Upregulation of RHOA and NKIRAS1 Genes in Lung Tumors Is Associated with Loss of Their Methylation as well as with Methylation of Regulatory miRNA Genes
E. A. Braga1,2*, V. I. Loginov1,2, I. V. Pronina1,2, D. S. Khodyrev3, S. V. Rykov4, A. M. Burdennyy1, M. V. Friedman5, T. P. Kazubskaya6, A. A. Kubatiev1, and N. E. Kushlinskii6
1Institute of General Pathology and Pathophysiology, 125315 Moscow, Russia; fax: +7 (495) 601-2366; E-mail: eleonora10_45@mail.ru2Research Center of Medical Genetics, Russian Academy of Medical Sciences, 115478 Moscow, Russia; fax: +7 (499) 324-0702; E-mail: karpukhin@med-gen.ru
3Federal Research Clinical Center of Specialized Types of Medical Care and Medical Technologies, Federal Medical and Biological Agency of Russia, 115682 Moscow, Russia; fax: +7 (495) 395-6430; E-mail: DmKh8@mail.ru
4State Research Institute of Genetics and Selection of Industrial Microorganisms, 117545 Moscow, Russia; fax: +7 (495) 315-0501; E-mail: genetika@genetika.ru
5Vavilov Institute of General Genetics, Russian Academy of Sciences, 117971 Moscow, Russia; fax: +7 (499) 135-6213; E-mail: iogen@vigg.ru
6Blokhin Russian Cancer Research Center, 115478 Moscow, Russia; fax: +7 (499) 324-6352; E-mail: biochimia@mtu-net.ru
* To whom correspondence should be addressed.
Received November 30, 2014; Revision received January 19, 2015
Methylation of CpG-islands in promoter regions as well as interaction of miRNAs with messenger RNAs of target genes are related to multilayer mechanisms regulating gene expression. The goal of this study was to assess a possibility for miRNA gene methylation to influence indirectly activation of their target genes in lung tumors. By using a unified collection of samples of non-small cell lung cancer, it was demonstrated that elevated levels of mRNA for RHOA and NKIRAS1 genes were significantly (Spearman rank correlation, P < 10–11) associated both with loss of methylation in their CpG-islands and methylation in a number of miRNA genes, which, according to the miRWalk database, were predicted to possess regulatory functions. Novel potential regulatory miRNAs for RHOA (miR-9-1/-3, -34b/c, -129-2, -125b-1, -375, -1258) and NKIRAS1 (miR-34b/c, -129-2, -125b-1, -193a, -124a-1/-2/-3, -212, -132) genes in lung cancer were identified.
KEY WORDS: CpG-islands, methylation, miRNA, target genes, RHOA, NKIRAS1, mRNA, lung cancerDOI: 10.1134/S0006297915040124
Abbreviations: AIA, allelic imbalance analysis; miRNA, microRNA; MSP, methylation-specific PCR; MSRA, methylation-sensitive restriction enzyme analysis; NSCLC, non-small cell lung cancer; RT-PCR, reverse-transcription polymerase chain reaction.
Among malignant tumors, lung cancer holds the first place in morbidity
and mortality rates. Annually, due to this disease, about 1.3 million
people become ill and 1.2 million people die worldwide. In Russia, more
than 40,000 people die every year due to lung cancer [1]. Five-year survival for patients with lung cancer,
stage I-II, comprises 57-67%, stage III – 5-25%, stage
IV – <1%. Notably, non-small cell lung cancer (NSCLC)
belongs to the most common types of lung cancer (>90%). These data
demonstrate relevance for investigating gene regulation in pathogenesis
of NSCLC, which is important to identify novel molecular markers and
targets for therapy of this socially significant disease.
Regulation of gene expression occurs at different levels, with various mechanisms being involved [2, 3]. Epigenetic mechanisms combined with genetic mechanisms (gene mutations and chromosomal aberrations) are responsible for regulation of gene activity and signaling pathways in cells. Methylation of promoter CpG-islands in protein-coding genes and effects from miRNAs binding to 3′-untranslated region within messenger RNA of target gene are epigenetic mechanisms. At the genomic level, methylation of promoter CpG-islands can result in gene inactivation, whereas interaction of regulatory miRNAs with mRNA occurs at posttranscriptional level and results in degradation of mRNA and impaired translation. Such inactivation pathway for protein-coding genes depends on the presence of particular miRNA, and its synthesis can be suppressed by a number of mechanisms including methylation of promoter region within the CpG-island of a particular miRNA gene. During the last decade, interest in identifying targets of methylation, methylome, and miRNA targets in tumors from different locations has sharply increased [4-7].
Broad target specificity is considered as an important property of miRNAs. Each miRNA can participate in regulation of hundreds of protein-coding genes, and conversely, a structural gene usually represents a target for numerous miRNAs (e.g. see miRWalk database [8]). According to bioinformatics data, miRNAs can be involved in regulation of more than half of protein-coding genes. In tumors, miRNA can exhibit oncogenic functions (miR-21) by inhibiting tumor growth suppressor genes as well as bear suppressor functions (miR-34) by inhibiting oncogenes and tumor progression genes [7]. Anticancer suppressor miRNAs can be inactivated by methylation in many types of cancer, which contributes to activation of oncogenes as targets for them. The current study was aimed at extending the range of suppressor miRNAs and their target genes. Two genes located on the short arm of human chromosome 3 (3p), RHOA (3p21.31) and NKIRAS1 (3p24.2), were examined as targets for miRNAs in NSCLC.
It is known that among other genomic regions, the 3p is an area with very frequent deletions as shown in epithelial tumors of the lung, kidneys, and other tissues. We were able to identify critical regions of the 3p containing numerous tumor growth suppressor genes; RASSF1A, RARB2, SEMA3B, CTDSPL, etc. are among the most studied tumor suppressor genes located on the 3p [9, 10].
However, in tumors the 3p was shown to contain not only deletions, but amplifications as well, and, consequently, genes exhibiting elevated expression and oncogenic activity. For instance, MECA3 (Major Epithelial CAncer region 3, 3p21.31) was found to have often loss of one allele in combination with amplification of the second allele [11-13]. Indeed, in this region several genes have been identified that exhibit oncogenic functions, e.g. the MST1R/RON and RHOA protooncogenes. In particular, of two alternative transcripts MST1R/RON (functional and oncogenic), in tumor cells the oncogenic transcript becomes activated [14].
Oncogenic potential and elevated RHOA expression were noted in epithelial tumors of different locations both at protein and mRNA levels [15-17]. Mutations in the RHOA gene in epithelial tumors were not found, so there should be some other factors that upregulate its expression during oncogenesis. Data on elevated transcriptional activity of the NKIRAS1 (3p24.2) gene homologous to the ras gene, which interacts with NF-κB transcription factor, were found in breast and lung cancer for the first time [18, 19]. This also suggests oncogenic function for NKIRAS1 [20, 21].
Here, we examined a role for methylation in regulating expression of RHOA (3p21.31) and NKIRAS1 (3p24.2) proto-oncogenes in NSCLC, in particular, a role for methylation of CpG-islands within intrinsic promoter regions and CpG-islands in a number of miRNA genes with regulatory functions predicted by the miRWalk database [8].
MATERIALS AND METHODS
Selection of miRNA genes overlapping with CpG-islands and related to tumor development that are predicted to regulate the RHOA (3p21.31) and NKIRAS1 (3p24.2) genes was performed by using the miRWalk database [8].
Samples of tumors and histologically intact lung tissue from 35 patients admitted for examination and treatment were collected and clinically evaluated at the Blokhin Russian Cancer Research Center. Patients with NSCLC who did not receive radiation or chemotherapy prior to surgery were recruited to the study. Tumors were evaluated according to the UICC TNM Classification, and were histologically examined according to the World Health Organization International Histological Classification of Tumors [22, 23]. For selection of samples with high numbers of tumor cells, tissue microsections (3-5 µm thick) were additionally analyzed after staining with hematoxylin and eosin. Samples containing at least 70% tumor cells were used for examination. Samples were kept at –70°C.
High molecular weight DNA was isolated from tumor samples and intact lung tissue according to a standard protocol by treating cells with proteinase K at 37°C overnight followed by phenol–chloroform extraction and ethanol precipitation. The DNA was kept at –20°C. After that, the DNA was quantitatively and qualitatively checked by running 0.8% agarose gel electrophoresis and using phage λ DNA (Fermentas–Thermo Fisher Scientific, USA) as a reference standard.
Total RNA was isolated from tumor samples and histologically intact lung tissues using a guanidinium thiocyanate–phenol–chloroform extraction protocol [24]. Before use, RNA samples were treated with RNase-free DNase. Aqueous RNA solution was kept at –40°C.
cDNA was synthesized using total messenger RNA, Moloney Murine Leukemia Virus Reverse Transcriptase (M-MuLV Reverse Transcriptase; Fermentas–Thermo Fisher Scientific), and random heptamers as primers according to the manufacturer’s protocol.
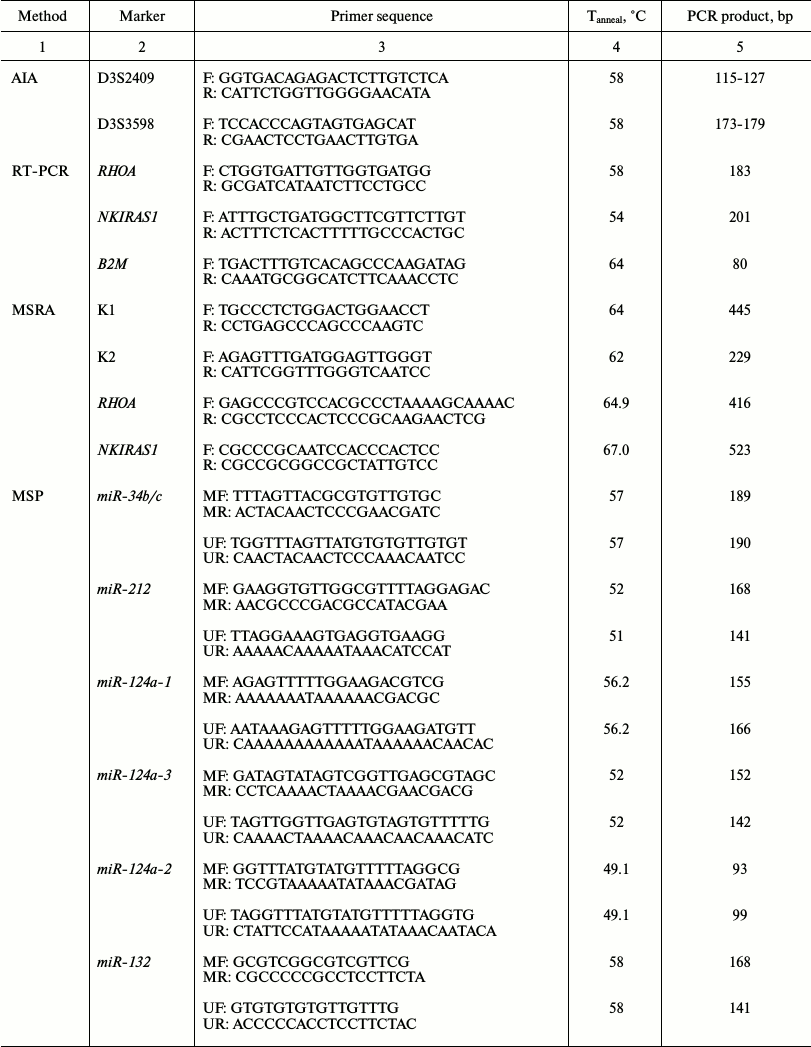
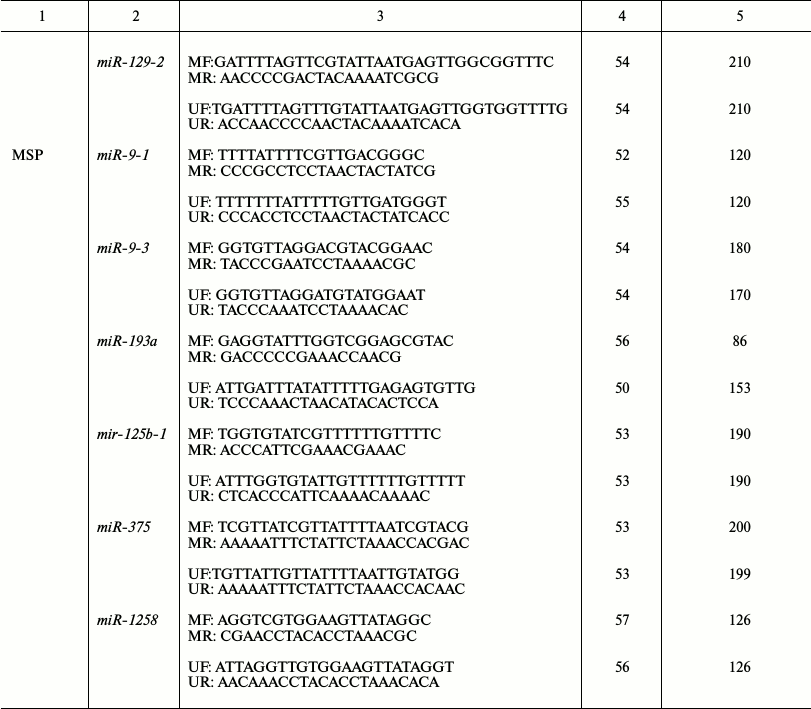
Amounts of RHOA and NKIRAS1 mRNAs were measured by semiquantitative RT-PCR in paired samples from the 35 patients with NSCLC. Primer sequences, annealing temperature (Tanneal), and size of PCR products are shown in the table. PCR was performed in 20 µl of reaction mixture containing 67 mM Tris-HCl, pH 8.8, 16.7 mM (NH4)2SO4, 0.01% Tween-20, supplemented with 0.2 mM of each dNTP, 0.2 µM of each primer, 2 µl of cDNA, and 1 U of recombinant thermostable Taq DNA polymerase (Fermentas–Thermo Fisher Scientific). The amplification reaction was performed as follows: 95°C for 5 min; 35 cycles (94°C for 15 sec, Tanneal (see table) for 25 sec, 72°C for 45 sec), and finally 72°C for 2 min using a DNA Engine Dyad Cycler (Bio-Rad, USA). Products of the PCR were analyzed by running 2% agarose gel supplemented with 0.5 µg/ml ethidium bromide. Then, gel snapshots were taken in transmitted UV-light using the Gel Imager-2 gel recording system (Helicon, Russia) followed by evaluation with Gel-Analysis software. Five-fold or more alterations of mRNA level in tumor samples in comparison with intact lung tissue were scored.
Primer sequence, annealing temperature (Tanneal), and size of
PCR products
Note: AIA for D3S2409 and D3S3598 markers was carried out by using
primer sequence and PCR conditions found in the GenBank Amplicon
database. Primer sequence used in RT-PCR for RHOA and in MSP for
miR-9-1, -9-3, -34b/c, -193a,
-124a-1/-2/-3, -129-2, -125b-1 were published
elsewhere [25-29]. Primer
sequence used in RT-PCR for B2M and NKIRAS1, in MSRA for
NKIRAS1 and RHOA, and in MSP for miR-212,
-375, and -1258 were earlier designed by using Primer Select
software from Lasergene7 suite software [20, 21, 30-32].
Primer sequence used in MSP for miR-132 was designed during the
current study. Oligonucleotide primers were synthesized and purchased
from Eurogene (Russia).
Methylation of promoter regions within RHOA and NKIRAS1 genes was analyzed by using methylation-sensitive HpaII (CCGG) and HhaI (GCGC) restriction enzymes (Fermentas–Thermo Fisher Scientific) according to the manufacturer’s protocol followed by PCR as published earlier [21]. Primer sequences, annealing temperature (Tanneal), and size of PCR products are presented in table. Reaction buffer contained 60 mM Tris-HCl, pH 8.5, 10 mM 2-mercaptoethanol, 25 mM KCl, and 0.1% Triton X-100. Recombinant thermostable Taq DNA polymerase (Fermentas–Thermo Fisher Scientific) was used together with dATP, dCTP, dGTP, and dTTP (SibEnzyme, Russia) during the amplification reaction. Completeness of DNA hydrolysis was assessed using a PCR fragment from the β-3A-adaptin gene (K1, 445 bp; ID GenBank AF247736.2; table) that contains regions for recognition by restriction enzymes used here with unmethylated CpG-dinucleotides (for normal and tumor tissues). DNA integrity before and after hydrolysis was evaluated using the RARB2 gene fragment that does not contain sites for these restriction enzymes (K2, 229 bp; table). Fragments of examined loci were amplified as follows: 95°C for 2 min; 35 cycles (92°C for 10 sec, Tanneal (see table) for 25 sec, 72°C for 25 sec), and finally 72°C for 3 min using the DNA Engine Dyad Cycler. PCR products from test and control samples were separated simultaneously by running 10% PAGE.
Bisulfite conversion of DNA and methylation-specific PCR (MSP) were applied to analyze methylation within 13 miRNA genes in paired samples from patients with NSCLC as published before [20]. After treatment with bisulfite, DNA was purified using a Centrifugal Filter Microcon Unit, Ultracel Discs YM-30 (Millipore, USA). Methylation for each miRNA gene was analyzed using two pairs of primers specific to methylated and unmethylated allele (table). Primer sequences, annealing temperature (Tanneal), and size of PCR products for 13 miRNA genes are given in the table. Fragments of examined miRNA genes were amplified as follows: 94°C for 3 min; 35 cycles (94°C for 10 sec, Tanneal (table) for 20 sec, 72°C for 30 sec); and finally 72°C for 3 min using the DNA Engine Dyad Cycler. Lack of PCR product for unconverted DNA was checked for each pair of primers. PCR products from test and control gene fragments were separated simultaneously by running 10% PAGE.
Allelic imbalance analysis (AIA) for D3S2409 and D3S3598 polymorphic markers in tumor DNA. Primer sequence, PCR conditions, and PCR products were taken from the GenBank Amplicon Database (table). The reaction buffer contained 60 mM Tris-HCl, pH 8.5, 10 mM 2-mercaptoethanol, 25 mM KCl, and 0.1% Triton X-100. PCR was performed as reported earlier [11, 16] by using the DNA Engine Dyad Cycler. PCR products were separated by running 10% PAGE followed by taking a snapshot in transmitted UV-light using the Gel Imager-2 gel recording system and further analyzed with the Gel-Analysis software.
Statistical analysis of data sets was performed by applying Fisher’s exact test. Significance level was set at 0.05. Concordance data for methylation and expression of RHOA and NKIRAS1 genes as well as correspondence of methylation data for sets of miRNA genes to expression of RHOA and NKIRAS1 target genes was assessed using the nonparametric Spearman rank correlation test and Student’s t-test:
with the number of degrees of freedom ν = N – 2, where N is sample size. Significance level was set at 10–6.
RESULTS
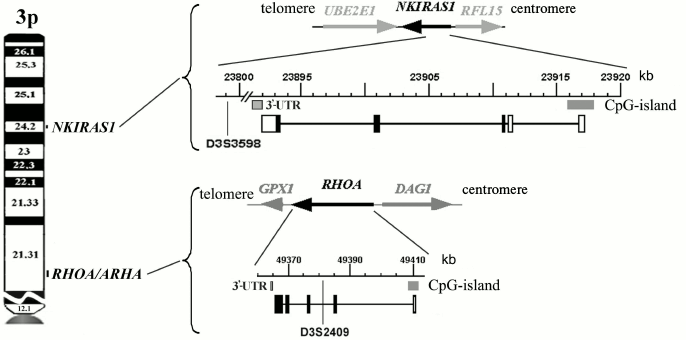
The location as well as structural and functional organization of RHOA and NKIRAS1 genes on the 3p are depicted in Fig. 1 showing that both genes have CpG-islands close to transcription start site and 3′-untranslated regions able to bind a number of miRNAs according to the miRWalk database [8].
Fig. 1. Genomic organization of the 3p RHOA and NKIRAS1 genes. Exon-intron structure, direction of transcription, position of CpG-islands, and 3′-untranslated regions (3′-UTR) involved in binding of regulatory miRNAs are depicted (NCBI, Release 106).
Elevated tumor expression of RHOA and NKIRAS1 genes in patients with NSCLC is associated with demethylation inside their promoter regions and amplification. By analyzing 35 paired NSCLC samples, it was found that the expression level for mRNA of RHOA and NKIRAS1 was significantly elevated (by ≥5-fold). In particular, ≥5-fold increase in expression of the NKIRAS1 gene was found in 21 out of 35 tumor samples compared to histologically intact lung tissue, whereas its decrease was observed only in 2 of 35 cases (Fisher’s exact test, P = 1.6·10–6). Moreover, this difference was even more significant for the RHOA gene (24/35 vs. 2/35; Fisher’s exact test, P = 4.56·10–8). Dominance of elevated expression for such genes in tumors of patients with NSCLC suggests oncogenic functions for RHOA and NKIRAS1 in this type of cancer.
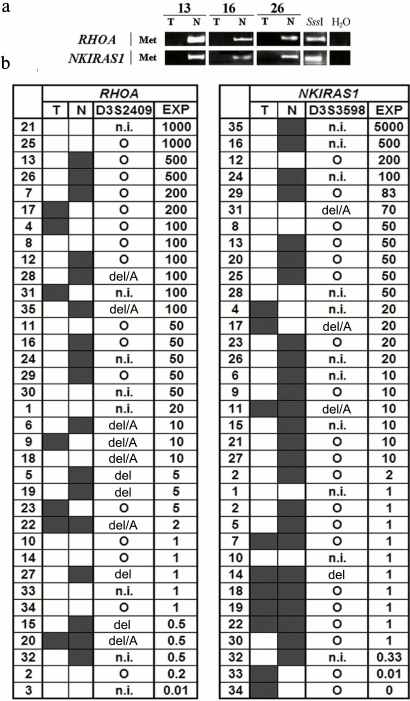
To evaluate the impact of genetic and epigenetic factors on altering expression of the above mentioned genes in NSCLC, the methylation status in promoter CpG-islands within RHOA and NKIRAS1 as well as allelic imbalances in D3S2409 (intron 2 of RHOA gene) and D3S3598 (located 92 kb away from the NKIRAS1 gene) were examined. Typical methylation patterns for these genes revealed in three paired samples are depicted on electrophoregrams (Fig. 2a), where intense methylation is evident in histologically intact lung tissue, but not in tumor samples.
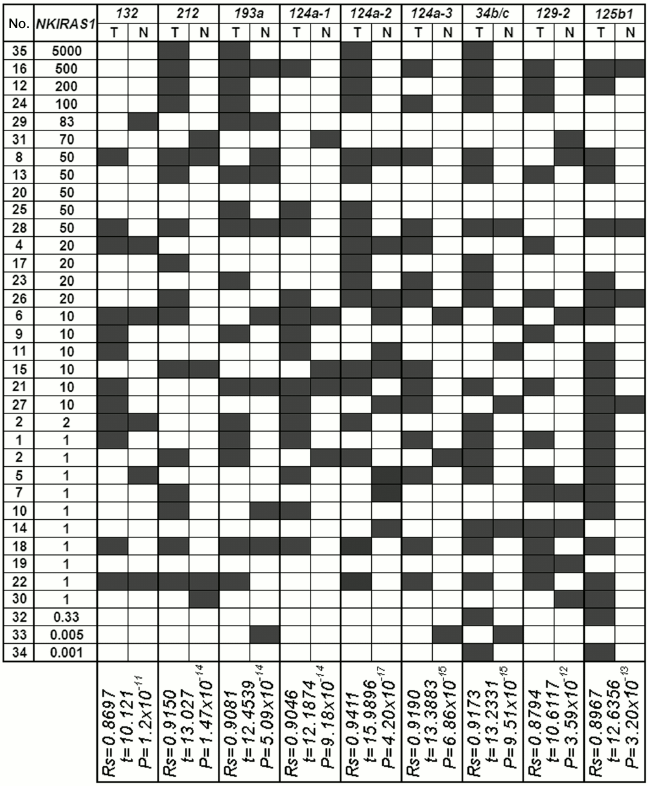
Fig. 2. a) Representative examples of MSRA (methylation-sensitive restriction enzyme analysis) amplified products for the RHOA and NKIRAS genes in three paired samples of NSCLC (13, 16, 26) are shown; T, tumor; N, relatively normal tissue; H2O, PCR in the absence of DNA; SssI, DNA sample from human fibroblast cell line L-68 treated by methyltransferase SssI (SibEnzyme, Russia). MSRA products were separated by running 3% agarose gel electrophoresis. PCR conditions and size of amplified fragments are presented in the table. b) Comparison of altered expression for RHOA and NKIRAS1 genes in tumors from patients with NSCLC having structural changes (methylation/demethylation and amplifications/deletions) in genes. The data are presented according to descending mRNA level of gene (EXP, right) in tumor (T) vs. histologically intact lung tissue (N). Left: results on evaluating gene methylation status by MSRA in samples of NSCLC (T/N) are shown. Center: AIA data for D3S2409 and D3S3598 markers are presented (see Fig. 1). Black rectangle, methylation (MSP product) found; open rectangle, methylation not found; A, amplification; del, deletion; del/A, deletion of one allele and amplification of the second allele; circle, preserved heterozygosity; n.i., non-informative case. Intensity of bands for RT-PCR products of RHOA and NKIRAS1 genes were normalized relative to intensity of the band for RT-PCR product of the B2M gene. PCR conditions and size of amplified fragments are presented in the table.
Data on altered mRNA level in tumors from 35 patients with NSCLC, on DNA methylation in tumors and paired histologically intact lung tissue as well as on allelic changes in adjacent polymorphic loci are summarized in Fig. 2b. It is obvious that elevated expression in tumors is usually associated with demethylation within the gene promoter region. In particular, for the RHOA gene in 24 NSCLC samples with elevated mRNA level (≥5-fold) methylation was only found in 12 samples (50%) of histologically intact lung tissue, while it was absent in paired tumor samples. As for the NKIRAS1 gene, demethylation was detected in 15 of 21 samples with elevated expression (71%). In two cases (samples No. 33 and 34) decline in NKIRAS1 expression is associated with gene methylation in tumor DNA.
Amplification or deletion of loci (according to the data for D3S2409 and D3S3598 markers) that overlap RHOA or NKIRAS1 can additionally contribute to altering expression of these genes. For instance, amplification of the second allele can facilitate elevated expression of RHOA in sample No. 18 and NKIRAS1 in sample No. 31 (Fig. 2b). There are examples when both methylation and amplification are associated with upregulated level of the RHOA gene (samples Nos. 28, 35, and 6). Using the nonparametric Spearman rank correlation test, it was found that altered mRNA level significantly correlated with genetic and epigenetic changes in the genes. In particular, for RHOA: Rs = 0.8629, t = 9.8079, p = 2.63·10–11; for NKIRAS1: Rs = 0.9103, t = 12.6322, p = 3.45·10–14.
However, there are many cases when changes at the genomic level cannot account for their upregulation during NSCLC (Fig. 2b). In particular, it was shown that 20-, 50-, and 1000-fold upregulation of mRNA for RHOA gene (sample Nos. 1, 30, 11, 8, 25, and 21) similar to 50- and 200-fold increase in mRNA level for NKIRAS1 (sample Nos. 28, 8, and 12) are not linked either to demethylation or their amplification.
Methylation in a set of miRNA genes is involved in activating expression of RHOA and NKIRAS1 genes in tumors of patients with NSCLC. Another pathway used to regulate genes with oncogenic function might be related to the impact from suppressor miRNAs. However, synthesis of miRNA genes in tumors can be inhibited via the same mechanism, i.e. methylation of promotor CpG-island, thus depriving a particular miRNA from participating in suppression of its target gene.
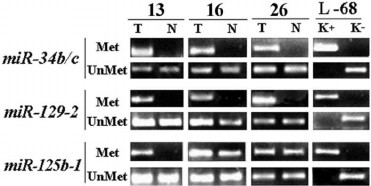
We examined methylation of promoter regions within 13 miRNA genes predicted to regulate the RHOA and/or NKIRAS1 genes (according to the miRWalk database [8]). As an example, a pattern of MSP products for three miRNAs (miR-34, -129, -125b) able to bind to 3′-untranslated region of mRNA for both the RHOA and NKIRAS1 genes is presented (Fig. 3). As for the miR-34b/c and miR-129-2 genes, PCR products with primers to methylated allele were observed in all three examined tumor samples in contrast to relatively normal lung tissues. For the miR-125b gene, a clear-cut difference between tumor and relatively normal lung tissues was evident in sample No. 13; however, PCR products with primers to methylated allele were detected in samples Nos. 16 and 26 in both tumor and histologically intact tissues. However, in sample No. 16 luminous intensity of product specific to methylated allele was found to be several times brighter in tumor than in adjacent histologically intact tissue.
Fig. 3. Representative examples of MSP product amplification for the miR-34b/c, miR-129-2, and miR-125b-1 genes in three paired samples of NSCLC (13, 16, 26) are shown. T, tumor; N, relatively normal tissue. MSP products with primers to methylated (Met) and unmethylated (UnMet) alleles are shown. L-68/K–, DNA sample from intact human fibroblast cell line L-68; L-68/K+, DNA sample from human fibroblast cell line L-68 treated by methyltransferase SssI (SibEnzyme, Russia). MSP products were separated by running 3% agarose gel electrophoresis. PCR conditions and size of amplified fragments are presented in the table.
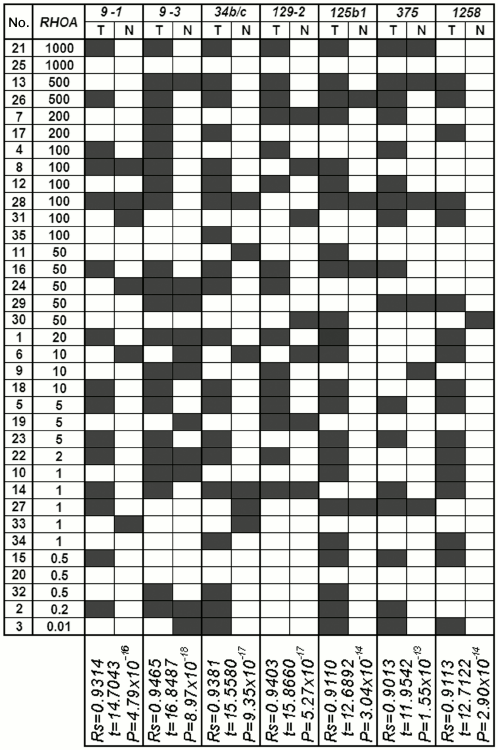
The results from our study regarding changes in level of mRNAs for the RHOA and NKIRAS1 genes were compared with the results of analyzed methylation in a group of miRNA genes with predicted regulatory functions (Figs. 4 and 5) [8]. A significant correlation between altered level of RHOA mRNA and methylation of seven genes was found for miR-9-1/-3, -34b/c, -129-2, -125b-1, -375b, -1258 (Spearman rank correlation test, P = 10–13-10–18; Fig. 4). Changes in the level of NKIRAS1 mRNA were related to methylation of nine genes such as miR-132, -212, -193a, -124a-1/-2/-3, -34b/c, -129-2, -125b-1 (Spearman rank correlation test, P = 10–11-10–17; Fig. 5).
Fig. 4. Comparison of data showing alterations (top-down) in mRNA level of RHOA in tumors from patients with NSCLC vs. data on altered miRNA gene methylation (miR-9-1, miR-9-3, miR-34b/c, miR-129-2, miR-125b-1, miR-375, miR-1258) in samples from tumors (T) and histologically intact lung tissue (N). Black rectangle, methylation (MSP product) found; open rectangle, methylation not found. By calculating Spearman’s rank correlation coefficient, a coincidence was considered for: (i) miRNA gene methylation in DNA from tumor found upon elevated expression level of potential target gene; (ii) miRNA gene demethylation in DNA from tumor found upon decreased expression level of potential target gene; (iii) lack of altered methylation and expression.
Fig. 5. Comparison of the data showing alterations (top-down) in mRNA level of NKIRAS1 in tumors from patients with NSCLC vs. data on altered miRNA gene methylation (miR-132, miR-212, miR-193a, miR-124a-1, miR-124a-2, miR-124a-3, miR-34b/c, miR-129-2, miR-125b-1) in samples from tumors (T) and histologically intact lung tissue (N). For comments, see legend to Fig. 4.
These results suggest that methylation of such miRNA genes inhibits their own expression and deprives them from capacity to suppress predicted target genes such as RHOA and/or NKIRAS1. Thus, in tumors of patients with NSCLC alterations in expression of oncogenic RHOA and NKIRAS1 genes were found that might result in both methylation of promoter regions within the genes as well as methylation of regulatory miRNAs. Analysis of allelic alterations in nearby polymorphic markers revealed cases of amplified RHOA and/or NKIRAS1 genes that also might result in elevated amount of mRNA gene.
DISCUSSION
Here we examined two levels of regulation for expression of the RHOA and NKIRAS1 genes in lung tumors. It was demonstrated that expression of these genes in tumors of patients with NSCLC might be altered at the genomic level via methylation/demethylation within promoter regions of these genes as well as due to amplification of loci that overlap them. We found that altered mRNA level significantly correlated with genetic and epigenetic changes in genes, particularly, in terms of demethylation and amplification impact on upregulated expression of the RHOA and NKIRAS1 genes. The results regarding elevated expression, demethylation, and amplification of the RHOA and NKIRAS1 genes in tumors of patients with NSCLC suggest their oncogenic functions in NSCLC. This is in agreement with numerous findings confirming oncogenic function for RHOA [15-17] and data about homology of NKIRAS1 with RAS1 oncogene as well as interaction of NKIRAS1 with NF-κB transcription factor [18, 19].
During the last decade, it has become evident that expression of protein-coding genes can be regulated by activity of miRNA that binds to 3′-untranslated region of mRNA and usually inhibits gene expression at posttranscriptional level. Moreover, miRNAs might be involved in regulation of more than half of protein-coding genes [8]. However, miRNA genes, especially suppressor miRNA genes (i.e. those that inhibit oncogenes), are also susceptible to epigenetic inactivation in tumors, even at higher rate than protein-coding genes [33]. Inactivation of suppressor miRNA genes associated with methylation makes them unable to inhibit target genes. By analyzing the main core of publications available in PubMed (http://www.ncbi.nlm.nih.gov/pubmed/) we found that as many as 90 miRNAs have been experimentally shown to be influenced by DNA methylation or demethylation on their expression in any given tumor type. A link between gene methylation and its expression was established for the majority of 13 miRNA genes examined here. For example, a role for methylation being involved in inactivation of miR-9-3, -124-1/-2/-3, -125b, -34b/c, -129-2, -193a, etc. genes in tumors of different locations was revealed [26-28, 34, 35]. Previously, we also found a link between methylation and amount of pri-miRNA miR-9-1 in NSCLC [20] and identified novel miRNA genes being methylated in tumors of patients with NSCLC, such as miR-125b-1, miR-137, miR-375, and miR-1258, which play an important role in cellular signaling pathways and pathogenesis of NSCLC [32].
In our study, we demonstrated that in NSCLC upregulated mRNA level for RHOA and NKIRAS1 is associated with methylation of seven and nine miRNA genes, respectively (miR-9-1/-3, -34b/c, -129-2, -125b-1, -375, -1258 as well as miR-132, -212, -193a, -124a-1/-2/-3, -34b/c, -129-2, -125b-1, respectively; Spearman rank correlation test, P < 10–11). Thereby, we determined novel potential regulatory miRNAs for RHOA (miR-9-1/-3, -34b/c, -129-2, -125b-1, -375, -1258) and NKIRAS1 (miR-34b/c, -129-2, -125b-1, -193a, -124a-1/-2/-3, -212, -132) proto-oncogenes in lung cancer. On the other hand, we show for the first time that RHOA and NKIRAS1 may be novel potential target genes for a number of miRNAs susceptible to methylation in NSCLC.
It is not surprising that among targets for examined suppressor miRNAs, mRNAs of typical oncogenic proteins were frequent. For example, miR-34b/c targets mRNA of MYC, CDK4, CDK6, E2F3, CREB, and MET [26, 36] whereas miR-193a targets well-known oncogenes such as K-ras, C-kit, and ERBB4 [28, 34, 37]. Later, it was demonstrated that methylation-associated inactivation of the miR-129-2 gene enhances expression of SOX4 oncogene linked to metastases [38]. The Sox5 oncogene and activator of epithelial–mesenchymal transition ZEB2 as well as CCNE1 (cyclin E1) are considered as targets for miR-132 [39-41]. Notably, miR-375 that was supposed to carry a specific function in regulating insulin expression turned out to be a regulator of several important oncogenic targets (AEG-1, YAP1, IGF1R, and PDK1) in many types of cancer [42].
According to our results as well as data published elsewhere, RHOA and NKIRAS1 can also be referred to oncogenes and genes of tumor progression. According to the miRWalk database [8], RHOA and NKIRAS1 can theoretically function as target genes for 13 miRNAs examined here; however, experimental data confirming this have not yet been obtained. In addition, data regarding regulatory miRNAs for poorly studied NKIRAS1 protooncogene are not available at all in peer-reviewed journals, and they are very limited for the RHOA oncogene. So far, it is known that RHOA mRNA is a target for miR-122, so this interacting pair plays an important role in invasion of hepatocellular carcinoma and epithelial–mesenchymal transition [43].
Thus, here we determined novel potential target genes and novel potential regulatory miRNAs involved in pathogenesis of NSCLC. We demonstrated the possibility of methylation of regulatory miRNA genes to influence indirectly activation of target genes in lung tumors. A systemic role of methylation in regulation of genes as well as their interactions was found.
This study was supported by the Russian Foundation for Basic Research (grant No. 13-04-00828).
REFERENCES
1.Davydov, M. I., and Aksel’, E. M. (2012)
Statistics of malignant neoplasms in Russia and CIS countries in 2010,
Vestnik RONTs im. N. N. Blokhina RAMN, 22, 54-61.
2.ENCODE Consortium (2012) Architecture of the human
regulatory network derived from ENCODE data, Nature, 489,
91-100.
3.ENCODE Consortium (2012) Landscape of transcription
in human cells, Nature, 489, 101-108.
4.Jones, P. A., and Baylin, S. B. (2007) The
epigenetics of cancer, Cell, 128, 683-692.
5.Jones, P. A. (2012) Functions of DNA methylation:
islands, start sites, gene bodies and beyond, Nat. Rev. Genet.,
13, 484-492.
6.Heller, G., Zielinski, C. C., and Zochbauer-Muller,
S. (2010) Lung cancer: from single-gene methylation to methylome
profiling, Cancer Metastasis Rev., 29, 95-107.
7.Sato, F., Tsuchiya, S., Meltzer, S. J., and
Shimizu, K. (2011) MicroRNAs and epigenetics, FEBS J.,
278, 1598-1609.
8.Dweep, H., Sticht, C., Pandey, P., and Gretz, N.
(2011) miRWalk-database: prediction of possible miRNA binding sites by
“walking” the genes of three genomes, J. Biomed.
Inform., 44, 839-847.
9.Dreijerink, K., Braga, E., Kuzmin, I., Geil, L.,
Duh, F.-M., Angeloni, D., Zbar, B., Lerman, M. I., Stanbridge, E. J.,
Minna, J. D., Protopopov, A., Li, J., Kashuba, V., Klein, G., and
Zabarovsky, E. (2001) The candidate tumor suppressor gene,
RASSF1A, from human chromosome 3p21.3 is involved in kidney
tumorigenesis, Proc. Natl. Acad. Sci. USA, 98,
7504-7509.
10.Zabarovsky, E. R., Senchenko, V., Loginov, V.,
Pavlova, T., Zabarovska, V., Dmitriev, A., Lung, M., Panda, C. K.,
Kashuba, V., Lerman, M. I., and Braga, E. A. (2011) Positional cloning
of tumor suppressor genes from 3p21.3 involved in major human cancers,
Horizons Cancer Res., 42, 103-127.
11.Braga, E., Senchenko, V., Bazov, I., Loginov, W.,
Ermilova, V., Kazubskaya, T., Garkavtseva, R., Mazurenko, N.,
Kisseljov, F., Liu, J., Kisselev, L., Lerman, M., Klein, G., and
Zabarovsky, E. (2002) Critical tumor-suppressor gene regions on
chromosome 3p in major human epithelial malignancies: allelotyping and
quantitative real time PCR, Int. J. Cancer, 100,
534-541.
12.Loginov, V. I., Bazov, V. I., Khodyrev, D. S.,
Pronina, I. V., Kazubskaya, T. P., Ermilova, V. D., Gar’kavtseva,
R. F., Zabarovskiy, E. R., and Braga, E. A. (2008) Regions of potential
suppressor genes in epithelial tumors of kidneys, mammary gland and
ovaries located on human chromosome 3, Genetika, 44,
250-256.
13.Braga, E., Loginov, W., Khodyrev, D., Pronina,
I., Kazubskaya, T., Bogatyrova, O., Kashuba, V. I., Senchenko, V. N.,
Klein, G., Lerman, M. I., Kisselev, L. L., and Zabarovsky, E. R. (2011)
A novel MECA3 region in human 3p21.3 harboring putative tumor
suppressor genes and oncogenes, Exp. Oncol., 33,
33-41.
14.Angeloni, D., Danilkovitch-Miagkova, A., Ivanova,
T., Braga, E., Zabarovsky, E., and Lerman, M. I. (2007)
Hypermethylation of Ron proximal promoter associates with lack of
full-length Ron and transcription of oncogenic short-Ron from an
internal promoter, Oncogene, 26, 4499-4512.
15.Pille, J. Y., Denoyelle, C., Varet, J., Bertrand,
J. R., Soria, J., Opolon, P., Lu, H., Pritchard, L. L., Vannier, J. P.,
Malvy, C., Soria, C., and Li, H. (2005) Anti-RhoA and anti-RhoC siRNAs
inhibit the proliferation and invasiveness of MDA-MB-231 breast cancer
cells in vitro and in vivo, Mol. Ther.,
11, 267-274.
16.Braga, E. A., Loginov, V. I., Klimov, E. A.,
Kilosanidze, G., Khodyrev, D. S., Kaganova, N. L., Kazubskaya, T. P.,
Ermilova, V. D., Gar’kavtseva, R. F., Pronina, I. V.,
Rud’ko, O. I., Zabarovskiy, E. R., Sulimova, G. E., and Kiselev,
L. L. (2006) Activation of RHOA gene transcription in epithelial
tumors maybe caused by amplification of gene copies and/or
demethylation of its promoter region, Mol. Biol. (Moscow),
40, 865-877.
17.Ma, L., Liu, Y. P., Geng, C. Z., Wang, X. L.,
Wang, Y. J., and Zhang, X. H. (2010) Overexpression of Rhoa is
associated with progression in invasive breast duct carcinoma,
Breast J., 16, 105-107.
18.Fenwick, C., Na, S. Y., Voll, R. E., Zhong, H.,
Im, S. Y., Lee, J. W., and Ghosh, S. (2000) A subclass of Ras proteins
that regulate the degradation of ΙκΒ, Science,
287, 869-873.
19.Chen, Y., Vallee, S., Wu, J., Vu, D., Sondek, J.,
and Ghosh, G. (2004) Inhibition of NF-κB activity by
IκBbeta in association with κB-Ras, Mol. Cell.
Biol., 24, 3048-3056.
20.Khodyrev, D. S., Pronina, I. V., Rykov, S. V.,
Beresneva, E. V., Fridman, M. V., Kazubskaya, T. P., Loginov, V. I.,
and Braga, E. A. (2012) Methylation of miRNA gene group is involved in
regulating expression of RAR-beta2 and NKIRAS1 target
genes upon lung cancer, Mol. Biol. (Moscow), 46,
773-785.
21.Loginov, V. I., Pronina, I. V., Burdennyy, A. M.,
Khodyrev, D. S., Kazubskaya, T. P., Braga, E. A., Kubatiev, A. A., and
Kushlinskiy, N. E. (2014) A role of methylation in regulating
expression of functionally significant genes on chromosome 3:
RHOA, GPX1, USP4, DAG1,
NKIRAS1 – in breast cancer, Mol. Med. (Moscow),
6, 30-37.
22.Sobin, L. Y., and Wittekind, Ch. N. Y. (2002)
UICC TNM Classification of Malignant Tumors, Wiley-Liss Inc.,
pp. 193-195.
23.Travis, W. D., Coby, T. V., Corrin, B.,
Shimosato, Y., and Brambilla, E. (1999) World Health Organization
International Histological Classification of Tumors; Histological
Typing of Lung and Pleural Tumors, Springer, Berlin.
24.Pronina, I. V., Loginov, V. I., Prasolov, V. S.,
Klimov, E. A., Khodyrev, D. S., Kazubskaya, T. P., Gar’kavtseva,
R. F., Sulimova, G. E., and Braga, E. A. (2009) Alteration of SEMA3B
gene expression levels in epithelial tumors, Mol. Biol. (Moscow), 43,
439-445.
25.Horiuchi, A., Imai, T., Wang, C., Ohira, S.,
Feng, Y., Nikaido, T., and Konishi, I. (2003) Up-regulation of small
GTPases, RhoA and RhoC, is associated with tumor progression in ovarian
carcinoma, Lab. Invest., 83, 861-870.
26.Lujambio, A., Calin, G. A., Villanueva, A.,
Ropero, S., Sanchez-Cespedes, M., Blanco, D., Montuenga, L. M., Rossi,
S., Nicoloso, M. S., Faller, W. J., Gallagher, W. M., Eccles, S. A.,
Croce, C. M., and Esteller, M. A. (2008) MicroRNA DNA methylation
signature for human cancer metastasis, Proc. Natl. Acad. Sci.
USA, 105, 13556-13561.
27.Ando, T., Yoshida, T., Enomoto, S., Asada, K.,
Tatematsu, M., Ichinose, M., Sugiyama, T., and Ushijima, T. (2009) DNA
methylation of microRNA genes in gastric mucosae of gastric cancer
patients: its possible involvement in the formation of epigenetic field
defect, Int. J. Cancer, 124, 2367-2374.
28.Bandres, E., Agirre, X., Bitarte, N., Ramirez,
N., Zarate, R., Roman-Gomez, J., Prosper, F., and Garcia-Foncillas, J.
(2009) Epigenetic regulation of microRNA expression in colorectal
cancer, Int. J. Cancer, 125, 2737-2743.
29.Soto-Reyes, E., Gonzalez-Barrios, R.,
Cisneros-Soberanis, F., Herrera-Goepfert, R., Perez, V., Cantu, D.,
Prada, D., Castro, C., Recillas-Targa, F., and Herrera, L. A. (2012)
Disruption of CTCF at the miR-125b1 locus in gynecological cancers,
BMC Cancer, 12, 40.
30.Angeloni, D., ter Elst, A., Wei, M. H., van der
Veen, A. Y., Braga, E. A., Klimov, E. A., Timmer, T., Korobeinikova,
L., Lerman, M. I., and Buys, C. H. (2006) Analysis of a new homozygous
deletion in the tumor suppressor region at 3p12.3 reveals two novel
intronic noncoding RNA genes, Genes Chromosomes Cancer,
45, 676-691.
31.Beresneva, E. V., Rykov, S. V., Khodyrev, D. S.
Pronina, I. V., Ermilova, V. D., Kazubskaya, T. P., Braga, E. A., and
Loginov, V. I. (2013) Methylation profile of group of miRNA genes in
clear cell renal cell carcinoma; involvement in cancer progression,
Genetika, 49, 366-375.
32.Rykov, S. V., Khodyrev, D. S., Pronina, I. V.,
Kazubskaya, T. P., Loginov, V. I., and Braga, E. A. (2013) Novel miRNA
genes methylated in lung tumors, Genetika, 49, 896-901.
33.Kunej, T., Godnic, I., Ferdin, J., Horvat, S.,
Dovc, P., and Calin, G. A. (2011) Epigenetic regulation of microRNAs in
cancer: an integrated review of literature, Mutat. Res.,
717, 77-84.
34.Gao, X. N., Lin, J., Li, Y. H., Gao, L., Wang, X.
R., Wang, W., Kang, H. Y., Yan, G. T., Wang, L. L., and Yu, L. (2011)
MicroRNA-193a represses c-kit expression and functions as a
methylation-silenced tumor suppressor in acute myeloid leukemia,
Oncogene, 30, 3416-3428.
35.Heller, G., Weinzierl, M., Noll, C.,
Babinsky, V., Ziegler, B., Altenberger, C., Minichsdorfer, C.,
Lang, G., Dome, B., End-Pfutzenreuter, A., Arns, B. M., Grin, Y.,
Klepetko, W., Zielinski, C. C., and Zochbauer-Muller, S. (2012)
Genome-wide miRNA expression profiling identifies miR-9-3 and miR-193a
as targets for DNA methylation in non-small cell lung cancers, Clin.
Cancer Res., 18, 1619-1629.
36.Cannell, I. G., and Bushell, M. (2010) Regulation
of Myc by miR-34c: a mechanism to prevent genomic instability, Cell
Cycle, 9, 2726-2730.
37.Yu, T., Li, J., Yan, M., Liu, L., Lin, H., Zhao,
F., Sun, L., Zhang, Y., Cui, Y., Zhang, F., Li, J., He, X., and Yao, M.
(2014) MicroRNA-193a-3p and -5p suppress the metastasis of human
non-small-cell lung cancer by downregulating the ERBB4/PIK3R3/mTOR/S6K2
signaling pathway, Oncogene, DOI: 10.1038/onc.2013.574.
38.Chen, X., Zhang, L., Zhang, T., Hao, M., Zhang,
X., Zhang, J., Xie, Q., Wang, Y., Guo, M., Zhuang, H., and Lu, F.
(2013) Methylation-mediated repression of microRNA 129-2 enhances
oncogenic SOX4 expression in HCC, Liver Int., 33,
476-486.
39.Renjie, W., and Haiqian, L. (2014) MiR-132,
miR-15a and miR-16 synergistically inhibit pituitary tumor cell
proliferation, invasion and migration by targeting Sox5, Cancer Lett.,
S0304-3835(14)00575-8; DOI: 10.1016/j.canlet.2014.10.003.
40.You, J., Li, Y., Fang, N., Liu, B., Zu, L.,
Chang, R., Li, X., and Zhou, Q. (2014) MiR-132 suppresses the migration
and invasion of lung cancer cells via targeting the EMT regulator ZEB2,
PLoS One, 9, e91827.
41.Wang, J., Xu, G., Shen, F., and Kang, Y. (2014)
miR-132 targeting cyclin E1 suppresses cell proliferation in
osteosarcoma cells, Tumour Biol., 35, 4859-4865.
42.Yan, J. W., Lin, J. S., and He, X. X. (2014) The
emerging role of miR-375 in cancer, Int. J. Cancer, 135,
1011-1018.
43.Wang, S. C., Lin, X. L., Li, J., Zhang, T. T.,
Wang, H. Y., Shi, J. W., Yang, S., Zhao, W. T., Xie, R. Y., Wei, F.,
Qin, Y. J., Chen, L., Yang, J., Yao, K. T., and Xiao, D. (2014)
MicroRNA-122 triggers mesenchymal-epithelial transition and suppresses
hepatocellular carcinoma cell motility and invasion by targeting RhoA,
PLoS One, 9, e101330.