REVIEW: ESR – A Retinal Protein with Unusual Properties from Exiguobacterium sibiricum
L. E. Petrovskaya1*, S. P. Balashov2, E. P. Lukashev3, E. S. Imasheva2, I. Yu. Gushchin4,5,6,7,8, A. K. Dioumaev2, A. B. Rubin3, D. A. Dolgikh1,3, V. I. Gordeliy4,5,6,7,8, J. K. Lanyi2, and M. P. Kirpichnikov1,3
1Shemyakin–Ovchinnikov Institute of Bioorganic Chemistry, Russian Academy of Sciences, 117997 Moscow, Russia; E-mail: lpetr65@yahoo.com2Department of Physiology and Biophysics, University of California, Irvine, 92697, USA; E-mail: balashov@uci.edu; eimashev@uci.edu; dioumaev@uci.edu; jklanyi@uci.edu
3Lomonosov Moscow State University, Biological Faculty, 119234 Moscow, Russia; E-mail: lukashev@biophys.msu.ru; rubin@biophys.msu.ru; dolgikh@nmr.ru; kirpichnikov@inbox.ru
4Institut de Biologie Structurale, Université Grenoble Alpes, 38044 Grenoble, France; E-mail: vangmf@gmail.com; valentin.gordeliy@gmail.com
5Institut de Biologie Structurale, Centre National de la Recherche Scientifique, 38044 Grenoble, France
6Institut de Biologie Structurale, Direction des Sciences du Vivant, Commissariat à l'Énergie Atomique, 38044 Grenoble, France
7Laboratory for Advanced Studies of Membrane Proteins, Moscow Institute of Physics and Technology, 141700 Dolgoprudniy, Moscow Region, Russia
8Institute of Complex Systems (ICS), ICS-6: Structural Biochemistry, Research Centre Jülich, 52425 Jülich, Germany
* To whom correspondence should be addressed.
Received January 12, 2015; Revision received March 10, 2015
This review covers the properties of a retinal protein (ESR) from the psychrotrophic bacterium Exiguobacterium sibiricum that functions as a light-driven proton pump. The presence of a lysine residue at the position corresponding to intramolecular proton donor for the Schiff base represents a unique structural feature of ESR. We have shown that Lys96 successfully facilitates delivery of protons from the cytoplasmic surface to the Schiff base, thus acting as a proton donor in ESR. Since proton uptake during the photocycle precedes Schiff base reprotonation, we conclude that this residue is initially in the uncharged state and acquires a proton for a short time after Schiff base deprotonation and M intermediate formation. Involvement of Lys as a proton donor distinguishes ESR from the related retinal proteins – bacteriorhodopsin (BR), proteorhodopsin (PR), and xanthorhodopsin (XR), in which the donor function is performed by residues with a carboxyl side chain. Like other eubacterial proton pumps (PR and XR), ESR contains a histidine residue interacting with the proton acceptor Asp85. In contrast to PR, this interaction leads to shift of the acceptor’s pKa to more acidic pH, thus providing its ability to function over a wide pH range. The presence of a strong H-bond between Asp85 and His57, the structure of the proton-conducting pathways from cytoplasmic surface to the Schiff base and to extracellular surface, and other properties of ESR were demonstrated by solving its three-dimensional structure, which revealed several differences from known structures of BR and XR. The structure of ESR, its photocycle, and proton transfer reactions are discussed in comparison with homologous retinal proteins.
KEY WORDS: retinal protein, proteorhodopsin, Exiguobacterium sibiricum, Schiff base, proton acceptor, proton donor, photocycleDOI: 10.1134/S000629791506005X
Abbreviations: BR, bacteriorhodopsin from Halobacterium salinarum; DDM, n-dodecyl-β-D-maltopyranoside; ESR, retinal protein from Exiguobacterium sibiricum; GPCR, G-protein-coupled receptors; GR, xanthorhodopsin from Gloeobacter violaceus; LPG, 1-palmitoyl-2-hydroxy-sn-glycero-3-phospho-(1′-rac-glycerol); PR, proteorhodopsin; XR, xanthorhodopsin from Salinibacter ruber.
In 2008, an article was published in BMC Genomics devoted to
deciphering the genome of Exiguobacterium sibiricum –
a microorganism isolated from permafrost aged about three million
years. Based on the data of coding sequences annotation, it was
suggested that this psychrotrophic bacterium is able to produce
rhodopsin [1]. Expression of the corresponding gene
in Escherichia coli revealed functional similarity of the
recombinant with other retinal proteins – proton
transporters [2]. By that time, it has already been
40 years since the first proton pump of this kind was discovered
– bacteriorhodopsin (BR) of purple membranes of the haloarchaea
Halobacterium salinarum [3]. During these
years, hundreds of reports focused on determination of the mechanism
that allows this protein to transform energy of absorbed light into
electrochemical proton gradient across the membrane.
The most important landmarks on this way were identification of the amino acid sequence of BR [4, 5], demonstration of electrogenicity of the proton transfer [6, 7], determination of the three-dimensional structure of the protein [8, 9], developing a mutagenesis system in H. salinarum [10], and obtaining of high resolution X-ray crystallography data [11]. It was established that BR molecules consist of two components: opsin (protein) and a chromophore (all-trans retinal linked to Lys216 residue through a Schiff base [4]). After light absorption, retinal isomerization into 13-cis configuration occurs [12, 13] with formation of the short-lived intermediate K [14, 15]. As a result of subsequent conformational changes in the chromophore returning it into all-trans conformation and accompanying structural rearrangements in the protein that proceeds though formation of intermediates L, M, N, and O, a proton is released at the outer surface of the protein and taken up from the cytoplasmic surface [16].
A key event in the BR photocycle is proton transfer from the Schiff base to the acceptor – the Asp85 [17] – that leads to formation of intermediate M (deprotonated Schiff base). It was shown that the system of hydrogen bonds connecting proton acceptor Asp85 with adjacent residues (Arg82, Asp212, etc.) is crucial for the functioning of the protein [11]. The Schiff base is reprotonated with participation of an intraprotein donor – the Asp96 residue [18]. It has been demonstrated that proton transport occurs due to sequential changes in pKa of the Schiff base and key amino acids due to conformational rearrangements [19]. Three-dimensional structures of different BR intermediates have been studied by X-ray crystallography at low temperatures [20-22]. On the whole, at present BR is one of the best-studied membrane proteins.
In 2000, the family of the retinal-containing proteins was supplemented by proteorhodopsin (PR), whose gene was discovered during sequencing of a metagenomic library obtained from marine microplankton [23]. It turned out that “green” (called this way due to location of its absorption maximum) proteorhodopsin (GPR) goes through the same key photocycle stages that are typical for BR. However, the pKa of the primary acceptor Asp97 in its molecule is significantly higher (7.5 against 2.5 in BR). Furthermore, proton release by the PR molecule occurs during the second part of photocycle, after its uptake from the cytoplasm.
Then, a vast number of BR structural analogs were discovered in Proteobacteria, Actinomycetales, Cyanobacteria, Fungi, and other microorganisms [24]. The amino acid sequences of these proteins are homologous to some extent. Their key positions are occupied by the same or homologous amino acids with analogous functional groups as in BR. Thus, aspartate is a proton acceptor in all the transport rhodopsin molecules, while the role of proton donor may be played by either aspartate or glutamate. Hence, finding a new retinal protein gene in 2008 was not a sensation anymore; so, there must have been substantial reasons to start isolating and studying this protein.
There were two reasons. First, the presence of the coding sequence of a retinal protein (we called it ESR after Exiguobacterium sibiricum rhodopsin) in the genome of a psychrotrophic soil bacterium was of interest [2]. All the known microbial rhodopsins belonged to halophilic Archaea, or marine microorganisms, while E. sibiricum was isolated from soil of freshwater origin. Second, the amino acid sequence alignment of ESR, BR, and PR revealed conservatism of all the key amino acid residues that are presumably involved in the proton transfer, excluding one. At the position that corresponds to the proton donor for the Schiff base, uniquely in ESR, instead of the typical carboxyl residue, there was a lysine (Lys96; Fig. 1). Later, the same residue at the corresponding position was found in proteorhodopsins belonging to related species, for example, Exiguobacterium AT1b living in hot springs of Yellowstone National Park [25], and in putative retinal proteins of oceanic proteobacteria [26]. The presence of such a unique structural feature raised a number of questions that we tried to answer during our studies of this interesting retinal protein.
Fig. 1. Alignment of amino acid sequences of ESR and homologous retinal proteins using CLUSTAL 2.1 software. Positions corresponding to amino acid residues Asp85, Lys96, and Lys225 in the ESR molecule are indicated with frames.
FEATURES OF ESR PHOTOCYCLE IN DIFFERENT MEDIA AND ITS RELATION TO
PROTON RELEASE
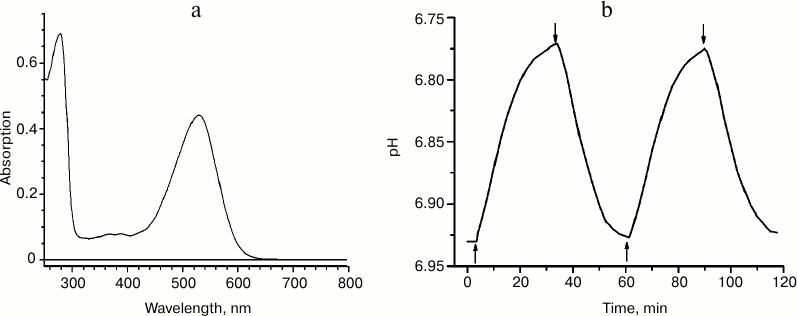
We expressed recombinant ESR having a C-terminal hexa-histidine tag in E. coli, extracted it from the membrane fraction in the presence of the detergent n-dodecyl-β-D-maltopyranoside (DDM), and purified it using immobilized-metal-affinity chromatography [2]. At pH 7.0, the absorption spectrum of the preparation had a maximum at 532 nm (Fig. 2a), shorter than that of BR and XR and closer to the absorption maximum of GPR. To prove the proton transfer activity of ESR, we tested light-induced pH changes in suspension of proteoliposomes with the incorporated protein or in E. coli cells expressing this protein. In response to illumination, the solution was acidified, which corresponds to transfer of protons from the cells or proteoliposomes containing ESR (Fig. 2b). The proton pump functioning was observed over a wide pH range – between 4.5 and 8.5. Based on these data, we concluded that the new retinal protein is a representative of the transport rhodopsin family that functions as proton pumps.
Fig. 2. Properties of recombinant ESR. a) Absorption spectrum for the sample in buffer: 50 mM sodium phosphate, 10 mM NaCl, 0.2% DDM, pH 8.0; b) light-induced proton release by ESR-containing proteoliposomes. The time points of turning the light on and off are indicated with arrows.
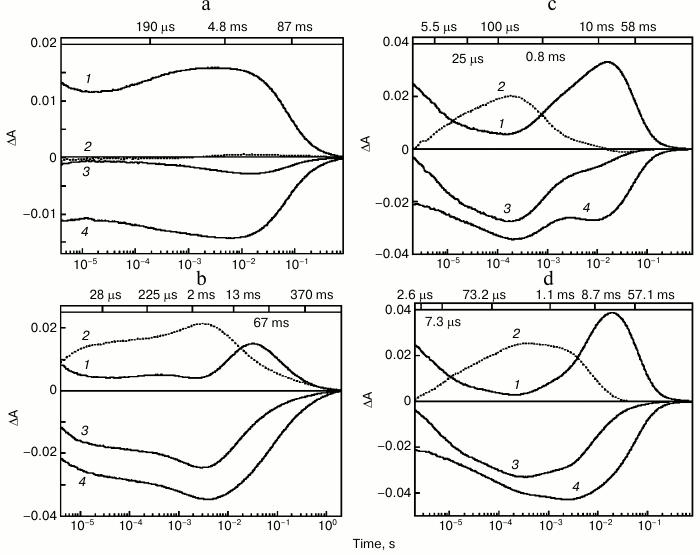
The photocycle of ESR was studied using laser flash spectroscopy (flash photolysis). It turned out that at neutral pH values in DDM micelles, in response to a flash ESR forms predominantly intermediates (K-, L-, and N/O-like) absorbing in the long wavelength region, while the quantity of an M-like intermediate, that corresponds to the deprotonated Schiff base having an absorption maximum at 410 nm, is very small (Fig. 3a). Increase of pH to above 9.0 results in fast emergence of an M intermediate that reaches its maximum 4 ms after the flash (Fig. 3b). The pKa of M formation in DDM micelles is ~8.8-9.0 [27].
Fig. 3. Photocycle of ESR in DDM micelles (a, b) and LPG (c, d) at pH 7.6 (a), 9.1 (b), 7.1 (c), 8.7 (d). 1) Absorption changes at 590 nm due to formation and decay of the K-, N-, and O-intermediates; 2) absorption changes at 410 nm due to M-intermediate formation and decay; 3) absorption changes at 550 nm due to return to the initial state and formation and decay of the K-, M-, and N-intermediates; 4) absorption changes at 510 nm due to formation of the L-intermediate and decay of the K- and O-intermediates. Numbers on the upper panel of the figure are time constants determined from kinetic analyses of the absorption changes. Adapted from [27] and [29] with permission from the American Chemical Society and American Society for Biochemistry and Molecular Biology.
After M decay via Schiff base protonation, the state analogous to N- or O-intermediate of the BR photocycle is formed. As shown with Fourier transform infrared spectroscopy (FTIR), retinal re-isomerization is one of the slowest stages and occurs at the very end of the ESR photocycle, which leads to the accumulation of mostly an N-type intermediate with retinal in the 13-cis configuration [28].
Studies on the photocycle of ESR in proteoliposomes or lipid-like detergent LPG (16:0 Lyso PG) [29] revealed the same pattern as in the protein solubilized in DDM micelles, but the M-intermediate in these media appeared already at neutral pH (the pKa of its formation was ~6.5) and decayed much faster (Fig. 3, c and d). As in DDM, the quantity of M-intermediate in the lipid environment increased along with pH. Thus, significant dependence of photocycle parameters on the environment is an interesting feature of ESR.
Experiments using a pH-sensitive dye, pyranine (pKa ~7.2), were carried out to study temporal events in the process of proton transfer by ESR. Increase in pyranine absorption at 460 nm in response to a flash corresponds to proton uptake from the environment, while decrease corresponds to proton release. Despite the small quantity of M-intermediate in the ESR photocycle in the DDM micelles, at pH 7.2 a substantial signal from pyranine was registered. In contrast to BR, in the ESR protein proton uptake occurs first ( ~ 12 ms) and then its release outward occurs (τ ~ 120 ms). A similar order of events is typical for the majority of eubacterial proton pumps. It is explained by the fact that residues homologous to Glu194 and Glu204 that form a proton release group in BR are absent in these molecules (for reviews see [30, 31]).
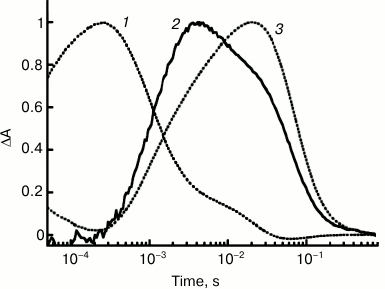
Study of the ESR preparation solubilized in LPG micelles has allowed comparison of the kinetics of proton transfer and of the protein photocycle in detail owing to presence of high levels of the M-intermediate at pH 7.2 already. Under these conditions, the proton uptake occurred at τ ~ 0.9 ms that was equal to the time of the M-intermediate decay and formation of the N-intermediate, i.e. Schiff base reprotonation. The proton release is featured by τ ~ 64 ms and correlates with decay of the long-wavelength intermediate and returning into the initial state (Fig. 4).
Fig. 4. Kinetics of light-induced proton uptake and release by ESR as compared to kinetics of M- and N-photocycle intermediates at pH 7.2: 1) absorption changes at 410 nm due to M-intermediate formation and its transition to N; 2) pyranine absorption changes at 455 nm due to its proton uptake and release; 3) absorption changes at 590 nm due to formation of intermediates N and O. Adapted from [29].
To determine if an intermediate donor–acceptor group is involved in H+ transfer, it was required to establish whether the proton uptake precedes reprotonation of the Schiff base or vice versa. For this, replacement of H2O with D2O was helpful [29]. It was known from BR studies that different photocycle stages are not slowed equally in heavy water. This kinetic effect appeared in our case as well: M-intermediate decay was 3-fold decelerated (τ ~ 3 ms), while the proton uptake rate was only 2.3-fold decelerated (τ ~ 2 ms). Hence, using heavy water distinguished in time the processes of proton uptake and Schiff base reprotonation more clearly and demonstrated that the first precedes the second. At first, the proton uptake occurs by a distinct group of the protein that does not function in the initial state. Then, after a short delay, the proton is transferred to the Schiff base. Thus, this group serves as the proton donor for the Schiff base in the ESR molecule. The nature of this group was studied in experiments with ESR mutants.
Is the M-intermediate present in the ESR photocycle at pH < pKa of its formation? The data that clarified the nature and the order of intermediates in the ESR photocycle under different conditions were obtained using Fourier transform infrared spectroscopy (FTIR) [28]. The band corresponding to the C=O stretch of protonated counter ion, i.e. deprotonated Schiff base, was found in these spectra already at pH ~ 5.0. Furthermore, in the K96A mutant photocycle in DDM, owing to low rate of the M-intermediate decay, even at pH 7.6 its slow formation (τ ~ 12 ms) is observed. One can hypothesize that the absence of M-intermediate accumulation in the wild-type protein photocycle in DDM at neutral pH values is for kinetic reasons, i.e. decay rate for this intermediate overrides its formation rate.
ROLE OF Lys96 RESIDUE IN SCHIFF BASE REPROTONATION
As mentioned above, an intriguing feature of the ESR amino acid sequence is the presence of a lysine residue at the position that corresponds to the intra-protein proton donor for the Schiff base. To characterize its functional role in the reprotonation process, we prepared and studied different mutant variants of this protein that have substitutions of this residue [29]. Mutant K96A, whose properties are considered in this section, was studied to the highest degree.
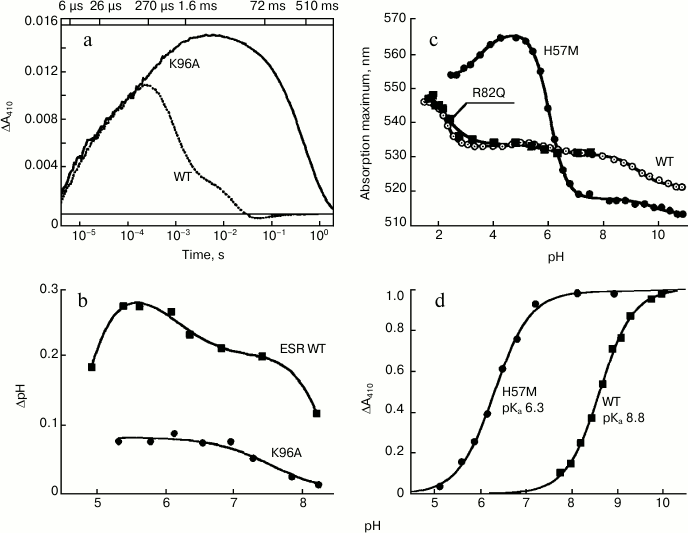
At pH 7.4, the rate of M formation is similar for K96A and the wild-type protein in LPG micelles. However, the rate of its decay, i.e. reprotonation of the Schiff base, is as much as 500-fold slower for the mutant (Fig. 5a). This leads to the apparent absence of the long-wavelength intermediates that follow M in the ESR photocycle and to a general deceleration of the photocycle of this mutant, for which the Schiff base reprotonation is a rate-limiting step. Since in the K96A mutant the Schiff base reprotonation happens directly from the environment, increasing pH from 7.0 to 9.0 is accompanied by linear deceleration of the M-intermediate decay, by more than one order of magnitude. Moreover, the mutant ESR in proteoliposomes or expressed in the E. coli cell membrane demonstrates decreased ability to transfer protons as compared to the wild-type protein (Fig. 5b).
Fig. 5. Properties of the ESR mutants. a) Light-induced absorption change at 410 nm in wild-type ESR as compared to K96A demonstrating deceleration of the Schiff base reprotonation in the mutant; b) light-induced pH changes in a suspension of E. coli cells expressing wild-type ESR and K96A mutant; c) pH-dependence of absorption maximum of H57M in comparison with wild type and R82Q mutant; d) pH-dependence of light-induced absorption changes at 410 nm (M-intermediate accumulation) of H57M as compared to wild-type protein. Adapted from [27] and [29].
Thus, we can state that this residue in the ESR molecule facilitates reprotonation, i.e. it functions as a proton donor for the Schiff base. Hence, ESR represents the first example of a proton-transfer rhodopsin in which this role is played by a residue having no carboxyl group. It was considered before that the presence of such group in a position corresponding to the proton donor as well as the carboxyl residue in the acceptor position of the retinal proteins is an essential condition for carrying out the proton transfer [32]. From the discovery and study of ESR, this rule was broken for the first time [29].
One can imagine two ways for the participation of Lys96 in reprotonation of the Schiff base in the ESR molecule. The donor function could be carried out directly by the ε-amino group of the lysine residue that temporarily acquires the proton after M-intermediate formation. Then, after transfer of the proton to the Schiff base, it returns to the initial non-protonated state. It is known that the pKa of lysine is very much dependent on the environment [33]. In the initial state of ESR, Lys96 is predominantly surrounded by hydrophobic residues that may decrease its pKa. On the formation of the M-intermediate, enlargement of the hydrophilic cavity in the cytoplasmic part of the molecule may occur, thus increasing the pKa of the lysine and causing capture of the proton.
At present, a second variant cannot be ruled out, the participation of the lysine side chain in a system of hydrogen bonds that involves water molecules which can serve as the proton donor. In this case, the protonation status of Lys96 would be unchanged.
Do carboxyl residues function as proton donors in the ESR molecule? To address this question, we made ESR mutants containing replacements of Lys96 with carboxylic residues (Asp or Glu) and planned to determine whether the Schiff base reprotonation in these mutants would occur as fast as in the wild-type protein.
ROLE OF His57 RESIDUE IN MODULATING pKa OF
PROTON ACCEPTOR Asp85
It is known that the function and the location of the absorption maxima for retinal proteins depend on protonation of an aspartic acid residue (Asp85 in BR), which is a proton acceptor from the Schiff base. Upon increase of the pH to a certain value, loss of a proton by the side chain of this residue occurs. As a result, it acquires the ability to carry out an acceptor function, i.e. to accept the proton from the Schiff base and to serve as a counter-ion for the Schiff base in the initial state. Deprotonation is accompanied by a shift in the absorption spectrum toward shorter wavelengths. Therefore, studying the pH-dependence of the absorption maxima of retinal proteins provides important information regarding the mechanism of their functioning under different conditions and the factors that affect the mechanism. Thus, the titration curve of the BR absorption maximum shows complex dependence with two pKa – 2.6 and 9.7 [34] – reflecting interaction of the proton acceptor Asp85 with residues that form a proton release group, Arg82, Glu204, and Glu194 [30]. For PR, the shift of the maximum in the 543-520 nm range is typical, which happens with pKa 7.1 and corresponds to deprotonation of the Asp97 residue [35]. The absorption maximum of both these proteins at low pH values coincided with the absorption maximum of mutant variants having replacements of aspartic acid residues to asparagine, which eliminates the negative charge of the counter-ion.
Dependence of the ESR absorption maximum on the pH is complex and includes three transitions with pKa 2.3, 6.0, and 9.1. The total shift of the maximum is 24 nm (from 545 to 521 nm) in the pH range 2.0-10.5 (Fig. 5c). Mutant variant D85N having Asp85 replaced with asparagine has an absorption maximum at 563 nm. Thus, at pH > 3.0 the majority of the ESR molecules contain the acceptor in the deprotonated state, which allows the protein to transport protons over a wide pH range. We studied the properties of some mutant variants of the protein to find the reason for the complex dependence of the spectral characteristics of ESR during titration.
It turned out that the pH-dependence of the absorption maximum for the R82Q mutant is identical to the curve obtained for the wild-type ESR (Fig. 5c). This suggests that in the ESR molecule, in contrast to BR [36, 37], the Arg82 residue is weakly bound to the proton acceptor from the Schiff base. A similar situation was observed for PR [38]. In contrast, replacement of His57 with a methionine residue (H57M) leads to substantial change in properties of the acceptor complex. At pH 5.0, the absorption maximum for this mutant is at 565 nm, which almost coincides with the absorption maximum for the D85N mutant. This means that at pH 5.0 the proton acceptor Asp85 in the H57M protein is fully protonated. Along with pH increase to 8.5, significant shift (47 nm) of the absorption maximum of the mutant to 517 nm was observed, with pKa 6.3 corresponding to pKa of Asp85 in H57M (Fig. 5c).
Such substantial increase in the Asp85 proton acceptor pKa in the H57M mutant as compared to the wild-type protein indicates that in the latter the His57 residue tightly interacts with the Asp85 residue, decreasing its pKa. As a result, acceptor Asp85 is deprotonated over a wide pH range. This interaction apparently causes the complex character of the pH-dependence of the wild-type ESR absorption maximum.
The presence of the histidine residue at the position that corresponds to His57 of ESR is a feature of eubacterial transport rhodopsins, including PR [39], XR [40], and GR [41]. At the same time, XR similarly to ESR, shows only marginal absorption maximum shift (3-5 nm) with pH increase from 4.0 to 10.0. Examination of the three-dimensional structure of XR revealed that the His62 and Asp96 residues are at a distance of ~2.5 Å from each other and form a strong hydrogen bond [40]. Unfortunately, data regarding properties of XR mutants having His62 replacements are not currently available due to the absence of an efficient system for expression of this protein in E. coli. Titration of PR in the pH range 6.0-10.0 leads to a more significant shift (23 nm) of its absorption maximum. In contrast to ESR, His75 replacements in this protein lead to shift of pH-dependence of the absorption maximum of the mutants toward the acidic region [39, 42]. Thus, we can state that structure of the acceptor complex in this protein apparently significantly differs from that of ESR.
Studying the H57M mutant photocycle at different pH values revealed that the appearance of the M-intermediate (deprotonated Schiff base) occurs at lower pH values as compared to the wild-type protein, the mutant having pKa 6.3 (Fig. 5d). This value corresponds to the pKa of the absorption maximum shift for H57M (see above), i.e. the pKa of the proton acceptor deprotonation in the initial state. In H57M, Schiff base deprotonation happens much faster than in the wild-type protein, which corresponds to higher pKa of the acceptor (Asp85). In general, the photocycle of this mutant features higher rate than the photocycle of the wild-type ESR, which is also typical for His75 mutants of PR [42].
Hence, the results obtained provide ample grounds to assume that at neutral and acidic pH values in the wild type protein His57 residue is protonated, interacts tightly with Asp85, and decreases pKa of the latter. In the process of photocycle, retinal isomerization, apparently, leads to disruption of a hydrogen bond between these residues and to deprotonation of His57 or to change of orientation of His57 relatively to Asp85 similar to what happens with Arg82 residue in BR [36, 43]. As a result of these events, pKa of the acceptor temporarily increases and it acquires an ability to accept the proton from the Schiff base.
THREE-DIMENSIONAL STRUCTURE OF ESR
The tertiary structure of ESR was studied by X-ray crystallography at 2.3 Å resolution [44]. According to the crystallographic data, ESR, like BR, belongs to the structural family of membrane proteins having seven α-helix segments that comprises all the currently known retinal proteins, including visual rhodopsin and G protein-coupled receptors (GPCR) [46-48].
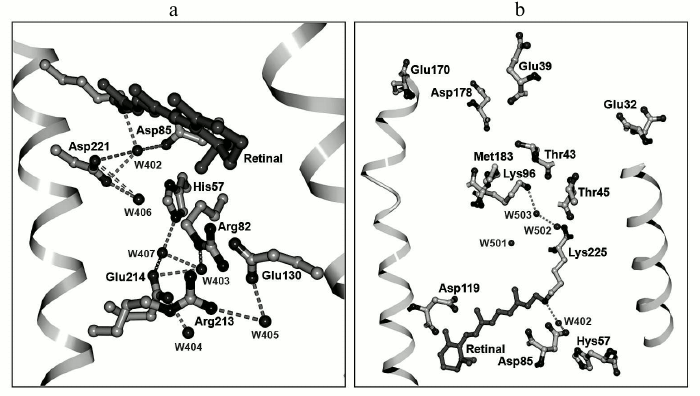
Similarly, like the other representatives of microbial transport rhodopsins, ESR contains a retinal residue in all-trans configuration covalently linked to the Lys225 residue. The protonated Schiff base is turned toward the extracellular part of the protein and is bound by hydrogen bonds through water molecule W402 to carboxylic residues that form the counter-ion – Asp85 and Asp221 (Fig. 6a). This configuration stabilizing positive charge of the Schiff base was found in the majority of known transport rhodopsins functioning as proton pumps. The side chain of the Asp85 residue in the ESR molecule is oriented just as in BR, in contrast to XR, where it is turned differently [40]. Similarly to BR, in the close vicinity of retinal there is another water molecule, W406, that forms a hydrogen bond with the Asp221 residue. While for BR in the initial state Arg82 is typically coupled to the proton acceptor Asp85, in the ESR molecule the side chain of this residue is directed outward from the Schiff base, similarly to its position in the M-intermediate of BR, and it is bound by a hydrogen bond to the Glu130 residue. This configuration explains the above-discussed absence of an influence of Arg82 on the functional state (protonation status) of the proton acceptor Asp85 in ESR [27]. ESR differs from the earlier transport rhodopsins by having a bend in α-helix F in the region of residue 185. The corresponding region of the helix is stabilized due to hydrogen bonding with side chains of Trp154 and Asn224 residues that are present at homologous positions in molecules of all the proteorhodopsins. The reason for such universality remains elusive.
Fig. 6. Three-dimensional structure of ESR: a) outer protein moiety; b) cytoplasmic protein moiety.
Explanation for the influence of the His57 residue on properties of the acceptor could be given based on the tertiary structure of ESR. The distance between these residues is very short, ~2.4 Å, which allows formation of a strong hydrogen bond. A similar bond is present in the XR structure [40], but its configuration differs significantly. In the ESR molecule, the side chain of the histidine is directed to Arg82 and is placed in a hydrophilic cavity containing several water molecules. In XR, the His62 side chain is turned to the opposite direction.
As stated above, the ESR molecule contains no homologs of Glu194 and Glu204 that in BR form, together with water molecules, the proton release group [49]. It is presumed that this is the reason for reverse (as compared to BR) order of proton transfer reactions: first, proton uptake occurs at the cytoplasmic side of the protein, then its release at the opposite side of the membrane by an unknown residue or a group of residues. The presence of the hydrophilic cavity that connects the extracellular part of ESR with the acceptor complex His57–Asp85 suggests their direct role as the proton source. Moreover, there are a large number of polar and charged residues associated by hydrogen bonds in this part of the protein that might also be the proton source. In particular, it is worth paying attention to the chain of hydrogen bonds connecting His57 with Asp214 through water molecule W407.
As discussed above, unlike the other representatives of the proton transport rhodopsin family, in ESR at the position that corresponds to the proton donor for the Schiff base there is a lysine residue rather than a carboxylic residue. Lys96 is surrounded primarily by hydrophobic residues; however, it is only separated from the hydrophilic cavity by the polar side chain of Thr43 residue (Fig. 6b). Such proximity of the proton donor to the environment distinguishes ESR from other proton pumps with known structures (BR, XR, and HmBR [50]) and may provide fast access of the protons from the cytoplasm in the reprotonation process. The side chain of the Leu93 residue as well as at least two water molecules are located between the Schiff base and Lys96.
Thus, studies on the three-dimensional structure of ESR have revealed the presence of structural elements common for the other representatives of the family of eubacterial proton pumps, XR [40] and BPR [51], but also distinct features of this protein related to the presence of the lysine residue as the proton donor and the complex system of hydrogen bonds between the Schiff base and the outer protein surface.
SUGGESTED SCHEME FOR THE FUNCTIONING OF ESR AS A PROTON PUMP IN
COMPARISON WITH OTHER TRANSPORT RHODOPSINS
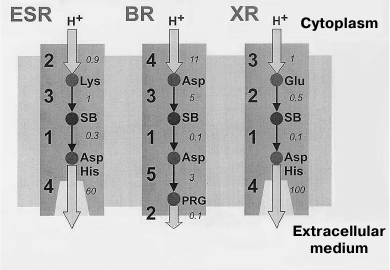
Based on spectroscopic and structural studies, the order of events accompanying light-dependent proton transfer by ESR can be described as follows. In the initial state of the protein, retinal is in all-trans configuration, the Schiff base is protonated (positively charged), proton acceptor Asp85 is deprotonated (negatively charged), and the proton donor is in the neutral state. The ion pair formed by the Schiff base and Asp85 is stabilized by water molecule W402 (Fig. 6a). Absorption of a light quantum leads to retinal isomerization from all-trans to 13-cis configuration and destabilization of this pair. As a result, the proton is transferred to the acceptor Asp85 that is bound to His57 by a hydrogen bond (L-M1 stage). The mechanism of the fast (femtosecond-microsecond range) primary processes in ESR has not been studied in detail; however, we expect that it is similar to those described for BR, PR, and XR [52-55]. During the M1–M2 transition, proton uptake from the environment takes place by the donor Lys96 residue, which acquires positive charge for a short while. The proximity of this residue to the hydrophilic cavity in the cytoplasmic moiety of the protein accelerates opening of the “channel” that leads to the increase in pKa of the lysine.
M2 decay is accompanied by proton transfer from donor to the Schiff base at the N1 stage. In the final stage of the photocycle, deprotonation of the acceptor group, proton release into the medium by an unidentified group X, and retinal re-isomerization into all-trans configuration occur. Thus, the protein returns to its initial state.
The main differences of the photocycle of ESR from the other proton-transport rhodopsins (BR, XR, and PR) are related to the presence of a lysine residue at the ESR position that corresponds to the position of the proton donor Asp96 in BR and also the absence of residues that form the proton release group in BR (Glu194 and Glu204). Both Lys96 of ESR and Asp96 of BR in the initial state are surrounded by hydrophobic residues, which facilitate maintaining their electroneutrality. However, while for the Asp96 residue (having initial pKa greater than 11) this means the presence of a proton on it, in the ESR molecule the Lys96 donor is not protonated. At this stage, its pKa is apparently about 6.0, and only later it increases to ~8.5. According to these differences, in the photocycle of BR the Schiff base reprotonation (M-intermediate decay) occurs during the M to N1 transition due to H+ transfer from Asp96 until the moment of proton capture by this residue. During the process of proton capture, the pKa of Asp96 drops to 7.5 [56, 57]. The Schiff base reprotonation occurs similarly in the XR and PR molecules, where the proton donor is a glutamic acid residue. On the contrary, Lys96 of ESR first obtains a proton from cytoplasm and only then transfers it to the Schiff base (Fig. 7).
Fig. 7. Proton transfer scheme in ESR, BR, and XR. The proton transfer stages are indicated with numbers, and typical proton transfer times in milliseconds are italicized. For ESR: 1) Schiff base deprotonation (formation of M-intermediate); 2) proton uptake from the cytoplasm; 3) Schiff base reprotonation (M-intermediate decay); 4) acceptor deprotonation and proton release. For BR: 1) Schiff base deprotonation (formation of M-intermediate); 2) proton release by PRG (proton release group); 3) Schiff base reprotonation (M-intermediate decay); 4) proton uptake from the cytoplasm; 5) acceptor deprotonation and proton transition to PRG. For XR: 1) Schiff base deprotonation (formation of M-intermediate); 2) Schiff base reprotonation (M-intermediate decay); 3) proton uptake from cytoplasm; 4) acceptor deprotonation and proton release.
The proton release at one of the last photocycle stages is an important feature that distinguishes ESR from BR. In the BR photocycle, it happens during the M1–M2 transition, i.e. at an earlier stage. However, mutant variants of BR having no Glu194 and Glu204 residues demonstrate so-called “late” proton release, like ESR [58, 59]. This order of events is also typical for XR and PR due to the absence of the proton release group in their molecules.
During last 15 years, the discovery of many new retinal proteins [60] – proteorhodopsins [61], xanthorhodopsins [62], channel rhodopsins [63, 64], chloride and sodium pumps [65, 66] – along with substantial progress in determining their tertiary structures [40, 50, 51, 67-69] has restored interest in studying the light-dependent ion transport started over 40 years ago by D. Oesterhelt and W. Stoeckenius [3]. Studying the new representatives of the family supplements our knowledge of the structural basis of such transport [66] and promotes their application in bio-optoelectronics as a basis for molecular memory elements and photoactive media in dynamic holography [70-72], and in a new field – optogenetics [73] – for regulation of neuronal activity [74-76]. The sensitivity of photocycle reaction kinetics and fluorescence of the proton pumps to electrochemical potentials [77, 78] revealed the possibility of using them as sensors for detection of intraprotein [79] and transmembrane changes of potentials [80].
Our studies on ESR – a new retinal protein from E. sibiricum – have broaden our knowledge of the mechanism of Schiff base reprotonation and of the nature of the amino acid residues that play the role of the intraprotein proton donor. The example of ESR is the first demonstration that a lysine residue can serve as the donor facilitating proton delivery from the cytoplasmic protein surface to the Schiff base. The interaction between Asp85 and His57 residues that leads to ESR proton acceptor pKa shift toward acidic pH that we discovered provides its ability to function over a broad pH range. The presence of the hydrogen bond between these residues and other features typical for proteorhodopsin and xanthorhodopsin families was demonstrated from the three-dimensional structure of ESR. Studies devoted to ESR are continuing, and they will undoubtedly lead to discovery of interesting new properties of this protein. It remains to be determined, which amino acid residues are responsible for the proton release under different conditions, how the ESR photocycle changes depending on lipid environment, and whether the proton transfer is accompanied by intramolecular charge transfer. It is possible that these studies will assist in addressing the question of evolutionary mechanisms by which light-sensitive retinal protein was encoded in the psychrotrophic bacterium genome from the Siberian tundra permafrost.
We thank the staff of the Soil Cryology Lab (headed by Dr. E. M. Rivkina) at the Institute of Physicochemical and Biological Problems of Soil Science for the opportunity to work with the genetic material from the permafrost.
This work was supported by the Russian Foundation for Basic Research grant 14-04-00499a, SS-1766.2014.4, Fulbright Visiting Scholar (for L. E. P.), and Russian Academy of Sciences program “Molecular and Cell Biology”. Work of J. K. L. and S. P. B. was supported by the National Institutes of Health grant (GM29498) and the Department of Energy grant (DEFG03-86ER13525). Determination of the ESR three-dimensional structure was supported by special agreement 5.1 CEA(IBS) – HGF(FZJ) STC, BMBF (PhoNa – Photonic Nanomaterials), Russian Foundation for Basic Research grant 13-04-91320, Federal target program “Research and Development” contract 14.587.21.0004, and program of the Ministry of Education and Science of the Russian Federation “5-100”; using the Grenoble Instruct Center (ISBG; UMS 3518 CNRS-CEA-UJF-EMBL) platform with support from FRISBI (ANR-10-INSB-05-02) and GRAL (ANR-10-LABX-49-01) within the framework of the Grenoble Partnership for Structural Biology (PSB). X-Ray diffraction data were collected on the ID23-1 and ID29 beam lines at the European Synchrotron Radiation Facility (ESRF), Grenoble, France.
REFERENCES
1.Rodrigues, D. F., Ivanova, N., He, Z., Huebner, M.,
Zhou, J., and Tiedje, J. M. (2008) Architecture of thermal adaptation
in an Exiguobacterium sibiricum strain isolated from 3 million
year old permafrost: a genome and transcriptome approach, BMC
Genomics, 9, 547.
2.Petrovskaya, L. E., Lukashev, E. P., Chupin, V. V.,
Sychev, S. V., Lyukmanova, E. N., Kryukova, E. A., Ziganshin, R. H.,
Spirina, E. V., Rivkina, E. M., Khatypov, R. A., Erokhina, L. G.,
Gilichinsky, D. A., Shuvalov, V. A., and Kirpichnikov, M. P. (2010)
Predicted bacteriorhodopsin from Exiguobacterium sibiricum is a
functional proton pump, FEBS Lett., 584, 4193-4196.
3.Oesterhelt, D., and Stoeckenius, W. (1973)
Functions of a new photoreceptor membrane, Proc. Natl. Acad. Sci.
USA, 70, 2853-2857.
4.Ovchinnikov, Y. A., Abdulaev, N. G., Feigina, M.
Y., Kiselev, A. V., and Lobanov, N. A. (1979) The structural basis of
the functioning of bacteriorhodopsin: an overview, FEBS Lett.,
100, 219-224.
5.Khorana, H. G., Gerber, G. E., Herlihy, W. C.,
Gray, C. P., Anderegg, R. J., Nihei, K., and Biemann, K. (1979) Amino
acid sequence of bacteriorhodopsin, Proc. Natl. Acad. Sci. USA,
76, 5046-5050.
6.Drachev, L. A., Jasaitis, A. A., Kaulen, A. D.,
Kondrashin, A. A., Liberman, E. A., Nemecek, I. B., Ostroumov, S. A.,
Semenov, A. Y., and Skulachev, V. P. (1974) Direct measurement of
electric current generation by cytochrome oxidase, H+-ATPase
and bacteriorhodopsin, Nature, 249, 321-324.
7.Kaulen, A. D. (2000) Electrogenic processes and
protein conformational changes accompanying the bacteriorhodopsin
photocycle, Biochim. Biophys. Acta, 1460, 204-219.
8.Henderson, R., and Unwin, P. N. (1975)
Three-dimensional model of purple membrane obtained by electron
microscopy, Nature, 257, 28-32.
9.Henderson, R., Baldwin, J. M., Ceska, T. A.,
Zemlin, F., Beckmann, E., and Downing, K. H. (1990) Model for the
structure of bacteriorhodopsin based on high-resolution electron
cryomicroscopy, J. Mol. Biol., 213, 899-929.
10.Ni, B., Chang, M., Duschl, A., Lanyi, J., and
Needleman, R. (1990) An efficient system for the synthesis of
bacteriorhodopsin in Halobacterium halobium, Gene,
90, 169-172.
11.Luecke, H., Schobert, B., Richter, H.-T.,
Cartailler, J.-P., and Lanyi, J. K. (1999) Structure of
bacteriorhodopsin at 1.55 Å resolution, J. Mol. Biol.,
291, 899-911.
12.Tsuda, M., Glaccum, M., Nelson, B., and Ebrey, T.
G. (1980) Light isomerizes the chromophore of bacteriorhodopsin,
Nature, 287, 351-353.
13.Mathies, R. A., Lin, S. W., Ames, J. B., and
Pollard, W. T. (1991) From femtoseconds to biology: mechanism of
bacteriorhodopsin light-driven proton pump, Annu. Rev. Biophys.
Biophys. Chem., 20, 491-518.
14.Lozier, R. H., Bogomolni, R. A., and Stoeckenius,
W. (1975) Bacteriorhodopsin: a light-driven proton pump in
Halobacterium halobium, Biophys. J., 15,
955-963.
15.Litvin, F. F., Balashov, S. P., and Sineshchekov,
V. A. (1975) The investigation of the primary photochemical conversions
of bacteriorhodopsin in purple membranes and cells of Halobacterium
halobium by the low temperature spectrophotometry method,
Bioorg. Khim., 1, 1767-1777.
16.Lozier, R. H., Niederberger, W., Bogomolni, R.
A., Hwang, S., and Stoeckenius, W. (1976) Kinetics and stoichiometry of
light-induced proton release and uptake from purple membrane fragments,
Halobacterium halobium cell envelopes, and phospholipid vesicles
containing oriented purple membrane, Biochim. Biophys. Acta,
440, 545-556.
17.Subramaniam, S., Greenhalgh, D. A., and Khorana,
H. G. (1992) Aspartic acid 85 in bacteriorhodopsin functions both as
proton acceptor and negative counter-ion to the Schiff base, J.
Biol. Chem., 267, 25730-25733.
18.Otto, H., Marti, T., Holtz, M., Mogi, T., Lindau,
M., Khorana, H. G., and Heyn, M. P. (1989) Aspartic acid-96 is the
internal proton donor in the reprotonation of the Schiff base of
bacteriorhodopsin, Proc. Natl. Acad. Sci. USA, 86,
9228-9232.
19.Subramaniam, S., Hirai, T., and Henderson, R.
(2002) From structure to mechanism: electron crystallographic studies
of bacteriorhodopsin, Phil. Trans. R. Soc. Lond. A, 360,
859-874.
20.Luecke, H., and Lanyi, J. K. (2003) Structural
clues to the mechanism of ion pumping in bacteriorhodopsin, Adv.
Protein Chem., 63, 111-130.
21.Schobert, B., Brown, L. S., and Lanyi, J. K.
(2003) Crystallographic structures of the M and N intermediates of
bacteriorhodopsin: assembly of a hydrogen-bonded chain of water
molecules between Asp96 and the retinal Schiff base, J. Mol.
Biol., 330, 553-570.
22.Wickstrand, C., Dods, R., Royant, A., and Neutze,
R. (2014) Bacteriorhodopsin: would the real structural intermediates
please stand up? Biochim. Biophys. Acta, 1850,
536-553.
23.Beja, O., Aravind, L., Koonin, E. V., Suzuki, M.
T., Hadd, A., Nguyen, L. P., Jovanovich, S. B., Gates, C. M., Feldman,
R. A., Spudich, J. L., Spudich, E. N., and DeLong, E. F. (2000)
Bacterial rhodopsin: evidence for a new type of phototrophy in the sea,
Science, 289, 1902-1906.
24.Brown, L. S., and Jung, K.-H. (2006)
Bacteriorhodopsin-like proteins of eubacteria and fungi: the extent of
conservation of the haloarchaeal proton-pumping mechanism,
Photochem. Photobiol. Sci., 5, 538-546.
25.Vishnivetskaya, T. A., Lucas, S., Copeland, A.,
Lapidus, A., Glavina del Rio, T., Dalin, E., Tice, H., Bruce, D. C.,
Goodwin, L. A., Pitluck, S., Saunders, E., Brettin, T., Detter, C.,
Han, C., Larimer, F., Land, M. L., Hauser, L. J., Kyrpides, N. C.,
Ovchinnikova, G., Kathariou, S., Ramaley, R. F., Rodrigues, D. F.,
Hendrix, C., Richardson, P., and Tiedje, J. M. (2011) Complete genome
sequence of the thermophilic bacterium Exiguobacterium sp. AT1b,
J. Bacteriol., 193, 2880-2881.
26.Iverson, V., Morris, R. M., Frazar, C. D.,
Berthiaume, C. T., Morales, R. L., and Armbrust, E. V. (2012)
Untangling genomes from metagenomes: revealing an uncultured class of
marine Euryarchaeota, Science, 335, 587-590.
27.Balashov, S. P., Petrovskaya, L. E., Lukashev, E.
P., Imasheva, E. S., Dioumaev, A. K., Wang, J. M., Sychev, S. V.,
Dolgikh, D. A., Rubin, A. B., Kirpichnikov, M. P., and Lanyi, J. K.
(2012) Aspartate-histidine interaction in the retinal Schiff base
counter-ion of the light-driven proton pump of Exiguobacterium
sibiricum, Biochemistry, 51, 5748-5762.
28.Dioumaev, A. K., Petrovskaya, L. E., Wang, J. M.,
Balashov, S. P., Dolgikh, D. A., Kirpichnikov, M. P., and Lanyi, J. K.
(2013) Photocycle of Exiguobacterium sibiricum rhodopsin
characterized by low-temperature trapping in the IR and time-resolved
studies in the visible, J. Phys. Chem. B, 117,
7235-7253.
29.Balashov, S. P., Petrovskaya, L. E., Imasheva, E.
S., Lukashev, E. P., Dioumaev, A. K., Wang, J. M., Sychev, S. V.,
Dolgikh, D. A., Rubin, A. B., Kirpichnikov, M. P., and Lanyi, J. K.
(2013) Breaking the carboxyl rule: lysine-96 facilitates reprotonation
of the Schiff base in the photocycle of a retinal protein from
Exiguobacterium sibiricum, J. Biol. Chem., 288,
21254-21265.
30.Balashov, S. P. (2000) Protonation reactions and
their coupling in bacteriorhodopsin, Biochim. Biophys. Acta,
1460, 75-94.
31.Gerwert, K., Freier, E., and Wolf, S. (2014) The
role of protein-bound water molecules in microbial rhodopsins,
Biochim. Biophys. Acta, 1837, 606-613.
32.Khorana, H. G. (1993) Two light-transducing
membrane proteins: bacteriorhodopsin and the mammalian rhodopsin,
Proc. Natl. Acad. Sci. USA, 90, 1166-1171.
33.Isom, D. G., Castaneda, C. A., Cannon, B. R., and
Garcia-Moreno, B. E. (2011) Large shifts in pKa
values of lysine residues buried inside a protein, Proc. Natl. Acad.
Sci. USA, 108, 5260-5265.
34.Balashov, S. P., Imasheva, E. S., Govindjee, R.,
and Ebrey, T. G. (1996) Titration of aspartate-85 in bacteriorhodopsin:
what it says about chromophore isomerization and proton release,
Biophys. J., 70, 473-481.
35.Dioumaev, A. K., Brown, L. S., Shih, J., Spudich,
E. N., Spudich, J. L., and Lanyi, J. K. (2002) Proton transfers in the
photochemical reaction cycle of proteorhodopsin, Biochemistry,
41, 5348-5358.
36.Balashov, S. P., Govindjee, R., Kono, M.,
Imasheva, E., Lukashev, E., Ebrey, T. G., Crouch, R. K., Menick, D. R.,
and Feng, Y. (1993) Effect of the arginine-82 to alanine mutation in
bacteriorhodopsin on dark adaptation, proton release, and the
photochemical cycle, Biochemistry, 32, 10331-10343.
37.Brown, L. S., Bonet, L., Needleman, R., and
Lanyi, J. K. (1993) Estimated acid dissociation constants of the Schiff
base, Asp85, and Arg82 during the bacteriorhodopsin photocycle,
Biophys. J., 65, 124-130.
38.Partha, R., Krebs, R., Caterino, T. L., and
Braiman, M. S. (2005) Weakened coupling of conserved arginine to the
proteorhodopsin chromophore and its counter-ion implies structural
differences from bacteriorhodopsin, Biochim. Biophys. Acta,
1708, 6-12.
39.Bergo, V. B., Sineshchekov, O. A., Kralj, J. M.,
Partha, R., Spudich, E. N., Rothschild, K. J., and Spudich, J. L.
(2009) His-75 in proteorhodopsin, a novel component in light-driven
proton translocation by primary pumps, J. Biol. Chem.,
284, 2836-2843.
40.Luecke, H., Schobert, B., Stagno, J., Imasheva,
E. S., Wang, J. M., Balashov, S. P., and Lanyi, J. K. (2008)
Crystallographic structure of xanthorhodopsin, the light-driven proton
pump with a dual chromophore, Proc. Natl. Acad. Sci. USA,
105, 16561-16565.
41.Tsukamoto, T., Kikukawa, T., Kurata, T., Jung, K.
H., Kamo, N., and Demura, M. (2013) Salt bridge in the conserved
His-Asp cluster in Gloeobacter rhodopsin contributes to trimer
formation, FEBS Lett., 587, 322-327.
42.Hempelmann, F., Holper, S., Verhoefen, M. K.,
Woerner, A. C., Kohler, T., Fiedler, S. A., Pfleger, N., Wachtveitl,
J., and Glaubitz, C. (2011) His75-Asp97 cluster in green
proteorhodopsin, J. Am. Chem. Soc., 133, 4645-4654.
43.Luecke, H., Schobert, B., Richter, H.-T.,
Cartailler, J.-P., and Lanyi, J. K. (1999) Structural changes in
bacteriorhodopsin during ion transport at 2 Å resolution,
Science, 286, 255-260.
44.Gushchin, I., Chervakov, P., Kuzmichev, P.,
Popov, A. N., Round, E., Borshchevskiy, V., Ishchenko, A., Petrovskaya,
L., Chupin, V., Dolgikh, D. A., Arseniev, A. A., Kirpichnikov, M., and
Gordeliy, V. (2013) Structural insights into the proton pumping by
unusual proteorhodopsin from nonmarine bacteria, Proc. Natl. Acad.
Sci. USA, 110, 12631-12636.
45.Palczewski, K., Kumasaka, T., Hori, T., Behnke,
C. A., Motoshima, H., Fox, B. A., Le Trong, I., Teller, D. C., Okada,
T., Stenkamp, R. E., Yamamoto, M., and Miyano, M. (2000) Crystal
structure of rhodopsin: a G-protein-coupled receptor, Science,
289, 739-745.
46.Pierce, K. L., Premont, R. T., and Lefkowitz, R.
J. (2002) Seven-transmembrane receptors, Nat. Rev. Mol. Cell
Biol., 3, 639-650.
47.Kobilka, B. K. (2011) Structural insights into
adrenergic receptor function and pharmacology, Trends Pharmacol.
Sci., 32, 213-218.
48.Katritch, V., Fenalti, G., Abola, E. E., Roth, B.
L., Cherezov, V., and Stevens, R. C. (2014) Allosteric sodium in class
A GPCR signaling, Trends Biochem. Sci., 39, 233-244.
49.Luecke, H., Schobert, B., Cartailler, J.-P.,
Richter, H.-T., Rosengarth, A., Needleman, R., and Lanyi, J. K. (2000)
Coupling photoisomerization of retinal to directional transport in
bacteriorhodopsin, J. Mol. Biol., 300, 1237-1255.
50.Shevchenko, V., Gushchin, I., Polovinkin, V.,
Round, E., Borshchevskiy, V., Utrobin, P., Popov, A., Balandin, T.,
Buldt, G., and Gordeliy, V. (2014) Crystal structure of Escherichia
coli-expressed Haloarcula marismortui bacteriorhodopsin I in
the trimeric form, PloS one, 9, e112873.
51.Ran, T., Ozorowski, G., Gao, Y., Sineshchekov, O.
A., Wang, W., Spudich, J. L., and Luecke, H. (2013) Cross-protomer
interaction with the photoactive site in oligomeric proteorhodopsin
complexes, Acta Crystallogr., D69, 1965-1980.
52.Mathies, R. A., Lin, S. W., Ames, J. B., and
Pollard, W. T. (1991) From femtoseconds to biology: mechanism of
bacteriorhodopsin light-driven proton pump, Annu. Rev. Biophys.
Biophys. Chem., 20, 491-518.
53.Lenz, M. O., Huber, R., Schmidt, B., Gilch, P.,
Kalmbach, R., Engelhard, M., and Wachtveitl, J. (2006) First steps of
retinal photoisomerization in proteorhodopsin, Biophys. J.,
91, 255-262.
54.Lanyi, J. K. (2006) Proton transfers in the
bacteriorhodopsin photocycle, Biochim. Biophys. Acta,
1757, 1012-1018.
55.Lanyi, J. K., and Balashov, S. P. (2011)
Xanthorhodopsin, in Halophiles and Hypersaline Environments
(Ventosa, A., Oren, A., and Ma, Y., eds.) Springer-Verlag,
Berlin-Heidelberg, pp. 319-340.
56.Balashov, S. P., Lu, M., Imasheva, E. S.,
Govindjee, R., Ebrey, T. G., Othersen III, B., Crouch, R. K., and
Menick, D. R. (1999) The proton release group in bacteriorhodopsin
controls the rate of the final step of its photocycle at low pH,
Biophys. J., 76, A147.
57.Zscherp, C., and Heberle, J. (1997) pH dependence
of proton transfer reactions within bacteriorhodopsin: a time resolved
ATR/FT-IR study, Biophys. J., 72, A206.
58.Brown, L. S., Varo, G., Needleman, R., and Lanyi,
J. K. (1995) Functional significance of a protein conformation change
at the cytoplasmic end of helix F during the bacteriorhodopsin
photocycle, Biophys. J., 69, 2103-2111.
59.Balashov, S. P., Imasheva, E. S., Ebrey, T. G.,
Chen, N., Menick, D. R., and Crouch, R. K. (1997) Glutamate-194 to
cysteine mutation inhibits fast light-induced proton release in
bacteriorhodopsin, Biochemistry, 36, 8671-8676.
60.Venter, J. C., Remington, K., Heidelberg, J. F.,
Halpern, A. L., Rusch, D., Eisen, J. A., Wu, D., Paulsen, I., Nelson,
K. E., Nelson, W., Fouts, D. E., Levy, S., Knap, A. H., Lomas, M. W.,
Nealson, K., White, O., Peterson, J., Hoffman, J., Parsons, R.,
Baden-Tillson, H., Pfannkoch, C., Rogers, Y.-H., and Smith, H. O.
(2004) Environmental genome shotgun sequencing of the Sargasso Sea,
Science, 304, 66-74.
61.Beja, O., Spudich, E. N., Spudich, J. L.,
Leclerc, M., and DeLong, E. F. (2001) Proteorhodopsin phototrophy in
the ocean, Nature, 411, 786-789.
62.Balashov, S. P., Imasheva, E. S., Boichenko, V.
A., Anton, J., Wang, J. M., and Lanyi, J. K. (2005) Xanthorhodopsin: a
proton pump with a light-harvesting carotenoid antenna, Science,
309, 2061-2064.
63.Sineshchekov, O. A., Jung, K.-H., and Spudich, J.
L. (2002) Two rhodopsins mediate phototaxis to low- and high-intensity
light in Chlamydomonas reinhardtii, Proc. Natl. Acad. Sci.
USA, 99, 8689-8694.
64.Nagel, G., Ollig, D., Fuhrmann, M., Kateriya, S.,
Mustl, A. M., Bamberg, E., and Hegemann, P. (2002) Channelrhodopsin-1:
a light-gated proton channel in green algae, Science,
296, 2395-2398.
65.Inoue, K., Ono, H., Abe-Yoshizumi, R., Yoshizawa,
S., Ito, H., Kogure, K., and Kandori, H. (2013) A light-driven sodium
ion pump in marine bacteria, Nat. Commun., 4, 1678; doi:
1610.1038/ncomms2689.
66.Beja, O., and Lanyi, J. K. (2014)
Nature’ s toolkit for microbial rhodopsin ion pumps,
Proc. Natl. Acad. Sci. USA, 111, 6538-6539.
67.Gordeliy, V. I., Labahn, J., Moukhametzianov, R.,
Efremov, R., Granzin, J., Schlesinger, R., Buldt, G., Savopol, T.,
Scheidig, A. J., Klare, J. P., and Engelhard, M. (2002) Molecular basis
of transmembrane signaling by sensory rhodopsin II–transducer
complex, Nature, 419, 484-487.
68.Moukhametzianov, R., Klare, J. P., Efremov, R.,
Baeken, C., Goppner, A., Labahn, J., Engelhard, M., Buldt, G., and
Gordeliy, V. I. (2006) Development of the signal in sensory rhodopsin
and its transfer to the cognate transducer, Nature, 440,
115-119.
69.Gushchin, I., Reshetnyak, A., Borshchevskiy, V.,
Ishchenko, A., Round, E., Grudinin, S., Engelhard, M., Buldt, G., and
Gordeliy, V. (2011) Active state of sensory rhodopsin II: structural
determinants for signal transfer and proton pumping, J. Mol.
Biol., 412, 591-600.
70.Hampp, N. (2000) Bacteriorhodopsin as a
photochromic retinal protein for optical memories, Chem. Rev.,
100, 1755-1776.
71.Vsevolodov, N. (1998) Biomolecular
Electronics. An Introduction via Photosensitive Proteins,
Birkhauser, Boston-Basel-Berlin.
72.Korchemskaya, E., Burykin, N., Bugaychuk, S.,
Maksymova, O., Ebrey, T., and Balashov, S. (2007) Dynamic holography in
bacteriorhodopsin/gelatin films: effects of light-dark adaptation at
different humidity, Photochem. Photobiol., 83,
403-408.
73.Deisseroth, K. (2011) Optogenetics, Nat.
Methods, 8, 26-29.
74.Boyden, E. S., Zhang, F., Bamberg, E., Nagel, G.,
and Deisseroth, K. (2005) Millisecond-timescale, genetically targeted
optical control of neural activity, Nat. Neurosci., 8,
1263-1268.
75.Deisseroth, K., Feng, G. P., Majewska, A. K.,
Miesenbock, G., Ting, A., and Schnitzer, M. J. (2006) Next-generation
optical technologies for illuminating genetically targeted brain
circuits, J. Neurosci., 26, 10380-10386.
76.Chow, B. Y., Han, X., Dobry, A. S., Qian, X. F.,
Chuong, A. S., Li, M. J., Henninger, M. A., Belfort, G. M., Lin, Y. X.,
Monahan, P. E., and Boyden, E. S. (2010) High-performance genetically
targetable optical neural silencing by light-driven proton pumps,
Nature, 463, 98-102.
77.Dubrovskii, V. T., Balashov, S. P., Sineshchekov,
O. A., Chekulaeva, L. N., and Litvin, F. F. (1982) Photoinduced changes
in the quantum yields of the photochemical cycle of conversions of
bacteriorhodopsin and transmembrane transport of protons in
Halobacterium halobium cells, Biokhimiya, 47,
1036-1046.
78.Kolodner, P., Lukashev, E. P., Ching, Y. C., and
Rousseau, D. L. (1996) Electric-field-induced Schiff-base deprotonation
in D85N mutant bacteriorhodopsin, Proc. Natl. Acad. Sci. USA,
93, 11618-11621.
79.Slouf, V., Balashov, S. P., Lanyi, J. K.,
Pullerits, T., and Polivka, T. (2011) Carotenoid response to retinal
excitation and photoisomerization dynamics in xanthorhodopsin, Chem.
Phys. Lett., 516, 96-101.
80.Kralj, J. M., Douglass, A. D., Hochbaum, D. R.,
Maclaurin, D., and Cohen, A. E. (2012) Optical recording of action
potentials in mammalian neurons using a microbial rhodopsin, Nat.
Methods, 90, 90-95.