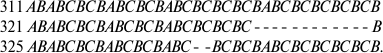Structure of Intergenic Spacer IGS1 of Ribosomal Operon from Schistidium Mosses
I. A. Milyutina1*, E. A. Ignatova2, M. S. Ignatov3, D. V. Goryunov1, and A. V. Troitsky1
1Belozersky Institute of Physico-Chemical Biology, Lomonosov Moscow State University, 119991 Moscow, Russia; fax: +7 (495) 939-3181; E-mail: iramilyutina@yandex.ru; bobr@belozersky.msu.ru2Lomonosov Moscow State University, Biological Faculty, 119991 Moscow, Russia; E-mail: arctoa@list.ru
3Main Botanical Garden, Russian Academy of Sciences, 127276 Moscow, Russia; E-mail: misha_ignatov@list.ru
* To whom correspondence should be addressed.
Received July 8, 2015
The structure of the intergenic spacer 1 (IGS1) of the ribosomal operon from 12 species of Schistidium mosses was studied. In the IGS1 sequences of these species, three conserved regions and two areas of GC- and A-enriched repeats were identified. All of the studied mosses have a conserved pyrimidine-enriched motif at the 5′-end of IGS1. Species-specific nucleotide substitutions and insertions were found in the conserved areas. The repeated units contain single nucleotide substitutions that make unique the majority of repeated units. The positions of such repeats in IGS1 are species-specific, but their number can vary within the species and among operons of the same specimen. The comparison of IGS1 sequences from the Schistidium species and from representatives of ten other moss genera revealed the presence of common conserved motifs with similar localization. Presumably, these motifs are elements of termination of the pre-rRNA transcription and processing of rRNA.
KEY WORDS: rDNA, IGS, intergenic spacer, conserved motifs, mossesDOI: 10.1134/S0006297915110103
Abbreviations: bp, nucleotide base pair; IGS1, intergenic spacer 1.
In eukaryotes, rRNA genes form multigenic families consisting of
tandemly located repeated units strongly varying in number (from
several hundreds to several thousands). A repeated unit includes the
genes of 18S, 5.8S, and 26S rRNAs separated by transcribed spacers
(ITS) 1 and 2 and by the intergenic spacer (IGS). Depending on
localization of the 5S rRNA genes, two types of ribosomal operon
organization are described [1]. If the 5S rRNA
genes form separate clusters, such type of ribosomal gene organization
is called S-type (separation). Early land plants, including mosses, are
characterized by L-type (linkage) of the ribosomal operon, which is
specified by presence of the 5S rRNA gene within the intergenic spacer.
The 5S rRNA gene divides the spacer into two parts – IGS1 located
between the genes of 26S and 5S rRNAs, and IGS2, which is located
between the genes of 5S and 18S rRNAs. IGS1 contains elements that are
necessary for termination of transcription and processing. An absolute
majority of studies on plant IGS has been done on angiosperm plants
with the S-type structure of the ribosomal operon [2-7]. In the majority of these
works, the 3′-terminal region of IGS, including the promoter and
5′-external transcribed spacer (5′-ETS), was studied. The
IGS region in mosses is poorly studied. Full sequences of the
intergenic spacer have been obtained for only two species of the
Funariaceae family. The IGS1 region has been studied in more detail,
and in GenBank (www.ncbi.nlm.nih.gov), sequences from 10 moss species
are deposited, although the structure of these sequences has not been
described in detail [1].
In the present work, we studied specific features of the IGS1 structure of the ribosomal operon from acrocarpous mosses of the Schistidium genus for which we studied earlier the ITS1 structure, as well as the phylogeny of the genus based on the ITS1-2 sequences and regions of the chloroplast genome [8, 9]. In this work, interspecific and intraspecific polymorphism of the IGS1 region in members of this genus was described, and comparative analysis of IGS1 was performed.
MATERIALS AND METHODS
Thirty-three sequences of IGS1 from 12 species of Schistidium were determined. Below the species are listed, the numbers in parentheses indicating the number of populations studied: S. apocarpum (3), S. elegantulum (2), S. canadense (2), S. lancifolium (1), S. holmenianum (1), S. andreaeopsis (2), S. boreale (3), S. dupretii (2), S. rivulare (2), S. pulchrum (8), S. frigidum (3), and S. succulentum (2).
DNA was isolated using the Nucleospin Plant Extraction Kit (Macherey-Nagel, Germany). Amplification was performed with primers complementary to the 3′-terminal region of the 26S rRNA gene and to the 5S rRNA gene – 26dR2 (forward primer) GAGATGAATCCTTTGCAGACG and 5S(r)R2 (reverse primer) GAGTTCTGATGGGATCYGGTG. The amplification conditions were as follows: 94°C – 3 min; then 30 cycles 94°C – 20 s, 62°C – 20 s, 72°C – 1 min 20 s; final elongation for 5 min. The amplification was performed with the Encyclo Plus PCR Kit (Evrogen, Russia). After preparative electrophoresis in 1% agarose gel, fragments of DNA were cut and purified with a MinElute Gel Extraction Kit (Qiagen, USA). Most of the PCR products were sequenced without cloning. The heterogeneous templates were cloned into pTZ57R/T vector with a Thermo Scientific Ins TAclone PCR Cloning Kit (Thermo Scientific, USA). For sequencing in addition to the terminal primers, the inner ones were used – 26(F3) ACTCCAGGGTGCCCGCCC, 26S(r) CCTATGTGATCTCATTCCAAA, and 26S(R2) AYANNYAYCTTATTTTYGAACMCC. DNA was sequenced using the ABI PRISM® BigDye™ Terminator v. 3.1 reagent kit (Life Technologies, USA) with subsequent analysis of the reaction products using an Applied Biosystems (USA) 3730 DNA Analyzer automatic sequencer in the Center of Collective Use “Genome” in the V. A. Engelhard Institute of Molecular Biology. The resulting sequences were deposited in GenBank (www.ncbi.nlm.nih.gov) under accession numbers KT246290-246292 and KT273589-273617.
RESULTS
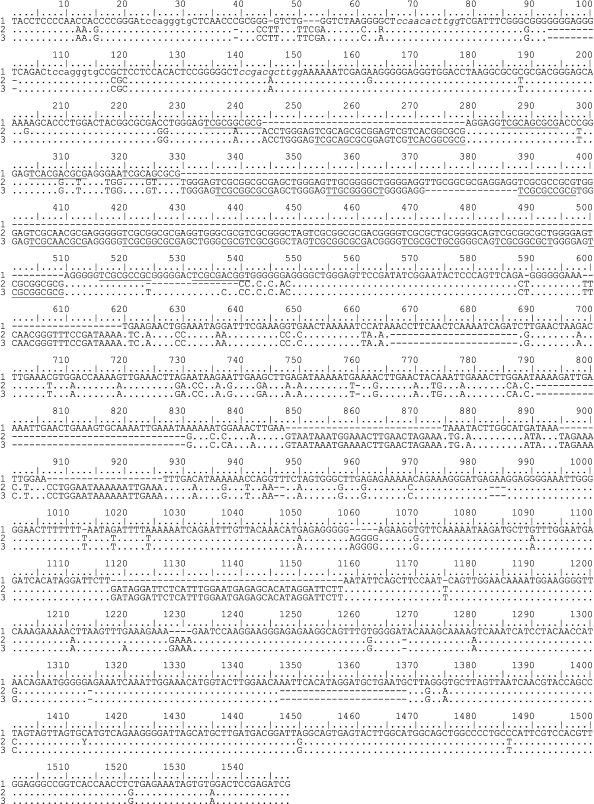
Structure of IGS1 sequences from Schistidium species. Two transversions (A→C and C→A) were detected at the 3′-end of the 26S rRNA gene from the Schistidium species in comparison with sequences of the 26S rRNA gene from other mosses. IGS1 begins from the pyrimidine-enriched motif TACCTCCCC(C) (Fig. 1, only three sequences of 33 are shown).
Fig. 1. Sequences of IGS1 from two Schistidium species: 1) S. apocarpum specimen 129; 2) S. pulchrum specimen 322; 3) S. pulchrum specimen 325. Lowercase letters indicate conserved motifs in the IGS1 from Schistidium found also in sequences from other mosses. Elements of B repeats are underlined.
The length of IGS1 sequences in the Schistidium species varies from 1120 to 1446 bp. In these sequences, five regions can be specified: three comparatively conserved ones slightly different in length, and two regions of repeats (Fig. 1).
The first conserved region adjoins to the 3′-end of the 26S rRNA gene (positions 1-224 of the alignment in Fig. 1). The length of this region varies insignificantly – from 214 to 226 bp. In this region, there are two exact copies of the TCCAGGGTGC motifs (positions 23-31 and 107-115) and the degenerated repeat CTCCARCACTTGG (positions 65-77)/CTCSSACRCTTGG (positions 138-150). After the second repeat, there is an oligo-A region consisting of 6 bp. The differences in size of the sequences in this region are determined by small insertions and by the length of mononucleotide motifs. The oligo-T sequence specific for regions of transcription termination is absent in this region, but there are two pyrimidine-enriched sites: CCCRYCCTCY at the distance of 105-113 bp from the IGS1 beginning and CTYTCTTTC at the distance of 44 bp from the IGS1 beginning in 21 of 33 sequences studied. The other specimens had in this site the sequence GTCTS.
The first conserved region is followed by a region of GC-enriched repeats. Its length varies from 104 to 350 bp that essentially determines the difference in the IGS1 lengths of different species. One specimen of S. andreaeopsis lacked the region of GC-enriched repeats. The content of GC in this region varies from 73.58 to 81.25%.
After the GC-enriched repeats, there is the second conserved region of 40-44 bp (positions 553-598 of the alignment in Fig. 1). In this region, a conserved motif CC(G/T)ATATCGG (positions 562-572) was detected. This motif is absent in the IGS1 sequences of three populations from S. frigidum and S. holmenianum (specimen 137).
Next to the second conserved region there is a region enriched with A-repeats. This region also varies significantly in different species (from 133 to 298 bp). The contents of A in the repeats vary from 42.86 to 51.37%.
The third conserved region with length of 544-598 bp (it begins from position 927 of the alignment in Fig. 1) is located between the A-enriched repeats and the 5S rRNA gene. The differences in the length of this region are associated with the presence of duplications of short nucleotide motifs (up to 24 bp), species-specific short insertions, and different numbers of mononucleotides. This region also contains a poly-T motif CTTTTTTT(T) that is unique in IGS1. In S. succulentum, this motif is shortened to CTT.
Structure of the region of GC-enriched repeats. Among GC-enriched repeats, there are three main repeated elements: A) 9-10-nucleotide motif ASSYGGG(G)AG; B) 10-nucleotide motif TYRYRVSRBR (in some sequences at the 5′-end the triplet TCG is added in variant B); C) 5-6-nucleotide purine-enriched motif RGGRRG.
In all studied sequences, the repeated elements B alternate with elements A or C (or with incomplete elements A and C). These short repeated elements are united in more elongated repeated units. Thus, IGS1 from S. apocarpum (Fig. 1) contains two 35-nucleotide repeats and one incomplete 16-nucleotide repeat:
(substitutions in repeats 2 and 3 in comparison with repeat 1 are
printed in bold, elements B are underlined).
Repeats 1 and 2 consist of alternating elements A-B-C-B, and repeat 3 contains the elements C-B. The repeated B and C elements have numerous substitutions. These substitutions are reproduced in sequences of different populations from the same species, in particular, from S. pulchrum (Fig. 1). The sequence of alternation of the initial elements A, B, and C can be different. Thus, in S. elegantulum (specimen 258), elements A and B alternate initially producing two 41-nucleotide repeats, then there are two 36-nucleotide repeats formed by the elements A-B-C-B, and then there are two A-B and four C-B repeats.
To study intraspecific polymorphism, the IGS1 sequences were determined in two, three, and even eight (for S. pulchrum) specimens from geographically remote populations. In the IGS1 sequences of three S. apocarpum populations from the Caucasus and from the Vologda and Chelyabinsk Regions, the repeats were identical in number of repeated units and nucleotide sequences, similarly to the S. canadense and S. boreale species. Single-nucleotide deletion in the region of GC-enriched repeats distinguishes the IGS1 sequence of one S. dupretii population (Perm Region) from another (Austria). Some species were different in the number of repeated elements. Thus, differences were found between two populations of S. frigidum (specimens 100 and 281) from the Anabar Plateau and from the Taimyr Peninsula:
Deletions in the repeats in the IGS1 sequences are also characteristic for different populations of S. pulchrum:
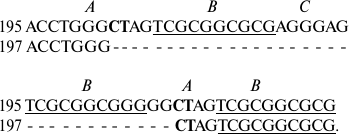
Usually deletions occur in parts of the sequence located between exact forward repeats. Thus, IGS1 of S. rivulare specimens from two populations (Karachaevo-Cherkessia, 195, and the Kuril Islands, 197) are different in deletion of four elements in the repeats, one of which is flanked by forward repeats with characteristic insertions of two nucleotides CT into element A:
In specimen 197, deletion of a part of the sequence between the forward
repeats in GGCTAG occurs.
Differences in the number of repeats in IGS1 between different operons were observed less frequently. Thus, in S. succulentum (specimen 106) after cloning the initially heterogeneous PCR-product, two IGS1 variants were found which differed in the number of GC-enriched repeats:
Numerous single nucleotide substitutions in the repeated elements B allow us to almost unequivocally align the repeats in the IGS1 sequences of different populations from the same species. Thus, single substitutions were found in 13 of 17 repeats of elements B in S. pulchrum. Due to their presence, the repeated elements B and C are unique, which allows us to more accurately determine what elements of the repeats are absent.
In the secondary structure, the GC-enriched repeats form hairpins because elements B are inverted repeats.
Region of A-enriched repeats. Every A-enriched repeat has in its beginning the triplet CTT (less often GTT or CTC). The length of the majority of the repeats is 19 bp. Because of numerous substitutions, the repeated elements of this region have nearly no exact repeats. However, in some repeats there are presise copies of A-enriched motifs. Thus, in the IGS1 sequence from S. liliputanum (specimen 286), there are two exact repeats of the motif CTTCAACTCAAAATCAAAA. In sequences from different Schistidium species, there are the same variants of repeats (identical or with insignificant number of substitutions), that makes alignment easier.
DISCUSSION
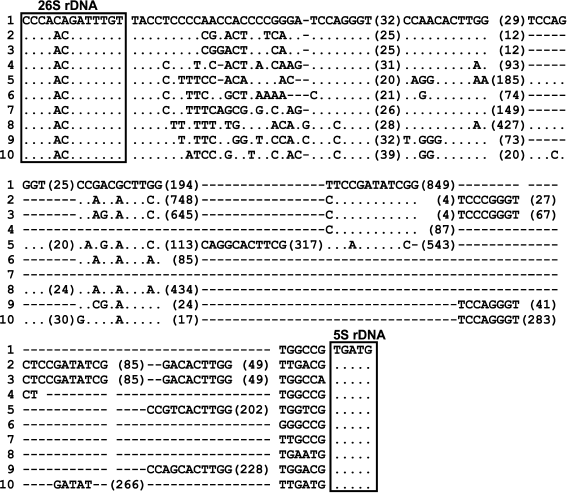
The set of sequenced IGS1 from mosses is not large, the database NCBI contains sequences from 10 species: Anacolia laevisphaera, Ancistrodes genuflexa, Bartramia rosea, Brachythecium rutabulum, Dicranella staphylina, Entosthodon obtusus, Funaria hygrometrica, Lembophyllum orbiculatum, Rhytidium rugosum, and Rigodium toxarion. These sequences are very different in their length: from 254 bp in Brachythecium rutabulum to nearly 1.5 kb in Ancistrodes genuflexa and Schistidium species. Nevertheless, in IGS1 of all species some conserved motifs and structure elements can be described (Fig. 2). The 5′-end of IGS1, TACCTCCCC(C), in Schistidium species is highly conserved. Other studied mosses have motif TACCY at the 5′-end of IGS1. Earlier, we found highly conserved motifs in the ITS1 sequences of mosses, and one of them, CACACA, is located at the 5′-end of ITS1 [9]. Thus, highly conserved motifs flank 3′-ends of the 18S and 26S rRNA genes in mosses. Functional sites of IGS1 of mosses are unknown, but the location of these motifs and their high conservatism can be related with processing of the ribosomal genes. A pyrimidine-enriched motif was also detected at the 5′-end of IGS in other plants [2, 3, 6, 10].
Fig. 2. Sequences of IGS1 from 10 moss species: 1) Schistidium apocarpum 129; 2) Anacolia laevisphaera FR694294; 3) Bartramia rosea FR694278; 4) Rigodium toxarion FR694310; 5) Ancistrodes genuflexa FR694319; 6) Brachythecium rutabulum FR695698; 7) Rhytidium rugosum FR694324; 8) Dicranella staphylina FR694295; 9) Funaria hygrometrica JQ736823; 10) Entosthodon obtusus JQ736824. The 3′-end of the 26S rRNA gene and the 5′-end of the 5S rRNA gene are framed. The conserved motifs of IGS1 common for all moss species are shown. The numerals show the number of base pairs between them.
The structure of IGS1 of Schistidium is similar to the structure of this region in other eukaryotes. For eukaryotes, the relatively conserved 5′- and 3′-terminal sequences are the same within genera and are adjoining to the genes of 26S and 5S rRNA (in Schistidium these sequences contain two short exact repeats at the 5′-end of IGS1). Unique terminal sequences of IGS1 were described for radish [3], Funaria hygrometrica [11], and Brassica [2]. In the latter work, the conservativeness of the 5′-terminal IGS sequences in the Cruciferae family was also shown.
Repeats in this region are not characteristic for all mosses. Thus, a relatively elongated region of repeats is described in IGS1 from Anacolia, Bartramia, and Schistidium, whereas such repeats are absent in Brachythecium and Funaria. Also, there is no GC-enriched repeat in the IGS1 sequence from S. andreaeopsis.
It seems that sites of transcription and processing termination are located in the conserved regions of the spacer. However, individual substitutions in the repeated elements of the region with GC-enriched repeats are also species-specific in IGS1 of Schistidium, and this can be useful for phylogenetic analysis together with the species-specific substitutions and insertions of the conserved regions.
We analyzed sequences of IGS1 of mosses from the database NCBI and found that in all sequences there is a highly conserved motif YCCMGGGT at a distance of 20-23 bp from the 5′-end of IGS1 (Fig. 2). The IGS1 of Schistidium species are found to have two such motifs. Two motifs were also found in the sequences from Ancistrodes, Funaria, and Dicranella, and three motifs were found in IGS1 from Entosthodon. Rigodium toxarion, Rhytidium rugosum, and Brachythecium rutabulum have one motif in IGS1.
Another short conserved IGS1 sequence in Schistidium species, CCRACRCTTGG, with slight substitutions, occurs 1-4 times in IGS1 of different mosses (Figs. 1 and 2). In Schistidium, there are two such sub-repeats at distances of 67-77 and 141-150 bp from the 5′-end of the spacer. In Anacolia laevisphaera, there are three degenerated repeats: CCAACACTTGG, CCAACACTTCG, and GACACTTGG. Sequences from Rigodium toxarion and Rhytidium rugosum were shown to have one motif. Two motifs are found in sequences from Brachythecium rutabulum and Dicranella staphylina. The potential secondary structure of these motifs is small hairpins.
In the majority of cases, the motif CCAACACTTGG is located at a distance of 30-40 bp from the motif YCCMGGGTR. Thus, the presence of the motif YCCMGGGTR at the distance of 23-25 bp from the 5′-end of the intergenic spacer and the location of the second motif CCAACACTTGG at the distance of 30-40 bp from the first motif is a conserved trait. Two such “pairs” are specific for Schistidium, Entosthodon, Funaria, and Dicranella, whereas species of Hypnales (Rigodium toxarion, Rhytidium rugosum, and Brachythecium rutabulum) have one pair. These conserved motifs are probably associated with transcription termination. Repeated structural elements of the transcription termination region were described in mice [12]. The only oligo-T region, which is an element of the transcription termination, is located in Schistidium at a distance of about 700-1100 bp from the 3′-end of the 26S rRNA gene (positions 1006-1012 in Fig. 1). In other mosses, poly-T is also located nearer to the 3′-end of IGS1. However, in the sequence of IGS1 from Brachythecium rutabulum, no poly-T regions longer than 4 bp have been found.
In eukaryotes, the transcription termination of RNA polymerase I is shown to be distant from the 3′-end of the 26S rRNA gene. Thus, in mouse, 565 more nucleotide residues of the spacer region are transcribed, whereas the end of the gene is produced as a result of processing [12]. Three sites of termination are found in IGS of Leishmania: one corresponds to the end of mature 28S rRNA, and two others form transcripts that are 185 and 576 nucleotide residues longer than the first [13].
In the moss Funaria hygrometrica with the whole IGS region sequenced, two sites of transcription initiation were shown to be located in IGS2 [11]. In angiosperm plants, the promoter is also known to be located at the 3′-end of the IGS. Nevertheless, in Schistidium species in the second conserved region of IGS1, a motif has been found similar to the sequence of the transcription initiation region in many angiosperm plants. This is a unique motif in IGS1 Schistidium (G/T)ATATCGG (positions 564-571 in Fig. 1). In the second intergenic spacer in Funaria hygrometrica, similar motifs GATAGGGGG and TATGTGGGGG were found located rather far from each other (distance of about 1700 bp) [11], the TATATAGGG sequence is specific for arabidopsis [4], and the sequence TATATAAGGG – for Brassica rapa [2]. The motif GATATCGG is present in IGS1 from Ancistrodes, Rigodium, Anacolia, and Bartramia (in the two last mosses this motif is duplicated), and with one substitution it occurs in Entosthodon. In other studied mosses in this aspect, this motif has not been found.
The comparison of IGS1 sequences of Schistidium and other mosses reveals that the 5′-end that has the oligopyrimidine sequence and the nucleotide motif YCCMGGGTR is more conserved, as well as the other conserved motif CCRACRCTTGG. The 3′-end is conserved in the limits of the Schistidium genus, and sequences of IGS1 from different species of the same genus are absent in the database NCBI.
Conserved regions of IGS1 from different Schistidium species can be aligned easily and have species-specific substitutions and insertions that can be useful for phylogenetic analysis.
This work was supported by the Russian Foundation for Basic Research (project No. 15-04-06027 – the experimental part) and by the Russian Science Foundation (project No. 14-50-00029 – the theoretical analysis).
REFERENCES
1.Wicke, S., Costa, A., Muñoz, J., and Quandt,
D. (2010) Restless 5S: the rearrangement(s) and evolution of the
nuclear ribosomal DNA in land plants, Mol. Phylogenet. Evol.,
61, 321-332.
2.Rocha, P. S., and Bertrand, H. (1995) Structure and
comparative analysis of the rDNA intergenic spacer of Brassica
rapa, Eur. J. Biochem., 229, 550-557.
3.Delcasso-Tremousaygue, D., Grellet, F., Panabieres,
F., Ananiev, E. D., and Delseny, M. (1988) Structural and
transcriptional characterization of the external spacer of a ribosomal
RNA nuclear gene from a higher plant, Eur. J. Biochem.,
172, 767-776.
4.Doelling, J. H., and Pikaard, C. S. (1995) The
minimal ribosomal RNA gene promoter of Arabidopsis thaliana includes a
critical element at the transcription initiation site, Plant J., 8,
683-692.
5.Doelling, J. H., Gaudino, R. J., and Pikaard, C. S.
(1993) Functional analysis of Arabidopsis thaliana rRNA gene and spacer
promoters in vivo and by transient expression, Proc. Natl. Acad. Sci.
USA, 90, 7528-7532.
6.Castiglione, M. R., Gelati, M. T., Cremonini, R.,
and Frediani, M. (2013) The intergenic spacer region of the rDNA in
Haplopappus gracilis (Nutt.) Gray, Protoplasma, 250, 683-689.
7.Garcia, S., Panero, J. L., Siroky, J., and Kovarik,
A. (2010) Repeated reunions and splits feature the highly dynamic
evolution of 5S and 35S ribosomal RNA genes (rDNA) in the Asteraceae
family, BMC Plant Biol., 10, 176.
8.Milyutina, I. A., Goryunov, D. V., Ignatov, M. S.,
Ignatova, E. A., and Troitsky, A. V. (2010) The phylogeny of
Schistidium (Bryophyta, Grimmiaceae) based on the primary and
secondary structure of nuclear rDNA internal transcribed spacers,
Mol. Biol., 44, 883-897.
9.Milyutina, I. A., and Ignatov, M. S. (2015)
Conserved motifs in the primary and secondary ITS1 structures of
bryophytes, Mol. Biol., 49, 348-357.
10.Fukunaga, K., Ichitani, K., Taura, S., Sato, M.,
and Kawase, M. (2005) Ribosomal DNA intergenic spacer sequence in
foxtail millet, Setaria italica (L.) P. Beauv. and its
characterization and application to typing of foxtail millet landraces,
Hereditas, 142, 38-44.
11.Capesius, I. (1997) Analysis of the ribosomal RNA
gene repeat from the moss Funaria hygrometrica, Plant Mol.
Biol., 33, 559-564.
12.Grummt, I., Maier, U., Ohrlein, A., Hassouna, N.,
and Bachellerie, J. P. (1985) Transcription of mouse rDNA terminates
downstream of the 3′-end of 28S RNA and involves interaction of
factors with repeated sequences in the 3′ spacer, Cell,
43, 801-810.
13.Abreu-Blanco, M. T., Ramirez, J. L.,
Pinto-Santini, D. M., Papadopoulou, B., and Guevara, P. (2010) Analysis
of ribosomal RNA transcription termination and 3′-end processing
in Leishmania amazonensis, Gene, 451, 15-22.