Formation of New Polysomes on Free mRNAs in a Cell-Free Translation Systems Is Accompanied by Partial Disassembly of Previously Formed Polysomes
E. A. Sogorin, S. Ch. Agalarov, and A. S. Spirin*
Institute of Protein Research, Russian Academy of Sciences, 142290 Pushchino, Moscow Region, Russia; fax: +7 (495) 514-0218; E-mail: spirin@vega.protres.ru* To whom correspondence should be addressed.
Received May 6, 2015
A method for detection of the fluorescence-labeled mRNA in translating ribosomal complexes has been developed. It is demonstrated that in the working cell-free translation system with preformed polysomes, formation of new polysomes on free mRNA takes place. For the first time, it is shown that the process is accompanied by partial disassembly of the previously formed polysomes. This result is interpreted as an indication of the direct relationship between processes of translation termination of polysomal ribosomes and translation initiation of free mRNAs.
KEY WORDS: eukaryotic polyribosomes, mRNA, cell-free translation system, fluorescenceDOI: 10.1134/S0006297915100144
Protein synthesis (translation) in living cells and in cell-free systems is carried out by ribosomes, which are organized into large associates with messenger RNA (mRNA) in such a way that some or even many ribosomes translate one chain of mRNA, moving along the chain one after another. The structure, in which the mRNA is associated with a number of translating ribosomes, is called polyribosomes or polysomes.
The development of cell-free translation systems with long lifetime, such as CECF (Continuous Exchange Cell-Free system [1]), allowed initiation of systematic study of the structural organization of polyribosome and their conformational changes during many successive rounds of translation [2]. For this purpose, cell-free systems based on wheat germ extracts were used [3, 4], since they do not contain endogenous mRNA, which allows monitoring the formation and structural changes of polysomes formed on added mRNA. It was found that during the hours of operation of the system, polysomes that are formed from the first minutes of the translation increase in size by recruiting new ribosomes simultaneously undergoing a series of sequential conformational transformations (“ontogenesis of polysomes”) [2].
In the present work, the question was raised whether new polyribosomes are formed on free (vacant) mRNAs in the already working translation system with previously formed polyribosomes. To clarify this, the 3′-end of mRNA was fluorescently labeled, and the labeled mRNA was added to already running translation system at different stages of protein synthesis. Thus, newly formed polyribosomes have to be fluorescently labeled and therefore easily recognized in the process of analysis of the polyribosomal profile after centrifugation of the translation mixture in a sucrose gradient.
The experiment showed that the newly added mRNA is involved in the formation of new polyribosomes at different stages of translation. However, it was unexpected that the formation of new polyribosomes on the newly added mRNAs was accompanied by decreasing size of the initially formed polysomes, i.e. their partial disassembly occurred. This result is interpreted as an indication of a direct relationship between the processes of translation termination of polysomal ribosomes and translation initiation of free mRNAs.
MATERIALS AND METHODS
Transcription of mRNA and its modification with fluorescent label. As mRNA, the hybrid polyribonucleotide sequence was used, which consisted of the following three parts: (1) 5′-untranslated region of RNA from tobacco mosaic virus (TMV), also referred to as “omega RNA”; (2) mRNA encoding (1650 nucleotides) firefly luciferase; (3) 3′-untranslated region of RNA from TMV. The whole construction is referred to as “5′UTRomega-Luc-3′UTRTMV”. The DNA construct for transcription of this mRNA (pTZ10omegaLUC) was described elsewhere [5]. Transcription in vitro was carried out according to the method described in [6], with slight modifications: the reaction mixture contained 400 mM Tris (pH 7.5, adjusted with acetic acid), 50 mM KCl, 111 mM Mg (OAc)2, 100 mM DTT, 100 mM β-mercaptoethanol, 10 mM spermidine, 0.05% Triton X-100, 1 mM EDTA, 4 mM of each nucleoside triphosphate (ATP, GTP, UTP, CTP), 15 ng/µl of linearized plasmid, 0.5 U/µl RNase inhibitor “RiboLock RNase inhibitor”, and 5 units/µl T7 RNA polymerase (Fermentas, Lithuania). The reaction was performed at 37°C for 2 h. Then the mixture was treated with acidic phenol (1 : 1 v/v), the aqueous phase was collected, NaOAc (pH 5.5) up to 300 mM and 3 volumes of ethanol were added. The mRNA was collected by centrifugation at 14,100g for 15 min, dried, and dissolved in deionized water. The solution was purified from nucleotides by precipitation in 3.5 M LiCl (30 min at 4°C). The precipitate was dissolved in deionized water and reprecipitated twice with ethanol in the presence of 2 M NH4OAc (first time) and 300 mM NaOAc (pH 5.5) (second time). The precipitate was washed with 80% ethanol and dissolved in water.
The mRNA was modified at the 3′-end as follows: 300 µg of mRNA was incubated for 50 min on ice in a solution containing 5 mM NaIO4 and 100 mM NaOAc (pH 5.5). After incubation, NaOAc (pH 5.5) up to 300 mM and 3 volumes of ethanol were added to the reaction mixture. Next, the 3′-end oxidized mRNA was incubated for 4.5 h on ice in a solution containing 7 mM of fluorescein-5-thiosemicarbazide (Invitrogen, USA; stock solution in DMSO) and 100 mM NaOAc (pH 5.5). The mRNA was reprecipitated as described above and purified by gel filtration through a MicroSpin™ G-50 spin-column (GE Healthcare, England).
Translation in vitro, analysis of polysomes, and detection of mRNA. The translation was carried out in the cell-free protein synthesis system based on wheat germ extracts in a stationary (batch) format [1]. The translation mixture was incubated at 25°C in a volume of 25 µl, and the reaction was terminated by addition of 50 µl buffer of the same composition as that for the preparation of sucrose gradient solution: 0.015 µg/ml cycloheximide, 20 mM Hepes-KOH, pH 8.0, 100 mM KOAc, 5 mM MgOAc, 0.1 mM EDTA. The mixture was applied to a 15-45% sucrose gradient. The sample was centrifuged for 2 h in a SW-41 rotor at 37,000 rpm and 4°C. Continuous registration of optical density in the gradient after centrifugation was performed at 260 nm using a flow-through cell. The fluorescence distribution in the sucrose gradient after centrifugation was also measured in a flow cell (cuvette volume 120 µl) on RF device 5103PC (Shimadzu, Japan) at excitation wavelength of 492 nm (slit width of 1.5 nm) and emission wavelength of 518 nm (slit width 15 nm). During the measurement time (~20 min), no bleaching of fluorescent label was observed.
RESULTS AND DISCUSSION
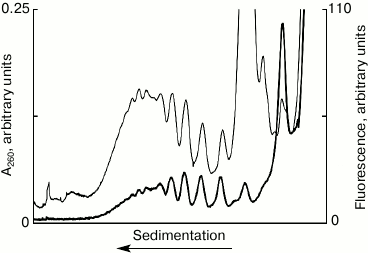
Detection of fluorescently labeled mRNA in polysomes. Figure 1 shows the results of the sedimentation analysis of the translation mixture after 10 min incubation with fluorescently labeled mRNA. The distribution profile of the optical density in the UV corresponds to the distribution of ribosomal material – polyribosomes, ribosomes, and ribosomal subunits after 10 min of translation system operation. The fluorescence profile is the distribution of the mRNA, using the system as run, after 10 min operation of the system, i.e. the actual distribution of polyribosomes was formed during these 10 min. The main conclusions that can be derived from Fig. 1 are as follows. First, the coincidence of the peak positions of the ultraviolet absorption and fluorescence clearly indicates the formation of polyribosomes on the labeled mRNA. Second, the discrete peaks make it possible to directly estimate the number of ribosomes, i.e. estimate the size of polysomes in each peak. Third, the distribution of the ultraviolet absorption and fluorescence shows clearly, what the maximum loading of ribosomes on the mRNA used in the experiment can be. The discrete nature of the distribution of peaks of optical density in the profile allows calculating the load of the mRNA by ribosomes – in this case, the maximum number of ribosomes per molecule of mRNA is equal to ten (“decasomes”). It is noteworthy that the distribution of peaks of fluorescence-labeled mRNA coincides with the distribution of peaks of optical density in the area of polysomes. It is evident, also, that about half of all related mRNA is associated with the fraction of polysomes, while only about 10% of mRNA bind to the 80S ribosomes. At the same time, a significant amount of mRNA (40%) is associated with the 40S subunit, which may indicate the presence of 48S ribosomal initiation complexes in the translational mixture. Thus, the proposed method recognizes mRNA within different ribosomal complexes and permits their quantitative assessment.
Fig. 1. Sedimentation profiles of the translation mixture: optical density (thin curve) and fluorescence (thick curve); 10 min translation with fluorescently labeled mRNA.
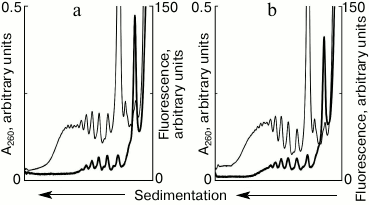
Formation of “new” polysomes in a running translational system. The described approach created an experimental base allowing clarification of the question: will new polysomes be formed on the newly added mRNA in the translation system in the presence of previously formed polysomes? To answer the question, the cell-free system of protein synthesis was started using unlabeled mRNA, and fluorescently labeled mRNA was added in the system at certain intervals of time. After 10 min of running the system, a significant moiety of ribosomes was involved in polysomes (Fig. 1). The results of the experiments, in which vacant mRNA was added after 10 min of incubation, are shown in Fig. 2. The presence of fluorescent peaks in the polyribosomal profile clearly indicates the formation of new polysomes on the added template. Within 5 min after the addition of new mRNA, new polysomes containing from two to five ribosomes per mRNA molecule were formed (Fig. 2a). Further increase of the incubation time to 10 min resulted in an increase in size and number of the new polysomes (Fig. 2b). Thus, the experiment shows that in the running cell-free translation system on the background of already formed polyribosomes, new polysomes continue to appear on free mRNAs, as well as “vacancies” in polysomes, which are not fully loaded with ribosomes, are filled in.
Fig. 2. Sedimentation profiles of translational mixture: optical density (thin curves) and fluorescence (thick curves). a) 10 min translation with unlabeled mRNA + 5 min translation after addition of fluorescently labeled mRNA; b) 10 min translation with unlabeled mRNA + 10 min translation after addition of fluorescently labeled mRNA.
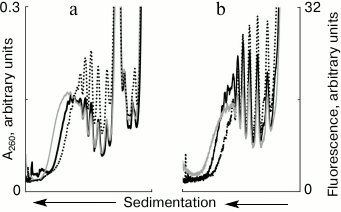
Partial disassembly of formed polysomes after addition of free mRNA. The cell-free translation system usually starts with a single addition of mRNA in the beginning of the process. In our experiments with the addition of free mRNA in the translation system, it was interesting to see how previously formed polysomes react to the appearance of a vacant template. To clarify this, the system with polysomes initially formed on the fluorescently labeled mRNAs was supplied with a new portion of unlabeled mRNAs (Fig. 3). This experiment allowed us to trace changes in the polyribosomal profile in the sucrose gradient, both by the distribution of optical density (Fig. 3a) and by the distribution of fluorescence (Fig. 3b). Within 10 min of translation, polysomes were formed in the system (Fig. 3, a and b, solid black curve), which with increasing incubation time continued to grow in size, i.e. the number of ribosomes on mRNA increased (Fig. 3, a and b, solid gray curve) if new mRNA was not added to the system. However, the increase in size of polysomes by attracting new ribosomes could be not only stopped, but also reversed. Exactly this effect was caused by the addition of new mRNA (Fig. 3, a and b, dotted curve). It is seen that after the addition of new mRNA, relatively “heavy” polysomes (8 to 10 ribosomes per molecule of the template) became less loaded (4 to 6 ribosomes in the template). Thus, the addition of free mRNA invoked not only the formation of new polysomes (Fig. 2), but also led to partial disassembly of the previously formed polysomes.
Fig. 3. Sedimentation profiles of translational mixture: a) optical density; b) fluorescence; 10 min translation with fluorescently labeled mRNA (solid black curves); 15 min translation with fluorescently labeled mRNA (solid gray curves); 10 min translation with fluorescently labeled mRNA + 5 min translation after addition of unlabeled mRNA (dotted curves).
The mechanism of partial disassembly of polysomes induced by free mRNA remains unclear. Obviously, the number of ribosomes disconnected from the mRNA begins to prevail over the amount of ribosomes re-joining the template. Normally, the ribosome dissociates from the mRNA upon completing the process of translation termination. One cannot exclude cases (especially in cell-free systems) when the ribosome prematurely leaves mRNA without completing the synthesis of a full-length protein. Our experiments do not distinguish between these two possibilities. Assuming the normal run of the process, it can be speculated that the free mRNA somehow stimulates translation termination, thereby ribosomes detached from the template, and partial disassembly of polysomes is observed. In the light of such interpretation, the direct relationship between the processes of translation termination of polysomal ribosomes and translation initiation of free mRNAs is possible.
This work was supported by the Russian Foundation for Basic Research (projects No. 15-04-01525-a and No. 13-04-40213-N) and by a grant of the Program “Molecular and Cell Biology” of the Presidium of the Russian Academy of Sciences.
REFERENCES
1.Shirokov, V. A., Kommer, A., Kolb, V. A., and
Spirin, A. S. (2007) in Methods in Molecular Biology, Vol. 375
(Grandi, G., ed.) Humana Press Inc., Totowa, NJ, pp. 19-55.
2.Afonina, Z. A., Myasnikov, A. G., Shirokov, V. A.,
Klaholz, B. P., and Spirin, A. S. (2015) Conformation transitions of
eukaryotic polyribosomes during multi-round translation, Nucleic
Acids Res., 43, 618-628.
3.Spirin, A. S., Baranov, V. I., Ryabova, L. A.,
Ovodov, S. Y., and Alakhov, Y. B. (1988) A continuous cell-free
translation system capable of producing polypeptides in high yield,
Science, 242, 1162-1164.
4.Madin, K., Sawasaki, T., Ogasawara, T., and Endo,
Y. (2000) A highly efficient and robust cell-free protein synthesis
system prepared from wheat embryos: plants apparently contain a suicide
system directed at ribosomes, Proc. Natl. Acad. Sci. USA,
97, 559-564.
5.Alekhina, O. M., Vassilenko, K. S., and Spirin, A.
S. (2007) Translation of non-capped mRNAs in a eukaryotic cell-free
system: acceleration of initiation rate in the course of polysome
formation, Nucleic Acids Res., 35, 6547-6559.
6.Pokrovskaya, I. D., and Gurevich, V. V. (1994)
In vitro transcription: preparative RNA yields in analytical
scale reactions, Anal. Biochem., 220, 420-423.