showed that physiological substrate of mcII-Pa metacaspase was the evolutionarily conserved multifunctional protein Tudor-SN [69]. Genes coding for Tudor-SN protein were found not only in plants, but also in animals including humans. The protein itself, being a component of many ribonucleoprotein complexes, is involved in a number of functional processes related to transcription, splicing, RNA interference, RNA editing, and RNA degradation [70-74]. Interestingly, during apoptosis in human cells Tudor-SN was recognized as a substrate by caspase-3 [69]. Both in plants and in animals, proteolytic cleavage of Tudor-SN led to its inactivation [69]. It was shown recently that not only plant metacaspases have common substrates with caspases from animal cells, but also metacaspases from fungi. Thus, PaMCA1 metacaspase of Podospora anserina is able to cleave in vivo a classical caspase substrate – PARP [75]. These data relaunched discussion of whether metacaspases can be considered as caspases. A number of researchers suppose that metacaspases can be considered as functional analogs of caspases, and RCD mechanisms are so conserved that common features can be found in RCD processes in different kingdoms of living organisms.
Papain-like enzymes. According to the mechanism of catalysis, papain-like endopeptidases are cysteine proteases. Taking into account their phylogenetic features, papain-like C1A proteases (C1 family of CA clan) were placed with enzymes similar to L-, B-, H-, and F-cathepsins from animals [24, 76].
Papain-like proteases are relatively stable proteins. They are often found in rather harsh environment, such as the apoplast, the vacuole, and lysosomes [77]. These proteases are globular proteins, composed of two domains, both of which are involved in substrate binding pocket formation. This pocket is able to bind substrates, and the radicals of the catalytic triad composed of Cys, His, and Asn residues are located in this pocket [78].
Activity and specificity of plant C1A peptidases in vitro were thoroughly studied by means of various tests using proteins, fluorogenic synthetic peptide substrates, and peptide inhibitors. Moreover, new bioinformatics methods allowing modeling of molecular interactions between endopeptidases and their substrates are being developed. Data accumulated so far enable us to conclude that plant papain-like proteases have rather low specificity. Nevertheless, studies of substrate specificity of these peptidases showed that their preferential substrate should have a nonpolar (including Pro) or aromatic amino acid residue in P2 position, and in some cases, it can be an Arg residue [79-83].
To be targeted to a specific intercellular compartment, protease precursors carry a signal peptide, whereas the presence of an autoinhibitory prodomain prevents premature activation of the enzyme [80, 84]. Upon translation, the polypeptide chain of the inactive proenzyme enters the ER lumen; the majority of C1A proteases is addressed through trans-Golgi into the vacuole, lysosomes, or is secreted into the apoplast [76]. However, some C1A proteases carrying a C-terminal signal for retention in the endoplasmic reticulum K/HDEL can be addressed into other specific compartments, such as ricinosomes, which are derivatives of the endoplasmic reticulum (ER) [79, 85, 86]. Further cleavage of the prodomain can occur in cis via intramolecular interactions or in trans via intermolecular interactions. Moreover, some data show that the prodomain can be cleaved by other proteases, which suggests that papain-like proteolytic enzymes can be a part of proteolytic cascades [80, 87].
A peculiar feature of plant papain-like proteases when compared to other proteolytic enzymes involved in RCD is that some of these enzymes can be reversibly inhibited by natural peptides, for example, by plant cystatins. Plant cystatins can interact with members of the papain-like C1A family of cysteine proteases; phytocystatins were placed in a separate subfamily within the cystatin family [24, 76]. Generally, plant cystatins are rather small proteins with molecular mass of 12-16 kDa. Nevertheless, high molecular weight phytocystatins with molecular mass of 85-87 kDa and containing several cystatin domains were also found [88-91]. Moreover, in vitro experiments showed that for inhibition of legumain-like C13 peptidases, phytocystatins should carry additional C-terminal extensions, which increase the molecular mass of the protein up to 23 kDa [92, 93]. As mentioned earlier, VPEs belong to the C13 family of cysteine peptidases, although there are no data so far that VPE activity can be regulated by protein inhibitors. Another peptide inhibitor of papain-like C1A cysteine proteases is a member of the serpin family, AtSerpin1, which is able to inhibit RD21A protease [94, 95].
Preproenzymes of plant papain-like proteases always consist of an N-terminal signal peptide, a prodomain, and a proteolytic domain containing the catalytic triad Cys-His-Asn. Especially for plants, an additional classification of papain-like proteases was proposed [80] that takes into account structural particularities of plant enzymes, such as an additional C-terminal granulin domain, proline-rich domain, C-terminal ER-retention signal (K/HDEL), vacuole localization signal NPIR in the beginning of the prodomain, and some others [79, 80, 96]. According to this classification, there are nine subfamilies of papain-like proteases in plants [80]. Data obtained so far have shown that members of at least seven subfamilies (CEP1-, RD19A-, RD21A-, XCP2-, XBCP3-, SAG12- and CTB3-like proteases) are implicated in the development of various types of RCD. However, there is no evidence of any special role of papain-like proteases belonging to the AALP- and THI1-like subfamilies in the development of RCD.
CEP1-like proteases. Members of CEP1-like subfamily of proteases are characterized by the presence of the C-terminal ER-retention signal KDEL. Interestingly, genes coding for homologs of KDEL-containing plant proteases were not found in the yeast genome or in animals [80, 97]. Proteases carrying ER-retention signal are often addressed into special structures, ricinosomes, which are the derivatives of ER but separated from it. Ricinosomes do not merge with the central vacuole, but as RCD is initiated they collapse and release proteases and hydrolases they contain [81, 85, 98]. Under biotic stress, CEP1 protease in A. thaliana is accumulated in ER and is probably further released in the apoplast [99].
Many members of CEP1-like protease subfamily were shown to participate in the development of RCD in various plants [100, 101]. The best-studied members of this subfamily are the CEP1, CEP2, and CEP3 proteases of A. thaliana, which are expressed in roots, stems, flowers, and green siliques [97]. Under biotic stress conditions, expression of CEP1 was also observed in leaves [99]. Participation of this protease in the development of regulated death of epidermal cells, caused by the infection with the ascomycete Erysiphe cruciferarum was studied in detail [99], as well as the role of CEP1 protease in RCD mechanisms during maturation of pollen in tapetum, a special layer that covers sporangia and anthers [101]. CEP2 protease was shown to participate in the development of RCD during the formation of a root cap [81]. Moreover, in other plant species KDEL-containing proteases participate in RCD processes involved in petal senescence in daylily (Hemerocallis sp.) [102], in orchid seed coat formation in Phalaenopsis sp. [103] and Jatropha curcas L. [104], in RCD in the endosperm of Ricinus communis L. [79], tomatoes [105], and J. curcas [104], as well as in the anther of tomatoes [106]. It should be emphasized that some KDEL-containing endopeptidases were able to cleave the hydroxyproline-rich glycoprotein extensin, which is a component of the plant cell wall [97].
Thus, a large experimental dataset has accumulated so far, showing the role of proteolytic CEP1-like enzymes in different types of RCD, including RCD mechanisms supporting normal plant development and providing defense against the pathogens.
RD21A- and XBCP3-like proteases. Proteolytic enzymes belonging to the RD21A- and XBCP3-like protease subfamilies (members of these two subfamilies differ mainly in primary structures of the prodomains) are characterized by the possible presence of an additional C-terminal fragment composed of a proline-rich domain followed by a granulin-like domain [80, 107]. In animals, granulins play a role as extracellular growth factors. They are expressed as pro-granulin, which comprises several copies of cysteine-rich granulin modules [108]. In plants, granulins were found only within proenzymes of some papain-like proteases. The function of granulins in plants remains unknown [80].
Sorting of A. thaliana RD21 protease occurs through the Golgi apparatus, where the protein is fucosylated, then the enzyme is accumulated in compartments derived from ER, and after fusion of these compartments with the vacuole the enzyme stays in the vacuole [87, 107, 109, 110]. It is generally thought that cleavage of the prodomain of RD21 in vivo can be catalyzed by one or several other proteases, which point out the possible implication of RD21 in proteolytic cascades. However, granulin-like domain cleavage occurs by autocatalysis [87]. Interestingly, RD21 protease purified from plants can be activated with SDS. This might indicate that RD21 protease is accumulated in vivo as a complex with a reversible endogenous inhibitor [87]. Moreover, it was shown that activity of RD21 could be inhibited by the irreversible endogenous inhibitor AtSerpin1, which belongs to the serpin family – a family of peptide inhibitors of proteases [94]. Within cells, AtSerpin1 is localized in the cytoplasm, so probably the inhibition of RD21 by AtSerpin1 can occur during translocation of the enzyme to the cytoplasm [110]. Elicitors of RCD in plants, such as benzothiadiazole (a salicylic acid agonist) or oxalic acid (a toxin of pathogenic fungi, such as Botrytis cinerea and Sclerotinia sclerotiorum), stimulate changes in the permeability of vacuole membranes, which can lead to the translocation of RD21 into the cytoplasm with formation of inactive RD21–AtSerpin1 complexes [110]. If the level of RD21 expression is decreased, or the level of AtSerpin1 expression is increased, development of elicitor-triggered RCD slows considerably, which led to the conclusion that RD21 is a stimulator of RCD, while AtSerpin1 reduces the activity of this effector [110].
NtCP14 protease from tobacco is closer to XBCP3 protease than to RD21A from the point of view of phylogeny. Nevertheless, it was shown that NtCP14 protease along with its inhibitor cystatin NtCYS has similar function to that of RD21 protease and its inhibitor AtSerpin1 [110, 111]. It was shown that in the majority of cells of tobacco embryos, NtCP14 protease is not active because it forms a complex with NtCYS, while in embryonic suspensor cells, where RCD starts to develop, the level of NtCYS decreases, leading to the appearance of NtCP14 protease molecules that are not bound to NtCYS and therefore active. The presence of active NtCP14 protease ensures further development of regulated death processes of suspensor cells [111].
Thus, we conclude that plant proteolytic enzymes bearing a granulin sequence within the proenzyme together with their inhibitors play a regulatory role in different types of RCD induced either to ensure ontogenesis or to assure the activation of defense mechanisms. Involvement of these enzymes in proteolytic cascades accompanying RCD cannot be excluded.
XCP2-like proteases. One of the main properties of plant C1A proteases belonging to the XCP2-like protease subfamily is the presence of a conserved putative glycosylation site in the protease domain [80]. Particularities of secondary structure of these proteolytic enzymes can be discussed since the crystal structure of papain, which also belongs to the XCP2-like protease subfamily, was defined some time ago [112]. The genome of A. thaliana contains genes coding for two members of this subfamily: XCP1 and XCP2. Genes of these two proteases along with a gene coding for the subtilisin-like serine protease XSP1 were initially identified in a xylem cDNA library [113]. It was shown later that during xylogenesis, XCP1 and XCP2 are transported into the central vacuole of the cell, where they participate in micro-autolysis preceding the macro-autolysis induced by tonoplast breakage [17, 114]. It is considered that after tonoplast breakage during formation of xylem tracheary elements, both enzymes continue further degradation of cellular content [17]. The hypothesis of implication of XCP1 and XCP2 enzymes in providing defense from pathogens that live and propagate in plant xylem was suggested recently [115].
CTB3(CathB)-like proteases. The primary structure of CTB3-like proteases is close to that of human cathepsin B. Their distinctive feature is the presence of four additional disulfide bonds and a conservative putative glycosylation site [80]. The genome of A. thaliana contains genes coding for three members of this subfamily: CTB1 (AtCathB1), CTB2 (AtCathB2), and CTB3 (AtCathB3) [80, 116]. The level of expression of these genes increases with senescence and during contacts with pathogens. Moreover, in mutant A. thaliana with deletion of all three genes of the CTB3-like proteases, significant delay in the development of senescence is observed, as well as the reduction of hypersensitive response induced by P. syringae [116]. In N. benthamiana plants, silencing of a CTB3-like protease (NbCathB) also leads to reduction in hypersensitive response to bacterial infections caused by Erwinia amylovora and P. syringae [117]. Moreover, NbCathB was found to be secreted into the apoplast, where it can be activated even in absence of pathogens [117]. Thus, one can conclude that plant cathepsin B-like proteolytic enzymes are implicated in processes related to RCD indispensable for ontogenesis, or for RCD providing resistance to pathogens.
RD19A-like proteases. RD19A-like proteases are characterized by the presence in the prodomain of an ERFNAQ motif and four additional cysteine residues, which are supposed to form disulfide bonds for additional stabilization of enzyme structure, as well as by the presence of a conserved putative glycosylation site [80]. As shown for A. thaliana, RD19 participates in biochemical pathways that assure resistance of the plant to bacterial infection caused by Ralstonia solanacearum. RD19 protease is translocated from the mobile structures associated with the vacuole into the nucleus [118]. However, direct implication of RD19 in RCD processes in A. thaliana has not been reported. On the other hand, data about the involvement of a close homolog of RD19, SmCP protease, in RCD processes in eggplant (Solanum melongena L.) was published. SmCP was also proposed to be implicated in RCD processes indispensable for ontogenesis, particularly during formation of xylem tracheary elements [119].
SAG12-like proteases. The majority of SAG12-like proteases contain an additional Cys residue before the catalytic Cys residue (motif CGCCWAFS) [80]. AtSAG12 protease of A. thaliana (from senescence-associated genes) was identified as one of the products encoded by genes associated with senescence [120]. It was shown that this proteolytic enzyme is localized in special vacuoles that are found in peripheral cytoplasm separately from the central vacuole in aging cells of the mesophyll. However, the phenotype of A. thaliana with deleted AtSAG12 gene did not differ from that of the wild type [121]. At the same time, expression of the gene coding for a homologous protease in rice (OsSAG12-1) was induced not only by senescence, but also during RCD supporting the response to a biotic stress [122]. Silencing of the OsSAG12-1 gene in rice led to accelerated senescence and increased RCD in case of infection with bacterial pathogen Xanthomonas oryzae, compared to the wild type plants, suggesting that OsSAG12-1 is an RCD suppressor [122].
Autophagins (ATG4). Describing the key role of cysteine proteases in regulated cell death in plants, it is not possible to omit autophagins or Atg4 – cysteine proteases that are key factors initiating autophagosome formation [123, 124]. It is important to note that autophagy in plants can be observed during different types of RCD, both before and after the point of no return [125, 126]. Thus, key factors of autophagy, including Atg4, can be considered as legitimate players and regulators of RCD processes in plants.
Atg4 proteases belong to the C54 family of cysteine proteases from the CA clan [24, 123]. The primary structure of these enzymes does not contain signal peptide, which defines their cytoplasmic localization. The main function of these enzymes is the processing of Atg8 protein, which gains thereafter the ability to bind phosphatidylethanolamine and to subsequently initiate autophagosome formation [124]. Unlike yeast, bearing one copy of Atg4 and one copy of Atg8 genes, mammals and plants have several homologous genes of Atg4 and Atg8 [127, 128]. Two genes coding for Atg4 (AtAtg4a and AtAtg4b) and nine genes coding for Atg8 (AtAtg8a-AtAtg8i) were found in the genome of A. thaliana [128]. One cannot exclude the possibility that two Atg4 genes of A. thaliana encode protein products with different functions. Indeed, in vitro experiments have shown that AtAtg4a demonstrated higher proteolytic activity than AtAtg4b, although the substrate specificity of the two enzymes was quite similar [128]. Such differences might have functional importance for regulation of a RCD program.
SERINE PROTEASES
Attempts to reveal the sources of caspase-like activity in plant cells during regulated cell death led to characterization of one more proteolytic RCD-related enzyme. It is a member of a subtilisin-like family of proteases named phytaspase after its source (from φυτό – plant in Greek) and manifested activity (aspartate specific protease) [129]. Phytaspase was shown to participate in the development of hypersensitive response in N. tabacum, bearing a resistance gene N, after infection of plants with tobacco mosaic virus [129]. Moreover, activity of this enzyme was also detected in response to mechanical damage in mono- and dicotyledonous plants [130].
Subtilisin-like proteases, also called subtilases, are serine proteases. These enzymes are characterized by the presence of a catalytic triad composed of Asp, His, and Ser residues [131]. According to the MEROPS classification of proteolytic enzymes, subtilisin-like enzymes belong to the S8 family, clan SB [80]. Plant genomes contain a large number of genes coding for various subtilases. For example, in the genome of A. thaliana there are 56 genes coding for these enzymes [132, 133]. Subtilases, like many other proteases, are translated as inactive zymogens composed of a signal peptide, prodomain, and peptidase domain, in which the protease-associated domain is localized [133-135]. Secondary structure of subtilases should resemble that of tomato SlSBT3 subtilase that was solved [136]. In plants, subtilases have various functions; some of them are related to the response to pathogen infection [135]. Substrate specificity for many of these enzymes is not very high [132, 133].
Unlike other characterized subtilases, phytaspases from rice and A. thaliana showed rather high specificity, recognizing, for example, caspase-6 classical cleavage site VEID. It turned out that phytaspases are also able to cleave a number of fluorescent peptide substrates cleaved by caspases, such as YVAD, VAD, IETD, LEHD, and some others. However, it should be noted that phytaspases were not able to cleave one classical caspase-3 substrate DEVD [129, 137], but cleaved with high efficiency IWLD peptide, which differs considerably from common caspase substrates [138]. Study of rice phytaspase using the positional scanning substrate combinatorial library (PS-SCL) [139] showed that unlike caspases, this enzyme prefers substrates containing hydrophobic amino acid residues in positions P4-P2 [138]. Moreover, it was shown that phytaspase cleaves more efficiently full-length proteins, which probably signifies that interactions between phytaspases and its substrate are not limited to the P4-P1 positions [138].
Overexpression of the phytaspase gene increased sensitivity of plants towards inducers of RCD, while silencing of this gene inhibited RCD, suggesting that phytaspase actively participates in processes of cell death [129]. However, the most remarkable feature of phytaspases is their localization. It was shown that constitutively expressed phytaspase zymogen is processed and the mature enzyme is secreted to the apoplast. When RCD is induced by biotic or abiotic factors, active phytaspase is translocated back to the cytoplasm, where it starts to hydrolyze proteins [129, 140]. Unfortunately, only one natural substrate of phytaspase – VirD2 protein from the plant pathogenic bacterium Agrobacterium tumefaciens – has been characterized so far. VirD2 is responsible for the delivery of a fragment of bacterial DNA in the nucleus of an infected cell. After being processed by phytaspase, VirD2 loses its C-terminal signal peptide, responsible for nuclear localization, which might be a part of the mechanism actively protecting the plant cell from undesirable transformation [141]. However, this process is not directly related to RCD. Thus, questions about natural phytaspase substrates that are processed during RCD and about functional consequences of this hydrolysis remain open.
It was shown that subtilisin-like AtSBT1.1 protease from A. thaliana is able to cleave the prepropeptide of the phytosulfokine AtPSK4 [142]. Active hormone produced upon this cleavage is able to stimulate differentiation of xylem tracheary elements [115]. In the cDNA library from xylem tissue of Arabidopsis, the gene of another subtilisin-like protease XSP1 was found [113]. However, functions of proteolytic enzyme XSP1 have not been investigated yet.
At least two more putative subtilisin-like functional analogs of animal caspases were found in oats. Both enzymes, named saspases (SAS-1 and SAS-2), showed high substrate specificity similar to that of caspases [135, 143]. These proteases were shown to be secreted into intercellular space during the development of RCD induced by Cochliobolus victoriae toxin victorin and accompanied by DNA fragmentation and mitochondrial dysfunction [143]. However, their functional role in RCD was not characterized. Their natural substrates also remain unknown.
ASPARTIC PROTEASES
The genome of A. thaliana contains more than fifty genes coding for aspartic proteases [144]. Despite of this large number of genes (the human genome codes for only eight aspartic proteases [145] and the genome of Caenorhabditis elegans – only twelve [146]), functions of corresponding protein products generally remain unknown. According to MEROPS classification of proteolytic enzymes, proteases from families A1, A3, A11, and A12 of AA clan and proteases from A22 family of AD clan were found in plants [80]. Generally, plant aspartic proteases are divided into three groups: typical aspartic proteases (group A), nucellin-like (group B), and atypical proteases (group C) [144, 147]. Typical aspartic proteases from plants usually contain an additional C-terminal domain that is cleaved during enzyme maturation. Nucellin-like aspartic proteases are homologs of nucellin – a protease found in cells of the nucellus in barley. Atypical aspartic proteases demonstrate various intermediate characteristics specific for typical and nucellin-like proteolytic enzymes [144, 147].
Data showing involvement of aspartic proteases in RCD of plant cells is rather limited. Nevertheless, it was shown that expression of genes coding for two atypical proteases (OsAP25 and OsAP37) in tapetal cells of rice was regulated by transcription factor EAT1. Proteolytic enzymes OsAP25 and OsAP37 are involved in mechanisms of regulated death of these cells [148]. Taking into account that expression of nucellin in barley is limited to nucellar cells in which RCD occurs, we can also hypothesize involvement of this representative of aspartic proteases in processes of cell death [149].
It should also be noted that among aspartic proteases, similarly to papain-like proteases, proteolytic enzymes capable of inhibiting the development of RCD were found. For example, in A. thaliana atypical aspartic protease PCS1 carries an N-terminal serine-rich fragment and is localized in ER. This enzyme can inhibit the development of RCD in some types of cells that normally die. Moreover, lack of expression of active PCS1 leads to massive RCD in gametophytes of both types, as well as during embryogenesis [150]. Another example from A. thaliana is the atypical protease UNDEAD, which can inhibit the development of programmed death of tapetal cells [151].
Thus, it was shown that some aspartic proteases participate in regulation of at least some types of RCD needed for normal ontogenesis. Moreover, these enzymes can act both as positive and negative regulators of RCD.
THREONINE PROTEASES
In eukaryotes, 26S proteasome is the main proteolytic component of the ubiquitin-dependent protein degradation system. The 26S proteasome (in plants as well) consists of two components: 20S proteasome and two 19S regulatory particles (RPs), which interact with ubiquitinated proteins assuring substrate specificity of 26S-complex. In A. thaliana, three β-subunits out of seven subunits of the 20S proteasome (PBA, PBB, and PBE) belong to the threonine peptidase family (T1 family, PB clan) and possess proteolytic activity [152]. In plant cells, there are both 26S proteasomes and free 20S proteasomes capable of ubiquitin-dependent and ubiquitin-independent cleavage of proteins, respectively [152, 153].
It was recently shown that 20S proteasomes are responsible for caspase-3-like activity detected during xylem development in A. thaliana and poplar [154]. Hatsugai with coauthors showed that proteolytically active β-subunit of 20S proteasome PBA1 is partially responsible for caspase-3-like activity that can be detected during the development of hypersensitive response of A. thaliana to bacterial infection caused by P. syringae [155]. Taking into account these data, proteasomes and/or their components were proposed to be involved in RCD mechanisms in plants.
METALLOPROTEINASES
Plant genomes also contain genes coding matrix metalloproteinases. For example, in the genome of A. thaliana there are five such enzymes [156, 157]. According to the MEROPS classification of proteolytic enzymes, these proteases belong to M10 family from MA clan [80]. As in other organisms, in plants matrix metalloproteinases are composed of a signal peptide, prodomain, and catalytic domain containing a zinc-binding motif. Activation of the enzyme occurs after physical separation of the prodomain from the catalytic site, which can be reached by means of proteolytic cleavage of the prodomain [158]. It is believed that plant matrix metalloproteinases are localized either in plasma membrane or in intercellular space [156, 157]. Moreover, At4-MMP metalloproteinase from A. thaliana bears non-cleavable N-terminal signal peptide that can target this enzyme to the ER [156]. Implication of plant matrix metalloproteinases in RCD processes was not demonstrated, except the fact that the gene coding for matrix metalloproteinase Cs1-MMP of cucumber is expressed de novo at final steps of aging processes in cotyledon before initiation of RCD [159]. This fact suggests that plant matrix metalloproteinases might be involved in RCD processes, although functions of these enzymes still need to be studied.
Plant genomes bear genes coding for hundreds of proteases. In A. thaliana, their number reaches 800 [24]. Among proteins encoded by plant protease genes, representatives of all five main classes of proteolytic enzymes are found: cysteine, serine, aspartic, threonine, and metalloproteinases. Now it is clear that representatives of all these classes of proteolytic enzymes are involved in developments of various types of regulated cell death (RCD) (table).
This review has summarized data about proteolytic enzymes of plants able to regulate initiation and further developments of regulated cell death caused by biotic and abiotic factors or by particularities of ontogenesis. However, this review does not discuss (with rare exceptions) participation of plant proteolytic enzymes in senescence, since it is still difficult to distinguish processes preceding regulated cell death and RCD itself. Moreover, the role of plant proteolytic enzymes in senescence was reviewed earlier [160, 161].
In fact, accumulation of data about the functional role of each proteolytic enzyme implicated in RCD mechanisms in plants has just started. Only a few physiological substrates of these enzymes have been identified so far. Obviously, we still need to characterize the majority of these substrates. There is some evidence that proteolytic cascades can be involved in initiation and regulation of RCD in plants, as well as in animals. Identification of particular endopeptidases involved in these cascades will undoubtedly be subject of further research. In plants, several types of RCD demanding participation of several proteolytic enzymes were already described; however, the question of regulation of their interactions remains without answer so far.
One of the reasons for intensive studies of RCD in humans and other animals is the direct connection of RCD in humans with processes of carcinogenesis. It is now clear that RCD in plants is tightly linked with processes of development, as well as with resistance mechanisms against various stresses and pathogens. Thus, studies of molecular mechanisms of regulated cell death in plants are not only important for basic research; they might provide new tools for regulation of defense mechanisms and ontogenesis in plants.
REFERENCES
1.Galluzzi, L., Bravo-San Pedro, J. M., Vitale, I.,
Aaronson, S. A., Abrams, J. M., Adam, D., Alnemri, E. S., Altucci, L.,
Andrews, D., Annicchiarico-Petruzzelli, M., Baehrecke, E. H., Bazan, N.
G., Bertrand, M. J., Bianchi, K., Blagosklonny, M. V., Blomgren, K.,
Borner, C., Bredesen, D. E., Brenner, C., Campanella, M., Candi, E.,
Cecconi, F., Chan, F. K., Chandel, N. S., Cheng, E. H., Chipuk, J. E.,
Cidlowski, J. A., Ciechanover, A., Dawson, T. M., Dawson, V. L., De
Laurenzi, V., De Maria, R., Debatin, K. M., Di Daniele, N., Dixit, V.
M., Dynlacht, B. D., El-Deiry, W. S., Fimia, G. M., Flavell, R. A.,
Fulda, S., Garrido, C., Gougeon, M. L., Green, D. R., Gronemeyer, H.,
Hajnoczky, G., Hardwick, J. M., Hengartner, M. O., Ichijo, H., Joseph,
B., Jost, P. J., Kaufmann, T., Kepp, O., Klionsky, D. J., Knight, R.
A., Kumar, S., Lemasters, J. J., Levine, B., Linkermann, A., Lipton, S.
A., Lockshin, R. A., Lopez-Otin, C., Lugli, E., Madeo, F., Malorni, W.,
Marine, J. C., Martin, S. J., Martinou, J. C., Medema, J. P., Meier,
P., Melino, S., Mizushima, N., Moll, U., Munoz-Pinedo, C., Nunez, G.,
Oberst, A., Panaretakis, T., Penninger, J. M., Peter, M. E.,
Piacentini, M., Pinton, P., Prehn, J. H., Puthalakath, H., Rabinovich,
G. A., Ravichandran, K. S., Rizzuto, R., Rodrigues, C. M., Rubinsztein,
D. C., Rudel, T., Shi, Y., Simon, H. U., Stockwell, B. R., Szabadkai,
G., Tait, S. W., Tang, H. L., Tavernarakis, N., Tsujimoto, Y., Vanden
Berghe, T., Vandenabeele, P., Villunger, A., Wagner, E. F., Walczak,
H., White, E., Wood, W. G., Yuan, J., Zakeri, Z., Zhivotovsky, B.,
Melino, G., and Kroemer, G. (2015) Essential versus accessory aspects
of cell death: recommendations of the NCCD 2015, Cell Death
Differ., 22, 58-73.
2.Lockshin, R. A. (2008) Early work on apoptosis, an
interview with Richard Lockshin, Cell Death Differ., 15,
1091-1095.
3.Kroemer, G., El-Deiry, W. S., Golstein, P., Peter,
M. E., Vaux, D., Vandenabeele, P., Zhivotovsky, B., Blagosklonny, M.
V., Malorni, W., Knight, R. A., Piacentini, M., Nagata, S., and Melino,
G. (2005) Nomenclature Committee on Cell Death. Classification of cell
death: recommendations of the Nomenclature Committee on Cell Death,
Cell Death Differ., 12, 1463-1467.
4.Galluzzi, L., Vitale, I., Abrams, J. M., Alnemri,
E. S., Baehrecke, E. H., Blagosklonny, M. V., Dawson, T. M., Dawson, V.
L., El-Deiry, W. S., Fulda, S., Gottlieb, E., Green, D. R., Hengartner,
M. O., Kepp, O., Knight, R. A., Kumar, S., Lipton, S. A., Lu, X.,
Madeo, F., Malorni, W., Mehlen, P., Nuñez, G., Peter, M. E.,
Piacentini, M., Rubinsztein, D. C., Shi, Y., Simon, H. U.,
Vandenabeele, P., White, E., Yuan, J., Zhivotovsky, B., Melino, G., and
Kroemer, G. (2012) Molecular definitions of cell death subroutines:
recommendations of the Nomenclature Committee on Cell Death 2012,
Cell Death Differ., 19, 107-120.
5.Crawford, E. D., and Wells, J. A. (2011) Caspase
substrates and cellular remodeling, Annu. Rev. Biochem.,
80, 1055-1087.
6.Van Doorn, W. G., Beers, E. P., Dangl, J. L.,
Franklin-Tong, V. E., Gallois, P., Hara-Nishimura, I., Jones, A. M.,
Kawai-Yamada, M., Lam, E., Mundy, J., Mur, L. A., Petersen, M.,
Smertenko, A., Taliansky, M., Van Breusegem, F., Wolpert, T.,
Woltering, E., Zhivotovsky, B., and Bozhkov, P. V. (2011) Morphological
classification of plant cell deaths, Cell Death Differ.,
18, 1241-1246.
7.Sanchez-Vallet, A., Mesters, J. R., and Thomma, B.
P. (2015) The battle for chitin recognition in plant–microbe
interactions, FEMS Microbiol. Rev., 39, 171-183.
8.Solovieva, A. D., Frolova, O. Yu., Solovyev, A. G.,
Morozov, S. Yu., and Zamyatnin, A. A., Jr. (2013) Effect of
mitochondria-targeted antioxidant SkQ1 on programmed cell death induced
by viral proteins in tobacco plants, Biochemistry (Moscow),
78, 1006-1012.
9.Lukhovitskaya, N. I., Yelina, N. E., Zamyatnin, A.
A., Jr., Schepetilnikov, M. V., Solovyev, A. G., Sandgren, M., Morozov,
S. Y., Valkonen, J. P., and Savenkov, E. I. (2005) Expression,
localization and effects on virulence of the cysteine-rich 8 kDa
protein of Potato mop-top virus, J. Gen. Virol.,
86, 2879-2889.
10.Ye, C. M., Chen, S., Payton, M., Dickman, M. B.,
and Verchot, J. (2013) TGBp3 triggers the unfolded protein response and
SKP1-dependent programmed cell death, Mol. Plant Pathol.,
14, 241-255.
11.Petrov, V., Hille, J., Mueller-Roeber, B., and
Gechev, T. S. (2015) ROS-mediated abiotic stress-induced programmed
cell death in plants, Front. Plant Sci, 6, 69.
12.Shahid, M., Pourrut, B., Dumat, C., Nadeem, M.,
Aslam, M., and Pinelli, E. (2014) Heavy-metal-induced reactive oxygen
species: phytotoxicity and physicochemical changes in plants, Rev.
Environ. Contam. Toxicol., 232, 1-44.
13.Danon, A., Rotari, V. I., Gordon, A., Mailhac,
N., and Gallois, P. (2004) Ultraviolet-C overexposure induces
programmed cell death in Arabidopsis, which is mediated by
caspase-like activities and which can be suppressed by caspase
inhibitors, p35 and defender against apoptotic death, J. Biol.
Chem., 279, 779-787.
14.Kim, Y., Wang, M., Bai, Y., Zeng, Z., Guo, F.,
Han, N., Bian, H., Wang, J., Pan, J., and Zhu, M. (2014) Bcl-2
suppresses activation of VPEs by inhibiting cytosolic Ca2+
level with elevated K+ efflux in NaCl-induced PCD in rice,
Plant Physiol. Biochem., 80, 168-175.
15.Li, Z., Yue, H., and Xing, D. (2012) MAP kinase
6-mediated activation of vacuolar processing enzyme modulates heat
shock-induced programmed cell death in Arabidopsis, New
Phytol., 195, 85-96.
16.Bagniewska-Zadworna, A., Arasimowicz-Jelonek, M.,
Smolinski, D. J., and Stelmasik, A. (2015) New insights into pioneer
root xylem development: evidence obtained from Populus
trichocarpa plants grown under field conditions, Ann. Bot.,
113, 1235-1247.
17.Avci, U., Petzold, H. E., Ismail, I. O., Beers,
E. P., and Haigler, C. H. (2008) Cysteine proteases XCP1 and XCP2 aid
micro-autolysis within the intact central vacuole during xylogenesis in
Arabidopsis roots, Plant J., 56, 303-315.
18.Del Pozo, O., and Lam, E. (1998) Caspases and
programmed cell death in the hypersensitive response of plants to
pathogens, Curr. Biol., 8, 1129-1132.
19.Bonneau, L., Ge, Y., Drury, G. E., and Gallois,
P. (2008) What happened to plant caspases? J. Exp. Bot.,
59, 491-499.
20.Sasaki, T. (1998) The rice genome project in
Japan, Proc. Natl. Acad. Sci. USA, 95, 2027-2028.
21.Dennis, C., and Surridge, C. (2000)
Arabidopsis thaliana genome. Introduction, Nature,
408, 791.
22.Hatsugai, N., Kuroyanagi, M., Yamada, K., Meshi,
T., Tsuda, S., Kondo, M., Nishimura, M., and Hara-Nishimura, I. (2004)
A plant vacuolar protease, VPE, mediates virus-induced hypersensitive
cell death, Science, 305, 855-858.
23.Rojo, E., Martin, R., Carter, C., Zouhar, J.,
Pan, S., Plotnikova, J., Jin, H., Paneque, M., Sanchez-Serrano, J. J.,
Baker, B., Ausubel, F. M., and Raikhel, N. V. (2004) VPEgamma exhibits
a caspase-like activity that contributes to defense against pathogens,
Curr. Biol., 14, 1897-1906.
24.Rawlings, N. D., Waller, M., Barrett, A. J., and
Bateman, A. (2014) MEROPS: the database of proteolytic enzymes, their
substrates and inhibitors, Nucleic Acids Res., 42,
503-509.
25.Hiraiwa, N., Nishimura, M., and Hara-Nishimura,
I. (1999) Vacuolar processing enzyme is self-catalytically activated by
sequential removal of the C-terminal and N-terminal
propeptides, FEBS Lett., 447, 213-216.
26.Kuroyanagi, M., Nishimura, M., and
Hara-Nishimura, I. (2002) Activation of Arabidopsis vacuolar
processing enzyme by self-catalytic removal of an auto-inhibitory
domain of the C-terminal propeptide, Plant Cell Physiol.,
43, 143-151.
27.Hara-Nishimura, I., and Hatsugai, N. (2011) The
role of vacuole in plant cell death, Cell Death Differ.,
18, 1298-1304.
28.Hatsugai, N., Yamada, K., Goto-Yamada, S., and
Hara-Nishimura, I. (2015) Vacuolar processing enzyme in plant
programmed cell death, Front. Plant Sci., 6, 234.
29.Hara-Nishimura, I., Hatsugai, N., Nakaune, S.,
Kuroyanagi, M., and Nishimura, M. (2005) Vacuolar processing enzyme: an
executor of plant cell death, Curr. Opin. Plant Biol., 8,
404-408.
30.Kuroyanagi, M., Yamada, K., Hatsugai, N., Kondo,
M., Nishimura, M., and Hara-Nishimura, I. (2005) Vacuolar processing
enzyme is essential for mycotoxin-induced cell death in Arabidopsis
thaliana, J. Biol. Chem., 280, 32914-32920.
31.Gauthier, A., Lamotte, O., Reboutier, D.,
Bouteau, F., Pugin, A., and Wendehenne, D. (2007) Cryptogein-induced
anion effluxes: electrophysiological properties and analysis of the
mechanisms through which they contribute to the elicitor-triggered cell
death, Plant Signal. Behav., 2, 86-95.
32.Kumar, D., Rampuria, S., Singh, N. K., Shukla,
P., and Kirti, P. B. (2015) Characterization of a vacuolar processing
enzyme expressed in Arachis diogoi in resistance responses
against late leaf spot pathogen, Phaeoisariopsis personata,
Plant Mol. Biol., 88, 177-191.
33.Qiang, X., Zechmann, B., Reitz, M. U., Kogel, K.
H., and Schafer, P. (2012) The mutualistic fungus Piriformospora
indica colonizes Arabidopsis roots by inducing an
endoplasmic reticulum stress-triggered caspase-dependent cell death,
Plant Cell, 24, 794-809.
34.Deng, M., Bian, H., Xie, Y., Kim, Y., Wang, W.,
Lin, E., Zeng, Z., Guo, F., Pan, J., Han, N., Wang, J., Qian, Q., and
Zhu, M. (2011) Bcl-2 suppresses hydrogen peroxide-induced programmed
cell death via OsVPE2 and OsVPE3, but not via OsVPE1 and OsVPE4, in
rice, FEBS J., 278, 4797-4810.
35.Kadono, T., Tran, D., Errakhi, R., Hiramatsu, T.,
Meimoun, P., Briand, J., Iwaya-Inoue, M., Kawano, T., and Bouteau, F.
(2010) Increased anion channel activity is an unavoidable event in
ozone-induced programmed cell death, PLoS One, 5,
e13373.
36.Yakimova, E. T., Kapchina-Toteva, V. M.,
Laarhoven, L. J., Harren, F. M., and Woltering, E. J. (2006)
Involvement of ethylene and lipid signaling in cadmium-induced
programmed cell death in tomato suspension cells, Plant Physiol.
Biochem., 44, 581-589.
37.Yakimova, E. T., Kapchina-Toteva, V. M., and
Woltering, E. J. (2007) Signal transduction events in aluminum-induced
cell death in tomato suspension cells, J. Plant Physiol.,
164, 702-708.
38.Kariya, K., Demiral, T., Sasaki, T., Tsuchiya,
Y., Turkan, I., Sano, T., Hasezawa, S., and Yamamoto, Y. (2013) A novel
mechanism of aluminum-induced cell death involving vacuolar processing
enzyme and vacuolar collapse in tobacco cell line BY-2, J. Inorg.
Biochem., 128, 196-201.
39.Nakaune, S., Yamada, K., Kondo, M., Kato, T.,
Tabata, S., Nishimura, M., and Hara-Nishimura, I. (2005) A vacuolar
processing enzyme, deltaVPE, is involved in seed coat formation at the
early stage of seed development, Plant Cell, 17,
876-887.
40.Radchuk, V., Weier, D., Radchuk, R., Weschke, W.,
and Weber, H. (2011) Development of maternal seed tissue in barley is
mediated by regulated cell expansion and cell disintegration and
coordinated with endosperm growth, J. Exp. Bot., 62,
1217-1227.
41.Tran, V., Weier, D., Radchuk, R., Thiel, J., and
Radchuk, V. (2014) Caspase-like activities accompany programmed cell
death events in developing barley grains, PLoS One, 9,
e109426.
42.Kinoshita, T., Yamada, K., Hiraiwa, N., Kondo,
M., Nishimura, M., and Hara-Nishimura, I. (1999) Vacuolar processing
enzyme is up-regulated in the lytic vacuoles of vegetative tissues
during senescence and under various stressed conditions, Plant
J., 19, 43-53.
43.Muller, G. L., Drincovich, M. F., Andreo, C. S.,
and Lara, M. V. (2010) Role of photosynthesis and analysis of key
enzymes involved in primary metabolism throughout the lifespan of the
tobacco flower, J. Exp. Bot., 61, 3675-3688.
44.Uren, A. G., O’Rourke, K., Aravind, L. A.,
Pisabarro, M. T., Seshagiri, S., Koonin, E. V., and Dixit, V. M. (2000)
Identification of paracaspases and metacaspases: two ancient families
of caspase-like proteins, one of which plays a key role in MALT
lymphoma, Mol. Cell, 6, 961-967.
45.Vercammen, D., Van de Cotte, B., De Jaeger, G.,
Eeckhout, D., Casteels, P., Vandepoele, K., Vandenberghe, I., Van
Beeumen, J., Inze, D., and Van Breusegem, F. (2004) Type II
metacaspases Atmc4 and Atmc9 of Arabidopsis thaliana cleave
substrates after arginine and lysine, J. Biol. Chem.,
279, 45329-45336.
46.Watanabe, N., and Lam, E. (2005) Two
Arabidopsis metacaspases AtMCP1b and AtMCP2b are
arginine/lysine-specific cysteine proteases and activate apoptosis-like
cell death in yeast, J. Biol. Chem., 280,
14691-14699.
47.Bozhkov, P. V., Suarez, M. F., Filonova, L. H.,
Daniel, G., Zamyatnin, A. A., Jr., Rodriguez-Nieto, S., Zhivotovsky,
B., and Smertenko, A. (2005) Cysteine protease mcII-Pa executes
programmed cell death during plant embryogenesis, Proc. Natl. Acad.
Sci. USA, 102, 14463-14468.
48.Tsiatsiani, L., Van Breusegem, F., Gallois, P.,
Zavialov, A., Lam, E., and Bozhkov, P. V. (2011) Metacaspases, Cell
Death Differ., 18, 1279-1288.
49.Acosta-Maspons, A., Sepulveda-Garcia, E.,
Sanchez-Baldoquin, L., Marrero-Gutierrez, J., Pons, T., Rocha-Sosa, M.,
and Gonzalez, L. (2014) Two aspartate residues at the putative p10
subunit of a type II metacaspase from Nicotiana tabacum L. may
contribute to the substrate-binding pocket, Planta, 239,
147-160.
50.Fagundes, D., Bohn, B., Cabreira, C., Leipelt,
F., Dias, N., Bodanese-Zanettini, M. H., and Cagliari, A. (2015)
Caspases in plants: metacaspase gene family in plant stress responses,
Funct. Integr. Genom., 15, 639-649.
51.Choi, C. J., and Berges, J. A. (2013) New types
of metacaspases in phytoplankton reveal diverse origins of cell death
proteases, Cell Death Dis., 4, e490.
52.Wen, S., Ma, Q. M., Zhang, Y. L., Yang, J. P.,
Zhao, G. H., Fu, D. Q., Luo, Y. B., and Qu, G. Q. (2013) Biochemical
evidence of key residues for the activation and autoprocessing of
tomato type II metacaspase, FEBS Lett., 587,
2517-2522.
53.Watanabe, N., and Lam, E. (2011)
Calcium-dependent activation and autolysis of Arabidopsis
metacaspase 2d, J. Biol. Chem., 286, 10027-10040.
54.Zhang, Y., and Lam, E. (2011) Sheathing the
swords of death: post-translational modulation of plant metacaspases,
Plant Signal. Behav., 6, 2051-2056.
55.Belenghi, B., Romero-Puertas, M. C., Vercammen,
D., Brackenier, A., Inze, D., Delledonne, M., and Van Breusegem, F.
(2007) Metacaspase activity of Arabidopsis thaliana is regulated
by S-nitrosylation of a critical cysteine residue, J. Biol.
Chem., 282, 1352-1358.
56.Huang, L., Zhang, H., Hong, Y., Liu, S., Li, D.,
and Song, F. (2015) Stress-responsive expression, subcellular
localization and protein–protein interactions of the rice
metacaspase family, Int. J. Mol. Sci., 16,
16216-16241.
57.Bollhoner, B., Zhang, B., Stael, S., Denance, N.,
Overmyer, K., Goffner, D., Van Breusegem, F., and Tuominen, H. (2013)
Post mortem function of AtMC9 in xylem vessel elements, New
Phytol., 200, 498-510.
58.Tsiatsiani, L., Timmerman, E., De Bock, P. J.,
Vercammen, D., Stael, S., Van de Cotte, B., Staes, A., Goethals, M.,
Beunens, T., Van Damme, P., Gevaert, K., and Van Breusegem, F. (2013)
The Arabidopsis metacaspase 9 degradome, Plant Cell,
25, 2831-2847.
59.Coll, N. S., Vercammen, D., Smidler, A., Clover,
C., Van Breusegem, F., Dangl, J. L., and Epple, P. (2010)
Arabidopsis type I metacaspases control cell death,
Science, 330, 1393-1397.
60.Coll, N. S., Smidler, A., Puigvert, M., Popa, C.,
Valls, M., and Dangl, J. L. (2014) The plant metacaspase AtMC1 in
pathogen-triggered programmed cell death and aging: functional linkage
with autophagy, Cell Death Differ., 21, 1399-1408.
61.Kim, S. M., Bae, C., Oh, S. K., and Choi, D.
(2013) A pepper (Capsicum annuum L.) metacaspase 9
(Camc9) plays a role in pathogen-induced cell death in plants, Mol.
Plant Pathol., 14, 557-566.
62.Wang, X., Wang, X., Feng, H., Tang, C., Bai, P.,
Wei, G., Huang, L., and Kang, Z. (2012) TaMCA4, a novel wheat
metacaspase gene functions in programmed cell death induced by the
fungal pathogen Puccinia striiformis f. sp. tritici,
Mol. Plant Microbe Interact., 25, 755-764.
63.Hao, L., Goodwin, P. H., and Hsiang, T. (2007)
Expression of a metacaspase gene of Nicotiana benthamiana after
inoculation with Colletotrichum destructivum or Pseudomonas
syringae pv. tomato, and the effect of silencing the gene on
the host response, Plant Cell Rep., 26, 1879-1888.
64.Watanabe, N., and Lam, E. (2011)
Arabidopsis metacaspase 2d is a positive mediator of cell death
induced during biotic and abiotic stresses, Plant J., 66,
969-982.
65.He, R., Drury, G. E., Rotari, V. I., Gordon, A.,
Willer, M., Farzaneh, T., Woltering, E. J., and Gallois, P. (2008)
Metacaspase-8 modulates programmed cell death induced by ultraviolet
light and H2O2 in Arabidopsis, J. Biol.
Chem., 283, 774-783.
66.Minina, E. A., Filonova, L. H., Fukada, K.,
Savenkov, E. I., Gogvadze, V., Clapham, D., Sanchez-Vera, V., Suarez,
M. F., Zhivotovsky, B., Daniel, G., Smertenko, A., and Bozhkov, P. V.
(2013) Autophagy and metacaspase determine the mode of cell death in
plants, J. Cell Biol., 203, 917-927.
67.Wrzaczek, M., Vainonen, J. P., Stael, S.,
Tsiatsiani, L., Help-Rinta-Rahko, H., Gauthier, A., Kaufholdt, D.,
Bollhoner, B., Lamminmaki, A., Staes, A., Gevaert, K., Tuominen, H.,
Van Breusegem, F., Helariutta, Y., and Kangasjarvi, J. (2015) GRIM
REAPER peptide binds to receptor kinase PRK5 to trigger cell death in
Arabidopsis, EMBO J., 34, 55-66.
68.Vercammen, D., Belenghi, B., Van de Cotte, B.,
Beunens, T., Gavigan, J. A., De Rycke, R., Brackenier, A., Inze, D.,
Harris, J. L., and Van Breusegem, F. (2006) Serpin1 of Arabidopsis
thaliana is a suicide inhibitor for metacaspase 9, J. Mol.
Biol., 364, 625-636.
69.Sundstrom, J. F., Vaculova, A., Smertenko, A. P.,
Savenkov, E. I., Golovko, A., Minina, E., Tiwari, B. S.,
Rodriguez-Nieto, S., Zamyatnin, A. A., Jr., Valineva, T., Saarikettu,
J., Frilander, M. J., Suarez, M. F., Zavialov, A., Stahl, U., Hussey,
P. J., Silvennoinen, O., Sundberg, E., Zhivotovsky, B., and Bozhkov, P.
V. (2009) Tudor staphylococcal nuclease is an evolutionarily conserved
component of the programmed cell death degradome, Nat. Cell
Biol., 11, 1347-1354.
70.Caudy, A. A., Ketting, R. F., Hammond, S. M.,
Denli, A. M., Bathoorn, A. M., Tops, B. B., Silva, J. M., Myers, M. M.,
Hannon, G. J., and Plasterk, R. H. (2003) A micrococcal nuclease
homologue in RNAi effector complexes, Nature, 425,
411-414.
71.Scadden, A. D. (2005) The RISC subunit Tudor-SN
binds to hyper-edited double-stranded RNA and promotes its cleavage,
Nat. Struct. Mol. Biol., 12, 489-496.
72.Hundley, H. A., and Bass, B. L. (2010) ADAR
editing in double-stranded UTRs and other noncoding RNA sequences,
Trends Biochem. Sci., 35, 377-383.
73.Zamyatnin, A. A., Jr., Lyamzaev, K. G., and
Zinovkin, R. A. (2010) A-to-I RNA editing: a contribution to diversity
of the transcriptome and an organism’s development, Biochemistry
(Moscow), 75, 1316-1323.
74.Gutierrez-Beltran, E., Moschou, P. N., Smertenko,
A. P., and Bozhkov, P. V. (2015) Tudor staphylococcal nuclease links
formation of stress granules and processing bodies with mRNA catabolism
in Arabidopsis, Plant Cell, 27, 926-943.
75.Strobel, I., and Osiewacz, H. D. (2013)
Poly(ADP-ribose) polymerase is a substrate recognized by two
metacaspases of Podospora anserine, Eukaryot. Cell,
12, 900-912.
76.Martinez, M., Cambra, I., Gonzalez-Melendi, P.,
Santamaria, M. E., and Diaz, I. (2012) C1A cysteine-proteases and their
inhibitors in plants, Physiol. Plant., 145, 85-94.
77.Trobacher, C. P., Senatore, A., and Greenwood, J.
S. (2006) Masterminds or minions? Cysteine proteinases in plant
programmed cell death, Can. J. Bot., 84, 651-667.
78.Turk, V., Turk, B., and Turk, D. (2001) Lysosomal
cysteine proteases: facts and opportunities, EMBO J., 20,
4629-4633.
79.Than, M. E., Helm, M., Simpson, D. J.,
Lottspeich, F., Huber, R., and Gietl, C. (2004) The 2.0 Å crystal
structure and substrate specificity of the KDEL-tailed cysteine
endopeptidase functioning in programmed cell death of Ricinus
communis endosperm, J. Mol. Biol., 336,
1103-1116.
80.Richau, K. H., Kaschani, F., Verdoes, M.,
Pansuriya, T. C., Niessen, S., Stuber, K., Colby, T., Overkleeft, H.
S., Bogyo, M., and Van der Hoorn, R. A. (2012) Subclassification and
biochemical analysis of plant papain-like cysteine proteases displays
subfamily-specific characteristics, Plant Physiol., 158,
1583-1599.
81.Hierl, G., Howing, T., Isono, E., Lottspeich, F.,
and Gietl, C. (2014) Ex vivo processing for maturation of
Arabidopsis KDEL-tailed cysteine endopeptidase 2 (AtCEP2)
pro-enzyme and its storage in endoplasmic reticulum derived organelles,
Plant Mol. Biol., 84, 605-620.
82.Liu, H., Chen, L., Li, Q., Zheng, M., and Liu, J.
(2014) Computational study on substrate specificity of a novel cysteine
protease 1 precursor from Zea mays, Int. J. Mol. Sci.,
15, 10459-10478.
83.Savvateeva, L. V., Gorokhovets, N. V., Makarov,
V. A., Serebryakova, M. V., Solovyev, A. G., Morozov, S. Y., Reddy, V.
P., Zernii, E. Y., Zamyatnin, A. A., Jr., and Aliev, G. (2015)
Glutenase and collagenase activities of wheat cysteine protease
Triticain-α: feasibility for enzymatic therapy assays, Int. J.
Biochem. Cell Biol., 62, 115-124.
84.Van der Hoorn, R. A. (2008) Plant proteases: from
phenotypes to molecular mechanisms, Annu. Rev. Plant Biol.,
59, 191-223.
85.Schmid, M., Simpson, D., Kalousek, F., and Gietl,
C. (1998) A cysteine endopeptidase with a C-terminal KDEL motif
isolated from castor bean endosperm is a marker enzyme for the
ricinosome, a putative lytic compartment, Planta, 206,
466-475.
86.Schmid, M., Simpson, D. J., Sarioglu, H.,
Lottspeich, F., and Gietl, C. (2001) The ricinosomes of senescing plant
tissue bud from the endoplasmic reticulum, Proc. Natl. Acad. Sci.
USA, 98, 5353-5358.
87.Gu, C., Shabab, M., Strasser, R., Wolters, P. J.,
Shindo, T., Niemer, M., Kaschani, F., Mach, L., and Van der Hoorn, R.
A. (2012) Post-translational regulation and trafficking of the
granulin-containing protease RD21 of Arabidopsis thaliana,
PLoS One, 7, e32422.
88.Madureira, H. C., Da Cunha, M., and Jacinto, T.
(2006) Immunolocalization of a defense-related 87 kDa cystatin in leaf
blade of tomato plants, Environ. Exp. Bot., 55,
201-208.
89.Martinez, M., and Diaz, I. (2008) The origin and
evolution of plant cystatins and their target cysteine proteinases
indicate a complex functional relationship, BMC Evol. Biol.,
8, 198.
90.Nissen, M. S., Kumar, G. N., Youn, B., Knowles,
D. B., Lam, K. S., Ballinger, W. J., Knowles, N. R., and Kang, C.
(2009) Characterization of Solanum tuberosum multicystatin and
its structural comparison with other cystatins, Plant Cell,
21, 861-875.
91.Green, A. R., Nissen, M. S., Kumar, G. N.,
Knowles, N. R., and Kang, C. (2013) Characterization of Solanum
tuberosum multicystatin and the significance of core domains,
Plant Cell, 25, 5043-5052.
92.Martinez, M., Diaz-Mendoza, M., Carrillo, L., and
Diaz, I. (2007) Carboxy terminal extended phytocystatins are
bifunctional inhibitors of papain and legumain cysteine proteinases,
FEBS Lett., 581, 2914-2918.
93.Margis-Pinheiro, M., Zolet, A. C., Loss, G.,
Pasquali, G., and Margis, R. (2008) Molecular evolution and
diversification of plant cysteine proteinase inhibitors: new insights
after the poplar genome, Mol. Phylogenet. Evol., 49,
349-355.
94.Lampl, N., Budai-Hadrian, O., Davydov, O., Joss,
T. V., Harrop, S. J., Curmi, P. M., Roberts, T. H., and Fluhr, R.
(2010) Arabidopsis AtSerpin1, crystal structure and in
vivo interaction with its target protease RESPONSIVE TO
DESICCATION-21 (RD21), J. Biol. Chem., 285,
13550-13560.
95.Fluhr, R., Lampl, N., and Roberts, T. H. (2012)
Serpin protease inhibitors in plant biology, Physiol. Plant.,
145, 95-102.
96.Ahmed, S. U., Rojo, E., Kovaleva, V.,
Venkataraman, S., Dombrowski, J. E., Matsuoka, K., and Raikhel, N. V.
(2000) The plant vacuolar sorting receptor AtELP is involved in
transport of NH(2)-terminal propeptide-containing vacuolar proteins in
Arabidopsis thaliana, J. Cell Biol., 149,
1335-1344.
97.Helm, M., Schmid, M., Hierl, G., Terneus, K.,
Tan, L., Lottspeich, F., Kieliszewski, M. J., and Gietl, C. (2008)
KDEL-tailed cysteine endopeptidases involved in programmed cell death,
intercalation of new cells, and dismantling of extensin scaffolds,
J. Bot., 95, 1049-1062.
98.Greenwood, J. S., Helm, M., and Gietl, C. (2005)
Ricinosomes and endosperm transfer cell structure in programmed cell
death of the nucellus during Ricinus seed development, Proc.
Natl. Acad. Sci. USA, 102, 2238-2243.
99.Howing, T., Huesmann, C., Hoefle, C., Nagel, M.
K., Isono, E., Huckelhoven, R., and Gietl, C. (2014) Endoplasmic
reticulum KDEL-tailed cysteine endopeptidase 1 of Arabidopsis
(AtCEP1) is involved in pathogen defense, Front. Plant Sci.,
5, 58.
100.Hierl, G., Vothknecht, U., and Gietl, C. (2012)
Programmed cell death in Ricinus and Arabidopsis: the
function of KDEL cysteine peptidases in development, Physiol.
Plant., 145, 103-113.
101.Zhang, D., Liu, D., Lv, X., Wang, Y., Xun, Z.,
Liu, Z., Li, F., and Lu, H. (2014) The cysteine protease CEP1, a key
executor involved in tapetal programmed cell death, regulates pollen
development in Arabidopsis, Plant Cell, 26,
2939-2961.
102.Valpuesta, V., Lange, N. E., Guerrero, C., and
Reid, M. S. (1995) Up-regulation of a cysteine protease accompanies the
ethylene-insensitive senescence of daylily (Hemerocallis)
flowers, Plant Mol. Biol., 28, 575-582.
103.Nadeau, J. A., Zhang, X. S., Li, J., and
O’Neill, S. D. (1996) Ovule development: identification of
stage-specific and tissue-specific cDNAs, Plant Cell, 8,
213-239.
104.Rocha, A. J., Soares, E. L., Costa, J. H.,
Costa, W. L., Soares, A. A., Nogueira, F. C., Domont, G. B., and
Campos, F. A. (2013) Differential expression of cysteine peptidase
genes in the inner integument and endosperm of developing seeds of
Jatropha curcas L. (Euphorbiaceae), Plant Sci.,
213, 30-37.
105.Trobacher, C. P., Senatore, A., Holley, C., and
Greenwood, J. S. (2013) Induction of a ricinosomal-protease and
programmed cell death in tomato endosperm by gibberellic acid,
Planta, 237, 665-679.
106.Senatore, A., Trobacher, C. P., and Greenwood,
J. S. (2009) Ricinosomes predict programmed cell death leading to
anther dehiscence in tomato, Plant Physiol., 149,
775-790.
107.Yamada, K., Matsushima, R., Nishimura, M., and
Hara-Nishimura, I. (2001) A slow maturation of a cysteine protease with
a granulin domain in the vacuoles of senescing Arabidopsis
leaves, Plant Physiol., 127, 1626-1634.
108.Bateman, A., and Bennett, H. P. (2009) The
granulin gene family: from cancer to dementia, BioEssays,
31, 1245-1254.
109.Carter, C., Pan, S., Zouhar, J., Avila, E. L.,
Girke, T., and Raikhel, N. V. (2004) The vegetative vacuole proteome of
Arabidopsis thaliana reveals predicted and unexpected proteins,
Plant Cell, 16, 3285-3303.
110.Lampl, N., Alkan, N., Davydov, O., and Fluhr,
R. (2013) Set-point control of RD21 protease activity by AtSerpin1
controls cell death in Arabidopsis, Plant J., 74,
498-510.
111.Zhao, P., Zhou, X. M., Zhang, L. Y., Wang, W.,
Ma, L. G., Yang, L. B., Peng, X. B., Bozhkov, P. V., and Sun, M. X.
(2013) A bipartite molecular module controls cell death activation in
the basal cell lineage of plant embryos, PLoS Biol., 11,
e1001655.
112.Kim, M. J., Yamamoto, D., Matsumoto, K., Inoue,
M., Ishida, T., Mizuno, H., Sumiya, S., and Kitamura, K. (1992) Crystal
structure of papain–E64-c complex. Binding diversity of E64-c to
papain S2 and S3 subsites, Biochem. J., 287, 797-803.
113.Zhao, C., Johnson, B. J., Kositsup, B., and
Beers, E. P. (2000) Exploiting secondary growth in Arabidopsis.
Construction of xylem and bark cDNA libraries and cloning of three
xylem endopeptidases, Plant Physiol., 123, 1185-1196.
114.Funk, V., Kositsup, B., Zhao, C., and Beers, E.
P. (2002) The Arabidopsis xylem peptidase XCP1 is a tracheary
element vacuolar protein that may be a papain ortholog, Plant
Physiol., 128, 84-94.
115.Petzold, H. E., Zhao, M., and Beers, E. P.
(2012) Expression and functions of proteases in vascular tissues,
Physiol. Plant., 145, 121-129.
116.McLellan, H., Gilroy, E. M., Yun, B. W., Birch,
P. R., and Loake, G. J. (2009) Functional redundancy in the
Arabidopsis cathepsin B gene family contributes to basal
defense, the hypersensitive response and senescence, New
Phytol., 183, 408-418.
117.Gilroy, E. M., Hein, I., van der Hoorn, R.,
Boevink, P. C., Venter, E., McLellan, H., Kaffarnik, F., Hrubikova, K.,
Shaw, J., Holeva, M., Lopez, E. C., Borras-Hidalgo, O., Pritchard, L.,
Loake, G. J., Lacomme, C., and Birch, P. R. (2007) Involvement of
cathepsin B in the plant disease resistance hypersensitive response,
Plant J., 52, 1-13.
118.Bernoux, M., Timmers, T., Jauneau, A., Briere,
C., De Wit, P. J., Marco, Y., and Deslandes, L. (2008) RD19, an
Arabidopsis cysteine protease required for RRS1-R-mediated
resistance, is relocalized to the nucleus by the Ralstonia
solanacearum PopP2 effector, Plant Cell, 20,
2252-2264.
119.Xu, F. X., and Chye, M. L. (1999) Expression of
cysteine proteinase during developmental events associated with
programmed cell death in brinjal, Plant J., 17,
321-327.
120.Lohman, K. N., Gan, S., John, M. C., and
Amasino, R. M. (1994) Molecular analysis of natural leaf senescence in
Arabidopsis thaliana, Physiol. Plant., 92,
322-328.
121.Otegui, M. S., Noh, Y. S., Martinez, D. E.,
Vila Petroff, M. G., Staehelin, L. A., Amasino, R. M., and Guiamet, J.
J. (2005) Senescence-associated vacuoles with intense proteolytic
activity develop in leaves of Arabidopsis and soybean, Plant
J., 41, 831-844.
122.Singh, S., Giri, M. K., Singh, P. K., Siddiqui,
A., and Nandi, A. K. (2013) Down-regulation of OsSAG12-1 results in
enhanced senescence and pathogen-induced cell death in transgenic rice
plants, J. Biosci., 38, 583-592.
123.Marino, G., Uria, J. A., Puente, X. S.,
Quesada, V., Bordallo, J., and Lopez-Otin, C. (2003) Human autophagins,
a family of cysteine proteinases potentially implicated in cell
degradation by autophagy, J. Biol. Chem., 278,
3671-3678.
124.Kaminskyy, V., and Zhivotovsky, B. (2012)
Proteases in autophagy, Biochim. Biophys. Acta, 1824,
44-50.
125.Escamez, S., and Tuominen, H. (2014) Programs
of cell death and autolysis in tracheary elements: when a suicidal cell
arranges its own corpse removal, J. Exp. Bot., 65,
1313-1321.
126.Minina, E. A., Bozhkov, P. V., and Hofius, D.
(2014) Autophagy as initiator or executioner of cell death, Trends
Plant Sci., 19, 692-697.
127.Li, M., Hou, Y., Wang, J., Chen, X., Shao, Z.
M., and Yin, X. M. (2011) Kinetics comparisons of mammalian Atg4
homologues indicate selective preferences toward diverse Atg8
substrates, J. Biol. Chem., 286, 7327-7338.
128.Woo, J., Park, E., and Dinesh-Kumar, S. P.
(2014) Differential processing of Arabidopsis ubiquitin-like
Atg8 autophagy proteins by Atg4 cysteine proteases, Proc. Natl.
Acad. Sci. USA, 111, 863-868.
129.Chichkova, N. V., Shaw, J., Galiullina, R. A.,
Drury, G. E., Tuzhikov, A. I., Kim, S. H., Kalkum, M., Hong, T. B.,
Gorshkova, E. N., Torrance, L., Vartapetian, A. B., and Taliansky, M.
(2010) Phytaspase, a relocalizable cell death promoting plant protease
with caspase specificity, EMBO J., 29, 1149-1161.
130.Chichkova, N. V., Galiullina, R. A., Taliansky,
M. E., and Vartapetian, A. B. (2008) Tissue disruption activates a
plant caspase-like protease with TATD cleavage specificity, Plant
Stress, 2, 89-95.
131.Dodson, G., and Wlodawer, A. (1998) Catalytic
triads and their relatives, Trends Biochem. Sci., 23,
347-352.
132.Rautengarten, C., Steinhauser, D., Bussis, D.,
Stintzi, A., Schaller, A., Kopka, J., and Altmann, T. (2005) Inferring
hypotheses on functional relationships of genes: analysis of the
Arabidopsis thaliana subtilase gene family, PLoS Comput.
Biol., 1, e40.
133.Schaller, A., Stintzi, A., and Graff, L. (2012)
Subtilases – versatile tools for protein turnover, plant
development, and interactions with the environment, Physiol.
Plant., 145, 52-66.
134.Yamagata, H., Masuzawa, T., Nagaoka, Y.,
Ohnishi, T., and Iwasaki, T. (1994) Cucumisin, a serine protease from
melon fruits, shares structural homology with subtilisin and is
generated from a large precursor, J. Biol. Chem., 269,
32725-32731.
135.Vartapetian, A. B., Tuzhikov, A. I., Chichkova,
N. V., Taliansky, M., and Wolpert, T. J. (2011) A plant alternative to
animal caspases: subtilisin-like proteases, Cell Death Differ.,
18, 1289-1297.
136.Ottmann, C., Rose, R., Huttenlocher, F.,
Cedzich, A., Hauske, P., Kaiser, M., Huber, R., and Schaller, A. (2009)
Structural basis for Ca2+-independence and activation by
homodimerization of tomato subtilase 3, Proc. Natl. Acad. Sci.
USA, 106, 17223-17228.
137.Chichkova, N. V., Galiullina, R. A.,
Beloshistov, R. E., Balakireva, A. V., and Vartapetian, A. B. (2014)
Phytaspases: aspartate-specific proteases involved in plant cell death,
Bioorg. Khim., 40, 658-664.
138.Galiullina, R. A., Kasperkiewicz, P.,
Chichkova, N. V., Szalek, A., Serebryakova, M. V., Poreba, M., Drag,
M., and Vartapetian, A. B. (2015) Substrate specificity and possible
heterologous targets of phytaspase, a plant cell death protease, J.
Biol. Chem., 290, 24806-24815.
139.Poreba, M., Szalek, A., Kasperkiewicz, P., and
Drag, M. (2014) Positional scanning substrate combinatorial library
(PS-SCL) approach to define caspase substrate specificity, Methods
Mol. Biol., 1133, 41-59.
140.Fomicheva, A. S., Tuzhikov, A. I., Beloshistov,
R. E., Trusova, S. V., Galiullina, R. A., Mochalova, L. V., Chichkova,
N. V., and Vartapetian, A. B. (2012) Programmed cell death in plants,
Biochemistry (Moscow), 77, 1452-1464.
141.Chichkova, N. V., Kim, S. H., Titova, E. S.,
Kalkum, M., Morozov, V. S., Rubtsov, Y. P., Kalinina, N. O., Taliansky,
M. E., and Vartapetian, A. B. (2004) A plant caspase-like protease
activated during the hypersensitive response, Plant Cell,
16, 157-171.
142.Srivastava, R., Liu, J. X., and Howell, S. H.
(2008) Proteolytic processing of a precursor protein for a
growth-promoting peptide by a subtilisin serine protease in
Arabidopsis, Plant J., 56, 219-227.
143.Coffeen, W. C., and Wolpert, T. J. (2004)
Purification and characterization of serine proteases that exhibit
caspase-like activity and are associated with programmed cell death in
Avena sativa, Plant Cell, 16, 857-873.
144.Faro, C., and Gal, S. (2005) Aspartic
proteinase content of the Arabidopsis genome, Curr. Prot.
Pept. Sci., 6, 493-500.
145.Dunn, B. M. (2002) Structure and mechanism of
the pepsin-like family of aspartic peptidases, Chem. Rev.,
102, 4431-4458.
146.Geier, G., Banaj, H. J., Heid, H., Bini, L.,
Pallini, V., and Zwilling, R. (1999) Aspartyl proteases in
Caenorhabditis elegans. Isolation, identification and
characterization by a combined use of affinity chromatography,
two-dimensional gel electrophoresis, microsequencing and databank
analysis, Eur. J. Biochem., 264, 872-879.
147.Guo, R., Xu, X., Carole, B., Li, X., Gao, M.,
Zheng, Y., and Wang, X. (2013) Genome-wide identification, evolutionary
and expression analysis of the aspartic protease gene superfamily in
grape, BMC Genom., 14, 554.
148.Niu, N., Liang, W., Yang, X., Jin, W., Wilson,
Z. A., Hu, J., and Zhang, D. (2013) EAT1 promotes tapetal cell death by
regulating aspartic proteases during male reproductive development in
rice, Nat. Commun., 4, 1445.
149.Chen, F., and Foolad, M. R. (1997) Molecular
organization of a gene in barley which encodes a protein similar to
aspartic protease and its specific expression in nucellar cells during
degeneration, Plant Mol. Biol., 35, 821-831.
150.Ge, X., Dietrich, C., Matsuno, M., Li, G.,
Berg, H., and Xia, Y. (2005) An Arabidopsis aspartic protease
functions as an anti-cell-death component in reproduction and
embryogenesis, EMBO Rep., 6, 282-288.
151.Phan, H. A., Iacuone, S., Li, S. F., and
Parish, R. W. (2011) The MYB80 transcription factor is required for
pollen development and the regulation of tapetal programmed cell death
in Arabidopsis thaliana, Plant Cell, 23,
2209-2224.
152.Kurepa, J., and Smalle, J. A. (2008) Structure,
function and regulation of plant proteasomes, Biochimie,
90, 324-335.
153.Kurepa, J., Wang, S., Li, Y., and Smalle, J.
(2009) Proteasome regulation, plant growth and stress tolerance,
Plant Signal. Behav., 4, 924-927.
154.Han, J. J., Lin, W., Oda, Y., Cui, K. M.,
Fukuda, H., and He, X. Q. (2012) The proteasome is responsible for
caspase-3-like activity during xylem development, Plant J.,
72, 129-141.
155.Hatsugai, N., Iwasaki, S., Tamura, K., Kondo,
M., Fuji, K., Ogasawara, K., Nishimura, M., and Hara-Nishimura, I.
(2009) A novel membrane fusion-mediated plant immunity against
bacterial pathogens, Genes Dev., 23, 2496-2506.
156.Maidment, J. M., Moore, D., Murphy, G. P.,
Murphy, G., and Clark, I. M. (1999) Matrix metalloproteinase homologues
from Arabidopsis thaliana. Expression and activity, J. Biol.
Chem., 274, 34706-34710.
157.Marino, G., Huesgen, P. F., Eckhard, U.,
Overall, C. M., Schroder, W. P., and Funk, C. (2014) Family-wide
characterization of matrix metalloproteinases from Arabidopsis
thaliana reveals their distinct proteolytic activity and cleavage
site specificity, Biochem. J., 457, 335-346.
158.Hadler-Olsen, E., Fadnes, B., Sylte, I.,
Uhlin-Hansen, L., and Winberg, J. O. (2011) Regulation of matrix
metalloproteinase activity in health and disease, FEBS J.,
278, 28-45.
159.Delorme, V. G., McCabe, P. F., Kim, D. J., and
Leaver, C. J. (2000) A matrix metalloproteinase gene is expressed at
the boundary of senescence and programmed cell death in cucumber,
Plant Physiol., 123, 917-927.
160.Roberts, I. N., Caputo, C., Criado, M. V., and
Funk, C. (2012) Senescence-associated proteases in plants, Physiol.
Plant., 145, 130-139.
161.Diaz-Mendoza, M., Velasco-Arroyo, B.,
Gonzalez-Melendi, P., Martinez, M., and Diaz, I. (2014) C1A cysteine
protease–cystatin interactions in leaf senescence, J. Exp.
Bot., 65, 3825-3833.