Natural Larval Diet Differently Influences the Pattern of Developmental Changes in DNA 5-Methylcytosine Levels in Apis mellifera Queens as Compared with Workers and Drones
A. Strachecka*, K. Olszewski, M. Bajda, and J. Demetraki-Paleolog
Department of Biological Basis of Animal Production, Faculty of Biology and Animal Breeding, University of Life Sciences in Lublin, 20-950 Lublin, Poland; E-mail: aneta.strachecka@up.lublin.pl; krzysztof.olszewski@up.lublin.pl; milena.bajda@wp.pl; jerzy.paleolog@up.lublin.pl* To whom correspondence should be addressed.
Received December 1, 2014; Revision received January 29, 2015
The principal mechanism of gene activation/silencing is DNA 5-methylcytosine methylation. This study was aimed at determining global DNA methylation levels in larvae, prepupae, pupae, and 1-day-old adults of Apis mellifera queens, workers and drones. The Imprint Methylated DNA Quantification Kit MDQ1 was used. Percentages of DNA 5-methylcytosine were low and relatively similar in the larvae of all the castes until 4th day of larval development (3-5%). However, they were higher in the drone and worker larvae than in the queen larvae. Generally, the developmental patterns of changes in the DNA methylation levels were different in the queens in comparison with the drones and workers. While methylation increased in the queens, it decreased in the drones and workers. Methylated DNA methylcytosine percentages and weights in the queen prepupae (15%, 9.18 ng) and pupae (21%, 10.74 ng) were, respectively, three and four times higher than in the worker/drone brood of the same age (2.5-4%, 0.03-0.07 ng). Only in the queens, after a substantial increase, did DNA methylation decrease almost twice between the pupal stage and queen emergence (from 21% and 10.74 ng to 12% and 6.78 ng). This finding seems very interesting, particularly for experimental gerontology.
KEY WORDS: Apis mellifera, DNA methylation, epigenetics, honeybee castes, 5-methylcytosine, ontogenesisDOI: 10.1134/S0006297915080076
In response to environmental changes, gene expression is regulated by several interacting systems operating at a number of levels, including epigenetic modifications of DNA [1, 2]. The principal gene-silencing mechanism is DNA methylation, which is a post-replication process involving the addition of a methyl group (-CH3) to DNA cytosine [3, 4]. S-Adenosyl-L-methionine is the donor of this group, and the transfer is activated by DNA-methyltransferases [5]. Many organisms display phenotypic plasticity, i.e. produce different phenotypes from the same genomes [6]. Female larvae of Apis mellifera receive different food to become queen or worker larvae, i.e. those in queen cells are provisioned with royal jelly (RJ), while those in worker cells are provided with worker jelly (WJ) that includes pollen [7-11]. However, up to 3 days after hatching, female larvae are totipotent and can still develop into normal queens or workers. Therefore, the 3rd and 4th day of larval development is the crucial period in female honeybee ontogenesis. On the other hand, drones develop from unfertilized eggs but also receive different larval food than worker and queen larvae. Can these differences in natural larval diets influence the developmental pattern of fluctuations in global DNA methylation in queens, drones, and workers in different ways?
Substances present in RJ reduced methyltransferase enzyme activities and methyltransferase gene expression, which consequently decreased methylation within the dynactin p62 gene, which, in turn, resulted in a significant increase in the number of queens and a reduction in the numerical strength of workers or intercastes [6, 11]. Larvae reared as queens or workers differed in their gene-expression patterns [12]. 4-4-Phenylbutyrate (PB), as well as its metabolite – phenylacetyl glutaminate (PG) – increased the expression of many genes involved in the apian development and lifespan when added to WJ [13]. Wheeler and Robinson [14] suggest that alternative carbohydrate sources (e.g. high fructose corn syrup or sucrose) elicited hundreds of differences in gene expression in adult workers. These changes in gene expression concerned protein metabolism and oxidation–reduction processes, including the expression of some genes involved in tyrosine and phenylalanine metabolism. Moreover, our previous studies indicated that such environmental factors as acaricides, caffeine, coenzyme Q10, and industrial air pollution influenced apian protein and antioxidant metabolism [15-20]. Furthermore, we showed that global methylation levels increased along with the age of the adult bees, and diet supplementation with caffeine significantly reduced DNA methylation levels in older workers [17], whereas amphotericin B treatment increased them [15]. All these findings suggest that environment-dependent changes in gene expression connected with changes in DNA methylation are crucial for honeybee development and survival.
Most of the studies concerning DNA 5-methylcytosine (m5C) levels were based on DNA isolated from the brains of adult honeybees [21]. In turn, DNA-methyltransferase (Dnmt) activities, as indicators of DNA methylation intensity, were determined in the worker and queen larvae, as well as the adult workers and queens [11]. In this research, we decided to take a more complex approach and study DNA m5C levels in larvae at different development stages as well as in prepupae, pupae, and newly emerged individual workers, queens, and drones. Kucharski et al. [6] found that intensive DNA replication was active in 3-5-day-old larvae, providing an opportunity for either adding or removing methyl tags to target loci. This confirms that this period is crucial in female honeybee ontogenesis. Shi et al. [22] showed that the numbers of differentially methylated genes in 2-, 4-, and 6-day-old queen and worker larvae amounted to 725, 3013, and 5049, respectively, and that in comparison to 4- and 6-day-old worker larvae, a large number of genes involved in development, reproduction, and metabolic regulation were downmethylated in the queen larvae. Therefore, among others, we examined exactly 3- and 4-day-old worker, queen, and drone larvae in our study. We also examined the prepupae and pupae because substantial changes in oxygen consumption and carbon dioxide output resulting from metamorphosis progress in these stages can be associated with changes in gene expression. There is no information in available literature about global DNA methylation levels in these stages. Large-scale physiological and behavioral changes occur in newly emerged bees as well. Therefore, these stages were also considered in our experiment.
We set the following hypotheses: 1) development-related patterns of changes in total DNA methylation levels are different in queens, workers, and drones; 2) global DNA methylation levels would increase during apian development, as they increase in the case of other animals. Therefore, the experiment was aimed at identifying the content of m5C in the DNA of queens, workers, and drones at different development stages (including larvae, prepupae, pupae, and emerged individual bees). The study of global DNA methylation changes is important to understand ongoing changes in gene expression during the development of different bee castes.
MATERIALS AND METHODS
Biological material. Ten queens were instrumentally inseminated, each with the semen of one drone, and separately introduced to 10 two-box (B1 + B2) queen-less colonies. After at least 1 month and after the beginning of egg laying, five of the queens (in five colonies) were separately caged within queen-excluder cages for 3 days in order to lay eggs. Each of the cages contained two empty combs (C1 and C2). Next, each of the five colonies was divided in half. One of the halves was maintained in the first box (B1), whereas the other, in the second box (B2). Each C1 was left in the B1 box, which also contained a queen, for rearing worker brood, whereas the 1-day-old larvae from the five C2 combs were transferred into plastic (Nicoplast, France) queen-cell cups and placed within five B2 queen-less boxes, respectively, to rear queens. On the 3rd and 4th day of female larval development and at the prepupal stage (the 11th day of apian ontogenesis), 20 brood samples were taken from each of the C1 combs and from the queen-cells from each of the five B2 boxes. On the 11th day, the queen cells were transferred from each of the B2 boxes into incubators, and then additional samples were taken on the 13th (pupae) and 16th (newly emerged queens) day. Twenty brood samples were also taken from each of the five B1 boxes (C1 combs) on the 16th (pupae) and 21st (newly emerged workers) day of apian ontogenesis. Altogether, 500 samples were collected (20 samples × 5 samplings × 5 colonies) and refrigerated at –25°C in sterile Eppendorf tubes for further analyses.
The five remaining of the 10 queens kept in the remaining five of the 10 colonies were caged separately within one-comb-cages containing one empty drone-cell comb (C3) each. On the 3rd or 4th day of larval development, 14th (prepupae), 18th (pupae), and 24th (newly emerged drones) day of drone ontogenesis, 20 samples were taken from each colony and refrigerated at –25°C in sterile Eppendorf tubes for further analyses.
DNA isolation. Quantitative and qualitative assessment of DNA. After defrosting, DNA was extracted from entire 3- and 4-day-old larvae, prepupae, and pupae, and also the heads and thoraces of the newly emerged 1-day-old insects using the DNeasy Tissue Kit 250 (Qiagen, Germany), according to the manufacturer’s protocol. DNA quantification was performed spectrophotometrically by measuring the absorbance at 230, 260, and 280 nm with a BioPhotometer (Eppendorf, USA). The DNA samples with A260/A280 ratio of 1.7-2.0 were used for methylation analyses. DNA quality was assessed via electrophoresis at 70 V for 60 min using a 1% agarose gel stained with ethidium bromide (EtBr) in an electrophoresis apparatus (Bio-Rad, USA). DNA molecules were visualized using a Syngene BTX 26M transilluminator. DNA concentration means and agarose gel application examples are shown in Table 1.
Table 1. Spectrophotometric assessment of
DNA isolated from the different development stages (I) and results of
the DNA electrophoresis in agarose gel (II)
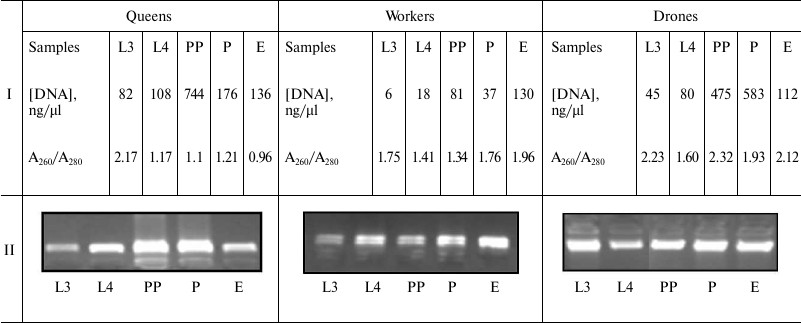
Note: L3, the 3rd day of larval development; L4, the 4th day of larval
development; PP, prepupae; P, pupae; E, newly emerged bee; Conc., DNA
concentration; A260/A280, DNA contamination.
Global DNA methylation analysis. Global DNA methylation analyses were performed using the Imprint Methylated DNA Quantification Kit MDQ1 (Sigma, USA) based on the ELISA principle. We used the 96-well plate format. DNA concentration was diluted to 150 ng/µl in the binding solution. DNA binding was achieved by incubating 30 µl diluted DNA at 37°C for 1 h. A 150-µl block solution was added, and the samples were incubated at 37°C for 30 min. Next, the DNA and block solutions were removed from all the wells, which were washed three times with 150 µl of 1× wash buffers. A 50-µl sample of diluted capture antibody was placed in each well and incubated at room temperature for 1 h. After removing the capture antibody and washing four times with the wash buffer, each well was filled with 50 µl of diluted detection antibody. The plates were incubated at room temperature for 30 min. The detection antibody was removed from the wells, which were then washed five times with the wash buffer. Each well was then filled with 100 µl of developing solution and incubated at room temperature for about 10 min for color change, and subsequently 50 µl of stop solution was added. The absorbance of each sample was measured five times at 450 nm. To calculate the percentages of methylated DNA cytosine relative to the Methylated Control DNA, in which 100% of cytosines are methylated (MC), the following equation was used: [(A450S – A450B)/(A450MC – A450B)] × 100%. Methylated DNA weights (in nanograms) were also computed with the following equation: [(A450S – A450B) – intercept]/slope. Here A450S is the average absorbance of the sample, A450MC is the average absorbance of the Methylated Control DNA, A450B is the average absorbance of the blank, and the intercept and slope are equal to 0.08208 and 2.68e–3, respectively.
Statistical analysis. Multivariate generalized linear model (GLM) was carried out (factors: caste, development stage, and colony). The colony impact proved to be insignificant. Therefore, only caste and development stage means were compared, using two-way ANOVA and Tukey’s test (SAS Institute v.9.13., 2002-2003 license 86636). Bliss transformation (y = arc sin (x/100)0.5) was used to obtain the percentages of DNA m5C.
RESULTS
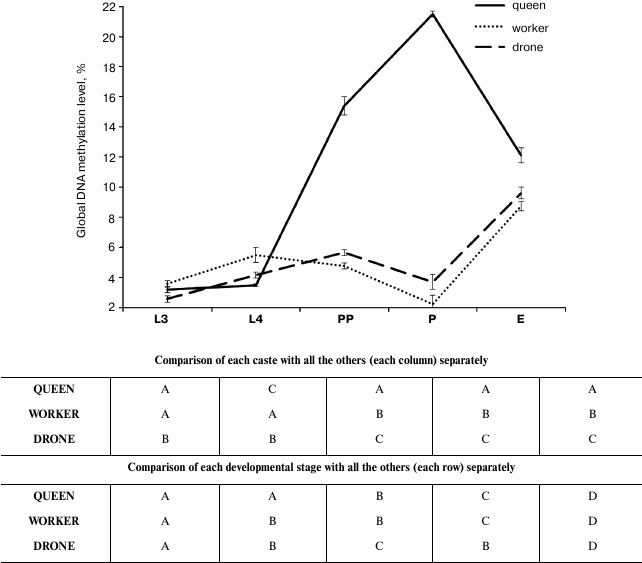
Percentages of global DNA methylation (DNA m5C percentage) slightly increased between the 3rd and 4th day of larval development (L3 and L4 points) in the workers and drones, but not in the queens (figure). The percentages were so low that they were not reflected in the methylated DNA weights (Table 2, rows L3 and L4). Next, DNA methylation levels markedly increased, but only in the queen prepupae and pupae (figure, points PP and P; Table 2, rows PP and P). Thus, in the queen prepupae and pupae the percentages/weights of methylated DNA were, on average, respectively three and four times higher than those in the worker brood of the same age. Subsequently, beginning from the pupal stage, DNA methylation levels, expressed both as DNA m5C percentages (figure) and methylated DNA weights (Table 2), markedly decreased in the queens but increased in the drones and workers. Consequently, the levels were similar in adult newly emerged drones, queens, and workers. The lowest DNA m5C percentage was identified in the worker and the highest in the queen pupae (figure). Concluding, the age-related, developmental patterns of changes in DNA methylation levels were different in the queens from those in the drones and workers. The changes in the methylation levels were about 3.5 times greater in the queens than in the workers and drones; especially the DNA of the queen pupae and prepupae was highly methylated.
Age-related developmental patterns of global DNA methylation levels (percentages of DNA m5C) in A. mellifera. Different upper case letters indicate differences significant at P ≤ 0.01. Results of comparisons made separately for averages within each caste and each developmental stage were given respectively in the upper and lower frame. L3, the 3rd day of larval development; L4, the 4th day of larval development; PP, prepupae; P, pupae; E, emergence
Table 2. Weights of methylated DNA (ng) at
the different developmental stages in A. mellifera
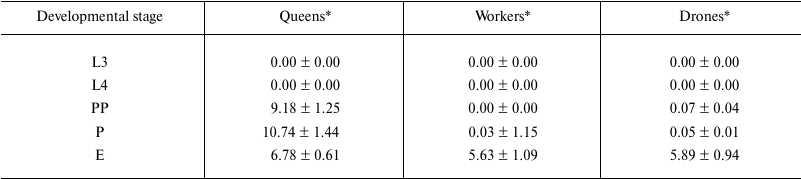
Note: L3, the 3rd day of larval development; L4, the 4th day of larval
development; PP, prepupae; P, pupae; E, newly emerged bees.
* Average values ± S.D.
DISCUSSION
Kucharski et al. [6], Cedar and Bergman [23], Miklos and Maleszka [24], and Lyko et al. [21] believed that environmental factors, particularly diet, influenced the expression of the apian genome and, consequently, the phenotype and behavior in different castes of honeybees. Our studies revealed that percentages of DNA m5C were low and more or less similar in the larvae of all the castes until the 4th day of larval development. This corresponds with the fact that larvae cease to be totipotent on the third-fourth day of their development. On the 4th day of larval development, juvenile hormone levels are higher in queens than in workers [22, 25]. About 150 ovarioles develop in the reproductive systems of queens from that day. In contrast, workers have their reproductive systems reduced to 2-20 ovarioles and are functionally sterile. Kucharski et al. [6] showed that cytosine-phosphate-guanosine (CpG) methylation by DNA methyltransferase 3 can be lower in queen larvae than in developing workers. Our study confirmed this with lower percentages of DNA m5C in the queen larvae than in the worker larvae at the initial and middle stages of their development (figure, points L3 and L4).
Most of the studies concentrated only on these larval periods and on comparing only queen and worker ontogenesis [9, 11, 21]. Our results provide one of the first insights into the fluctuations of general DNA methylation at the prepupal and pupal stages; it is of note that our study included not only females but also drones. The results showed that, during these later developmental periods, age-related developmental patterns of changes in the global DNA methylation levels were markedly different in the queens from those in the drones and workers, in which the patterns were similar (figure and Table 2). This similarity was observed despite the fact that a fertilized egg develops into a female, whereas an unfertilized one into a drone [26] and that drones anatomically and behaviorally significantly differ from workers and are fed a different larval diet. The striking thing is that while methylation levels rise in queens, they simultaneously decrease in drones and workers. Nevertheless, fluctuations in juvenile hormone (JH) contents during the larval development of drones were found to have a similar pattern as in the case of the female larvae, but the drone titers are intermediate between the high titers in the queens and low titers in the workers [27]. The rates/levels of JH synthesis in the corpora alta of drone larvae, however, were almost the same as those in the worker larvae. Analyses of the ecdysteroid titers have confirmed that endocrine activity in drone larvae is very similar to that in worker larvae (for more information see [7]). Thus, the similar developmental changes (fluctuations) in DNA methylation levels observed during worker and drone ontogenesis in our research (figure) may correspond with similar developmental patterns of fluctuations in endocrine activities noticed in these two castes in other studies. Developmental physiology of drones has received little attention when compared to queens and workers. In this context, the similar regularities on the biochemical and epigenetic levels may help better understanding the mechanisms of gene expression regulation. Moreover, the idea that workers appeared evolutionarily later than drones and queens [27] does not have to be true, in view of the similarities in the epigenetic and biochemical changes between the drones and workers observed during their ontogenesis. Perhaps “royal super-females” were formed later than “normal females” and drones because of natural selection, or workers (not fully functional females) and queens (super-females) have evolved at the same time.
Evans and Wheeler [28] and Barchuk et al. [7] identified hundreds of genes that are differentially expressed in queen and worker larvae. Queen larvae show upregulation of genes involved in metabolism and nutrient sensing, including the key components of the insulin/insulin-like signaling (IIS) and target of rapamycin (TOR) pathways [7, 29]. Decreasing the expression of the TOR kinase and insulin receptor substrate via RNA interference in the larvae causes individuals intended to become queens to develop worker-like traits [8, 29, 30]. Rascon et al. [31] suggested that suppression of IIS and TOR is accompanied with decreased JH titers and increased DNA methylation levels, and these may be associated with the development towards queens after the 7th day of ontogenesis. All the biochemical differences between queen and worker larvae are shaped particularly between the 3rd and 5th day of larval development. These differences may explain the fact that the methylation level was about 3.5 times higher in the queens than in the workers and drones at the prepupal/pupal stage in our study (Table 2 and figure). The high levels of DNA methylation in queen prepupae and pupae might also result from the methylation of genes responsible for worker behavior, polyethism, and physiology [32, 33], but we believe that the whole issue requires further study. It seems surprising that the percentage of m5C in the DNA of the newly emerged queens was higher than that in the workers, because the expected queen lifespan is longer than that of workers and aging is connected with a slow increase in DNA methylation [6, 9, 34, 35].
The most striking finding is, however, that in queens, after the methylation level increased to attain the maximal values at the pupal stages, it decreased almost 2-fold in a few days and became similar in the newly emerged queens, workers, and drones. This finding seems to be very interesting, particularly for experimental gerontology [6, 36, 37]. Moreover, if the decrease in DNA methylation level in queens and the corresponding increase in workers and drones is continued during the 1 or 2 days after emergence, the problem mentioned in the last sentence of the previous paragraph might find its explanation.
The worldwide honeybee depopulation has led to the development of genetic studies of the main parasites of bees [38-40]. The regular ontogenetic fluctuations of DNA methylation patterns observed in our studies in honeybees suggests that more epigenetic studies in their parasites are needed.
By enriching the knowledge of the ontogenetic pattern of global methylation level changes during drone ontogenesis and at prepupal and pupal stages in all the apian castes, our studies set new directions for further research. Particularly, the following questions ought to be answered. a) Why did the queen prepupae and pupae have very high global DNA methylation levels in comparison with the worker and drone prepupae and pupae? b) Why did the newly emerged queens have higher percentages of m5C in their DNA than the workers and drones? c) Why did the percentage of m5C in the DNA significantly decrease between the pupal and the newly emerged adult individual stages in the queens?
The idea that workers appeared evolutionarily later than drones and queens need not be true, considering the similarities in the epigenetic changes between the drones and workers observed during their ontogenesis. Perhaps “royal super-females” were formed later than “normal (worker) females”.
This work was carried out as part of the research project ZKB/MN/5 2012-2014, financed by the Ministry of Science and Higher Education in Poland.
REFERENCES
1.Foret, S., Kucharski, R., Pittelkow, Y., Lockett,
G., and Maleszka, R. (2009) Epigenetic regulation of the honeybee
transcriptome: unravelling the nature of methylated genes, BMC
Genom.; http://dx.doi:10.1186/1471-2164-10-472.
2.Bocklandt, S., Lin, W., Sehl, M. E., Sanchez, F.
J., Sinsheimer, J. S., Horvath, S., and Vilain, E. (2011) Epigenetic
predictor of age, PLoS, 6, 1-6.
3.Bird, A. (2002) DNA methylation patterns and
epigenetic memory, Gene Dev., 16, 6-21.
4.Suzuki, M. M., Kerr, A. R. W., De Sousa, D., and
Bird, A. (2007) CpG methylation is targeted to transcription units in
an invertebrate genome, Genome Res., 17, 625-631.
5.Rogalska, S. M., Achrem, M., and Wojciechowski, A.
(2010) Chromatyna. Molekularne Mechanizmy Epigenetyczne [in
Polish], Wydawnictwo UP, Poznan, pp. 15-80.
6.Kucharski, R., Maleszka, J., Foret, S., and
Maleszka, R. (2008) Nutritional control of reproductive status in
honeybees via DNA methylation, Science, 319,
1827-1830.
7.Barchuk, A. R., Cristino, A. S., Kucharski, R.,
Costa, L. F., Simoes, Z. L., and Maleszka, R. (2007) Molecular
determinants of caste differentiation in the highly eusocial honeybee
Apis mellifera, BMC Dev. Biol., 7, http://dx.doi:10.1186/1471-213X-7-70.
8.Patel, A., Fondrk, M. K., Kaftanoglu, O., Emore,
C., Hunt, G., Frederic, K., and Amdam, G. (2007) The making of a queen:
TOR pathway is a key player in diphenic caste development, PLoS
One, 2; http://dx.doi:10.1371/journal.pone.0000509.
9.Maleszka, R. (2008) Epigenetic integration of
environmental and genomic signals in honeybees, Epigenetics,
3, 188-192.
10.Kamakura, M. (2011) Royal actin induces queen
differentiation in honeybees, Nature, 473, 478-483.
11.Shi, Y. Y., Huang, Z. Y., Zeng, Z. J., Wang, Z.
L., Wu, X. B., and Wei, Y. Y. (2011) Diet and cell size both affect
queen–worker differentiation through DNA methylation in honey
bees (Apis mellifera, Apidae), PLoS One, 6; http://dx.doi:10.1371/journal.pone.0018808.
12.Evan, J., and Wheeler, D. (2000) Expression
profiles during honeybee caste determination, Genome Biol.,
2; http://dx.doi:10.1186/gb-2000-2-1-research0001.
13.Burzynski, S., Paleolog, J., Patii, S.,
Ilkowska-Musial, E., Borsuk, G., Olszewski, K., Chittur, S., Gupta, V.,
Sarangi, R., and Strachecka, A. (2013) Changed gene expression and
longevity in honeybees (Apis mellifera) fed with phenylbutyrate-
and phenylacetylglutaminate-supplemented diet, Med. Weter.,
69, 753-759.
14.Wheeler, M., and Robinson, G. (2014)
Diet-dependent gene expression in honeybees: honey vs. sucrose or high
fructose corn syrup, Sci. Rep., 4, 5726.
15.Strachecka, A., Borsuk, G., Olszewski, K.,
Paleolog, J., Gagos, M., Chobotow, J., Nawrocka, A., Gryzinska, M., and
Bajda, M. (2012) The effect of amphotericin B on the lifespan, body
surface protein concentrations and DNA methylation level of the
honeybees (Apis mellifera), J. Apic. Sci., 56,
107-113.
16.Strachecka, A., Gryzinska, M., and Krauze, M.
(2010) The influence of environmental pollution on the protective
proteolytic barrier of the honeybee Apis mellifera mellifera,
Pol. J. Environ. Stud., 19, 855-859.
17.Strachecka, A., Krauze, M., Olszewski, K.,
Borsuk, G., Paleolog, J., Merska, M., Chobotow, J., Bajda, M., and
Grzywnowicz, K. (2014) Unexpectedly strong effect of caffeine on the
vitality of western honeybees (Apis mellifera), Biochemistry
(Moscow), 79, 1192-1201.
18.Strachecka, A., Olszewski, K., Krauze, M.,
Paleolog, J., Borsuk, G., Merska, M., Bajda, M., and Chobotow, J.
(2014) Coenzyme Q10 treatments influence the lifespan and key
biochemical resistance systems in the honeybee, Apis mellifera,
Arch. Insect Biochem. Physiol., 86, 165-179.
19.Strachecka, A., Paleolog, J., Borsuk, G., and
Olszewski, K. (2012) Influence of formic acid on the body surface
proteolytic system in different developmental stages of Apis
mellifera L. workers, J. Apic. Res., 51, 252-262.
20.Strachecka, A., Paleolog, J., Borsuk, G.,
Olszewski, K., and Bajda, M. (2012) DNA methylation in the honeybee
(Apis mellifera) and its importance for biological research,
Med. Weter., 68, 391-396.
21.Lyko, F., Foret, S., Kucharski, R., Wolf, S.,
Falckenhayn, C., and Maleszka, R. (2010) The honeybee epigenomes:
differential methylation of brain DNA in queens and workers, PloS
Biol., 8, 1-12.
22.Shi, Y., Yan, W., Huang, Z., Wang, Z., Wu, X.,
and Zeng, Z. (2013) Genomewide analysis indicates that queen larvae
have lower methylation levels in the honey bee (Apis mellifera),
Naturwiss., 100, 193-197.
23.Cedar, H., and Bergman, Y. (2009) Linking DNA
methylation and histone modification: patterns and paradigms, Nat.
Rev. Genet., 10, 295-304.
24.Miklos, G., and Maleszka, R. (2011) Epigenomic
communication systems in humans and honeybees: from molecules to
behavior, Horm. Behav., 59, 399-406.
25.Hartfelder, K., Tozetto, S., and Rachinsky, A.
(1993) Sex-specific developmental profiles of juvenile hormone
synthesis in honeybee larvae, Roux’s Arch. Dev. Biol.,
202, 176-180.
26.Winston, M. (1987) The Biology of
Honeybee, Harvard University Press, Cambridge, pp. 46-213.
27.Harfelder, K., and Engels, W. (1998) Social
insect polymorphism: hormonal regulation of plasticity in development
and reproduction in the honeybee, Curr. Top. Dev. Biol.,
40, 45-77.
28.Evans, J., and Wheeler, D. (1999) Differential
gene expression between developing queens and workers in the honey bee
Apis mellifera, Proc. Natl. Acad. Sci. USA,
96, 5575-5580.
29.Wheeler, D., Buck, N., and Evans, J. (2006)
Expression of insulin pathway genes during the period of caste
determination in the honey bee Apis mellifera, Insect Mol.
Biol., 15, 597-602.
30.Wolschin, F., Mutti, N. S., and Amdam, G. V.
(2011) Insulin receptor substrate influences female caste development
in honeybees, Biol. Lett., 7, 112-115.
31.Rascon, B., Mutti, N., Tolfsen, C., and Amdam, G.
(2011) in Mechanisms of Life History Evolution. Honeybee Life
History Plasticity: Development, Behavior, and Aging (Flatt, T.,
and Heyland, A., eds.) Vol. 1, Oxford University Press Inc., New York,
pp. 253-266.
32.Page, R., Scheiner, R., Erber, J., and Amdam, G.
(2006) The development and evolution of division of labor and foraging
specialization in a social insect (Apis mellifera L.), Curr.
Top. Dev. Biol., 74, 253-286.
33.Amdam, G., Ihle, K., and Page, R. (2009) in
Hormones, Brain and Behavior. Regulation of Honeybee (Apis
mellifera) Life-Histories by Vitellogenin (Pfaff, D., Arnold, A.,
Fahrbach, S., Etgen, A., and Rubin, R., eds.) Vol. 4, Elsevier Academic
Press, San Diego, pp. 1003-1025.
34.Beye, M., Hunt, G. J., Page, R. E., Fondrk, M.,
Grohmann, L., and Moritz, R. (1999) Unusually high recombination rate
detected in the sex locus region of the honeybee (Apis
mellifera), Genetics, 153, 1701-1708.
35.Ikeda, T., Furukawa, S., Nakamura, J., Sasaki,
M., and Sasaki, T. (2011) CpG methylation in the hexamerin 110 gene in
the European honeybee Apis mellifera, J. Insect Sci.,
74, 1-11.
36.Lyko, F., and Maleszka, R. (2011) Insect as
innovative models for functional studies of DNA methylation, Trends
Genet., 27, 127-164.
37.Shao, X., He, S., Zhauang, X., Fan, Y., Li, Y.,
and Yao, Y. (2014) mRNA expression and DNA methylation in three key
genes involved in caste differentiation in female honeybees (Apis
mellifera), Zool. Res., 35, 92-98.
38.Cornman, R., Schatz, M., Johnston, J., Chen, Y.,
Pettis, J., Hunt, G., Bourgeois, L., Elsik, C., Anderson, D.,
Grozinger, C., and Evans, J. (2010) Genomic survey of the ectoparasitic
mite Varroa destructor, a major pest of the honeybee Apis
mellifera, BMC Genom., 11, http://dx.doi:10.1186/1471-2164-11-602.
39.Fries, I. (2010) Nosema ceranae in
European honeybees (Apis mellifera), J. Invert. Pathol.,
103, 573-579.
40.Ptaszynska, A. A., Borsuk, G., Wozniakowski, G.,
Gnat, S., and Malek, W. (2014) Loop-mediated isothermal amplification
(LAMP) assays for rapid detection and differentiation of Nosema
apis and N. ceranae in honeybees, FEMS Microbiol. Lett.
Fed. Europ. Microbiol. Sci., 357, 40-48.