REVIEW: Alternative Oxidase: Distribution, Induction, Properties, Structure, Regulation, and Functions
A. G. Rogov1*, E. I. Sukhanova1, L. A. Uralskaya1, D. A. Aliverdieva2, and R. A. Zvyagilskaya1
1Bach Institute of Biochemistry, Russian Academy of Sciences, Leninsky pr. 33, 119071 Moscow, Russia; E-mail: lloss@rambler.ru2Caspian Institute of Biological Resources, Dagestan Scientific Center, Russian Academy of Sciences, ul. M. Gadzhieva 45, 367025 Makhachkala, Russia; E-mail: pibrdncran@mail.ru
* To whom correspondence should be addressed.
Received September 9, 2014
The respiratory chain in the majority of organisms with aerobic type metabolism features the concomitant existence of the phosphorylating cytochrome pathway and the cyanide- and antimycin A-insensitive oxidative route comprising a so-called alternative oxidase (AOX) as a terminal oxidase. In this review, the history of AOX discovery is described. Considerable evidence is presented that AOX occurs widely in organisms at various levels of organization and is not confined to the plant kingdom. This enzyme has not been found only in Archaea, mammals, some yeasts and protists. Bioinformatics research revealed the sequences characteristic of AOX in representatives of various taxonomic groups. Based on multiple alignments of these sequences, a phylogenetic tree was constructed to infer their possible evolution. The ways of AOX activation, as well as regulatory interactions between AOX and the main respiratory chain are described. Data are summarized concerning the properties of AOX and the AOX-encoding genes whose expression is either constitutive or induced by various factors. Information is presented on the structure of AOX, its active center, and the ubiquinone-binding site. The principal functions of AOX are analyzed, including the cases of cell survival, optimization of respiratory metabolism, protection against excess of reactive oxygen species, and adaptation to variable nutrition sources and to biotic and abiotic stress factors. It is emphasized that different AOX functions complement each other in many instances and are not mutually exclusive. Examples are given to demonstrate that AOX is an important tool to overcome the adverse aftereffects of restricted activity of the main respiratory chain in cells and whole animals. This is the first comprehensive review on alternative oxidases of various organisms ranging from yeasts and protists to vascular plants.
KEY WORDS: respiratory chain, alternative oxidase, distribution, induction, properties, biogenesis, genes, structure, regulation, functionsDOI: 10.1134/S0006297914130112
Abbreviations: AOX, alternative oxidase; PTOX, plastid terminal oxidase; ROS, reactive oxygen species.
Mitochondria of all plants, most fungi, algae, and some protists
examined to date contain, in addition to the canonical cytochrome
oxidase of the respiratory chain, a cyanide-insensitive terminal
oxidase called “alternative oxidase” (AOX). The AOX is an
integral protein (32-36 kDa) of the inner mitochondrial membrane. It is
encoded by the nuclear genome, is localized on the matrix side of the
mitochondrial inner membrane, and is known to catalyze the
four-electron oxidation of ubiquinol (reduced form of ubiquinone) by
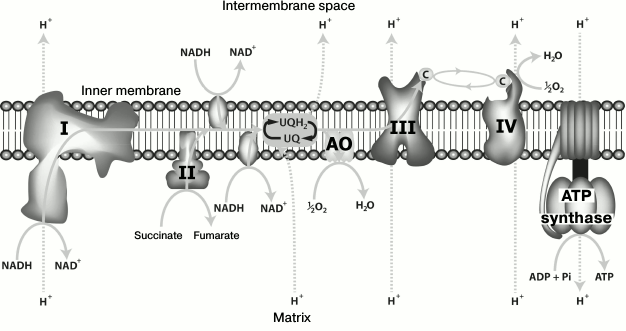
oxygen to water (Fig. 1). Unlike complexes I, III,
and IV of the respiratory chain, where electron transport is
accompanied by translocation of protons across the inner mitochondrial
membrane and by subsequent utilization of the proton gradient for ATP
synthesis, electron transport through AOX is not coupled with ATP
synthesis and energy accumulation; thus, the energy of ubiquinol
oxidation by oxygen is released as heat [1, 2]. Elucidating the structure, properties, and the
especially regulation of AOX is a matter of keen interest because of
the important role of AOX in fundamental biological processes, such as
thermogenesis in thermogenic plant organs, survival of parasitic
protists, adaptation of organisms to numerous biotic and abiotic stress
factors, regulation of photosynthesis in plants, protection of the
photosynthetic apparatus against oxidative destruction, and regulation
of plant–pathogen interactions and the extent of pathogenicity of
pathogenic fungi.
Fig. 1. Structure of the respiratory chain in plants and some fungi.
HISTORY OF THE DISCOVERY OF AOX
The first indications that cells of some algae and fungi retain respiration in the presence of cyanide, an inhibitor of the main mitochondrial respiratory chain, were obtained at the beginning of the last century (reviewed in [3]). Later, cyanide-resistant respiration was also found in plants and some protists. Several hypotheses were proposed to explain its nature. The role of AOX was ascribed tentatively to several candidates, such as a flavoprotein with low affinity to oxygen, the a-type cytochrome, cytochrome b7, and other compounds. The possibility was not excluded that the activity of the cyanide-resistant alternative pathway could be explained by radical reactions involving the main components of the respiratory chain [3]. In 1971, Bendal and Bonner (see [3]) demonstrated the weakness of these assumptions. Moreover, they proved that AOX, located in mitochondria, obtains reducing equivalents from the main respiratory chain at the substrate side of the antimycin A-sensitive locus, and that AOX activity is inhibited by chelating agents that bind variable valence cations, Fe in particular. Based on these findings, the authors proposed that nonheme iron is the most likely candidate for the role of the terminal cyanide-resistant oxidase. The discovery that hydroxamic acids specifically inhibit AOX at concentrations not affecting the activity of the main cytochrome pathway [4], together with the development of sophisticated methods of analysis, promoted rapid advances in this research area. In 1978, a highly purified preparation of AOX was obtained from the active thermogenic tissues of Arum maculatum plants [5]. In 1986-1987, a highly purified AOX was isolated from Sauromatum guttatum thermogenic plants, and its activity was found to correlate with the content of a 35.5-36-kDa protein [6, 7]. Monoclonal antibodies to this protein was obtained [8] and spread quickly around the world. The antibodies were successfully applied to identify AOX in mitochondria of the fungus Neurospora crassa [9].
Two years later [10], cDNA encoding a 38.9-kDa protein was isolated from the DNA library of S. guttatum; this protein was identified as a precursor of one or two AOX forms in this organism. Since then, highly and partially purified AOX preparations were isolated from the alga Chlamydomonas reinhardtii [11], the bloodstream form of Trypanosoma brucei [12], and thermogenic tissue of A. maculatum [13]. The genes encoding AOX have been isolated from soybean [14], tobacco [15], methylotrophic yeast Pichia pastoris [16], and the thermogenic tissue of Symplocarpus renifolius [17]. Since isolation of highly purified AOX samples from plants encounters certain difficulties, recombinant strains were used in some cases to determine the structure, regulation, and the functional role of AOX [18-22]. It became clear that AOX is widespread in nature and apparently is quite conserved.
TAXONOMIC DISTRIBUTION OF AOX
Bioinformatics methods have revealed that AOX is widespread among organisms at different levels of organization and is not confined to the plant kingdom [23]. AOX is not found only in archaea, mammals, some yeast, and protists. In plants, AOX acting as a terminal oxidase was found in all angiosperms examined to date; it was also found in gymnosperms, mosses including Physcomitrella patens, liverworts, Lycopodium, and ferns [24].
AOX has been found in many yeast species including Rhodotorula glutinis [25], Candida lipolytica (now Yarrowia lipolytica) [26, 27], C. parapsilosis [28-30], C. albicans, C. krusei [31], Pichia anomala (Hansenula anomala) [32], P. pastoris [16], P. stipitis [33], Debaryomyces hansenii [34, 35], the yeast Cryptococcus neoformans that is pathogenic for humans [36], and others. It is supposed that AOX is inherent to all yeasts with pronounced aerobic type of metabolism that are incapable of supporting their metabolism by glycolysis alone [37]. AOX is usually found in yeast species where complex I of the respiratory chain is operative. One exception is, perhaps, the yeast Debaryomyces (previously Endomyces) magnusii, where AOX is normally lacking, while complex I is functional at all growth stages. Remarkably, AOX is synthesized in this yeast upon disturbance of basic cytochrome-mediated electron transport [38]. No AOX was found in the yeasts Saccharomyces cerevisiae and Schizosaccharomyces pombe lacking complex I of the respiratory chain [37]. This circumstance is not entirely surprising, because otherwise uncontrolled uncoupling of mitochondria would occur [39, 40]. It was proposed that yeasts adopted aerobic fermentation or AOX-mediated respiration as an alternative to the complete phosphorylating cytochrome respiratory chain. Both strategies confer metabolic flexibility on yeasts, which allows them to respond to changes in growth conditions and ensures their antioxidant protection.
AOX has been found in some fungi, including Ustilago ayclis [41], U. maydis [42], Tapesia acuformis [43], Aspergillus niger [44], A. fumigatus [19], Emericella nidulans [45], Ajellomyces capsulatus [46], Podospora anserina [47, 48], Magnaporthe grisea [49], Phycomyces blakesleeanus [50], and the osmophilic yeast-like fungus Moniliella tomentosa [51].
AOX has been found in a series of pathogens: in phytopathogenic fungi Sclerotinia sclerotiorum ([52] and references therein) and Septoria tritici [13], the insect pathogenic fungus Metarhizium anisopliae [53], the human pathogenic yeast C. neoformans [36], the thermal dimorphic human pathogenic fungus Paracoccidioides brasiliensis [54], fungal spores of the intracellular parasite Antonospora (Paranosema) locustae [55] (this species with reduced metabolism is devoid of canonical mitochondria and the tricarboxylic acid cycle but retains the adenine nucleotide translocase to take up ATP from the host), the hemibiotrophic pathogenic fungus Moniliophthora perniciosa causing cocoa disease [56], and the endemic dimorphic pathogenic fungus Histoplasma capsulatum [57]. The pathogens have acquired the ability to express AOX in host cells; AOX plays an important role in pathogen survival, especially when the host cells are exposed to various stress factors [36].
AOX is present in mitochondria of algae [46, 58-60], drosophila [61], protists including Acanthamoeba castellanii [62, 63], A. polyphaga, the slime mold Dictyostelium discoideum [62, 64, 65], and in mitochondria of several parasitic pathogens, of which the most well-studied is T. brucei, the causative agent of African sleeping sickness. Because the mitochondria of bloodstream parasites are deprived of cytochrome oxidase, AOX is the only terminal oxidase, which is very important for their survival in host organisms [1]. The occurrence of only one terminal oxidase is helpful as a precondition for the use of AOX inhibitors to cure the disease caused by T. brucei [1, 20, 66]. AOX is also found in other protozoan pathogens, such as the causative agent of tropical malaria Plasmodium falciparum [67], Philasterides dicentrarchi causing systemic disease in turbot [68], as well as in Cryptosporidium parvum, Blastocystis hominis, T. congolense, and T. evansi [69].
Among animals, AOX or AOX-encoding genes have been found in sponges, Placozoa, Cnidaria, the annelids Arenicola marina, Nereis pelagica, and Marenzelleria, the sipunculans Sipunculus nudus, the oysters C. gigas and C. virginica, the mollusks Mytilus californianus Conrad, M. mercenaria, and M. galloprovincialis, the gastropods A. californica, Ilyanassa obsoleta, Lottia gigantea Sowerby, and Lymnaea stagnalis, the nematodes (roundworms), echinoderms, protostomes, deuterostomes (see [70]), and the ascidian Ciona intestinalis [71].
Bioinformatics methods have recently unveiled more than 25 AOX-expressing animal species including the lower chordates Branchiostoma floridae, C. savignyi, and Molgula tectiformis [70]. It is believed that a specific trait of AOX in animals is a C-terminal motif N-P-[YF]-X-P-G-[KQE], which is untypical for AOXs in other kingdoms [70].
Finally, a homolog of eukaryotic AOX was discovered in the α-proteobacterium Novosphingobium aromaticivorans [72]. The gene of N. aromaticivorans presumably encoding a bacterial AOX was cloned and expressed in Escherichia coli. The expression level depended on oxygen concentration and the carbon source in the medium. The AOX of proteobacteria is especially similar to the AOX of terrestrial plants and red algae [69].
Cyanobacteria and chloroplasts of higher plants were found to contain the genes of the plastid terminal oxidase (PTOX), a distantly related homolog of the mitochondrial AOX that oxidizes plastoquinol rather than ubiquinol. With this function, PTOX operates in the photosynthetic electron transport chain. Both proteins contain a diiron cluster and 20 conserved amino acid residues, six of which are involved in the binding of iron atoms [73]. The existence of these proteins in mitochondria and plastids, respectively, suggests that AOX and PTOX had a common bacterial ancestor that was a diiron-carboxylate protein whose function might have been associated with the binding and reduction of molecular oxygen from the environment, but the evolution of these proteins diverged at a later stage. The AOX became operative in bacteria with heterotrophic type of metabolism, whereas PTOX emerged in autotrophic bacteria capable of oxygenic photosynthesis. Divergent evolution of these proteins and the process of endosymbiosis led to the appearance of AOX in mitochondria and PTOX in chloroplasts [1, 74].
Detailed consideration of bacterial homologs of AOX and PTOX is beyond the scope of this review, which is focused on mitochondrial AOX. We present these data only with the aim to show that the natural distribution of AOX is much wider than we believed 10 or even 5 years ago.
The existence of numerous AOX sequences found in evolutionarily distant organisms might be useful in determining the amino acids or domains playing important roles in catalysis, the topography of AOX in the membrane, and posttranslational regulation of AOX, especially if such data are accompanied by structural studies.
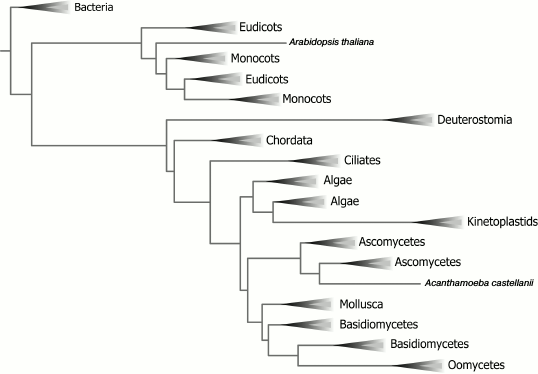
Considering that AOX is widespread among various organisms, it is interesting to trace the distribution of AOX protein in various taxa and reconstruct the possible evolution of AOX. For this purpose, we applied bioinformatics methods to determine the primary structures of AOX in representatives of different taxonomic groups and, based on multiple sequence alignment, a phylogenetic tree reproducing the possible evolution of AOX was constructed (Fig. 2).
Fig. 2. Phylogenetic tree of AOX based on analysis of primary structures (see text for details). The search of sequences was performed by the BLAST program [75] (algorithm blastp, cDNA database); the AOX sequence of T. brucei was used as the query [76]. The sample was composed in such a way as to collect as many AOX sequences as possible from various organisms. However, when the organism contained multiple AOX isoforms, only one of them having the highest degree of homology to AOX from T. brucei was used. The multiple sequence alignment was performed on the BLAST server according to the CLUSTALW2 algorithm [77], after which the phylogenetic tree was constructed. The phylogenetic tree is visualized using the web server iTOL [78].
PROPERTIES OF AOX
AOX is located in the inner mitochondrial membrane; its activity is insensitive to cyanide and is not inhibited by azide, CO, antimycin A, and myxothiazol. The AOX pathway is branched from the main respiratory chain at the level of ubiquinone (Fig. 1). The enzyme AOX catalyzes four-electron oxidation of reduced ubiquinone (ubiquinol) associated with reduction of oxygen to water; this reaction is independent of the main respiratory chain [16, 79]. As already mentioned above, electron transport through AOX is not coupled to ATP synthesis and energy accumulation; the energy of ubiquinol oxidation by oxygen is released as heat [1, 2, 80]. The AOX activity depends on the nature of oxidation substrate, the total concentration of ubiquinone, and its redox state in the membrane, as well as on intracellular oxygen concentration [81]. A characteristic feature of the alternative pathway is its effective inhibition (Ki = 1.5 mM) by aromatic hydroxamic acids (salicylhydroxamate is a typical example). Although it is now clear that hydroxamic acids are not specific “one-enzyme” inhibitors, they still remain the most convenient and commonly used tool for detecting the cyanide-resistant oxidase. Other inhibitors of AOX have also been described: 2,5-dibromo-3-methyl-6-isopropyl-p-benzoquinone, disulfiram, 5-decyl-6-hydroxy-4,7-dioxobenzothiazole, propyl gallate, various hydroxamic acids [37], and askofuranone [20].
Data on the affinity of AOX to oxygen are controversial [38]. It is assumed that the critical oxygen concentration for the alternative pathway is slightly higher than that for the main respiratory chain [1, 2]. Interestingly, the replacement of Thr179 or Cys172 in the AOX of S. guttatum resulted in a twofold increase in the enzyme affinity to oxygen [82].
The purified recombinant preparation of AOX from T. brucei, obtained after expression of the enzyme in a heme-deficient strain of E. coli, oxidized ubiquinol-1 according to the classical Michaelis–Menten kinetics (Km = 338 μM and Vmax = 601 μmol/min per mg protein [20]).
INDUCTION OF AOX
In several yeast species with aerobic metabolism including C. parapsilosis, Debaryomyces occidentalis, and C. albicans, the active AOX was present at all growth stages, and in the yeast D. hansenii AOX was not found only at the earliest stages of growth [37]. In contrast, in the yeasts D. magnusii (E. magnusii) [83], P. pastoris [16], and P. angusta (Hansenula polymorpha) [84], as well as in the fungus N. crassa [85] and some other organisms under normal conditions (i.e. under non-stress functioning of the main phosphorylating respiratory chain), the AOX activity was either low or indiscernible. However, the activity increased significantly when the terminal part of the main respiratory chain was inhibited by the following treatments: (a) growing the organisms on media deficient in the content of iron ions, sulfur, or copper; (b) growing or incubation of resting cells in the presence of antimycin A, cyanide, or azide; (c) growing or incubation of resting cells treated with the inhibitors of mitochondrial transcription and translation; (d) mutational changes in the nuclear or mitochondrial genomes; (e) inhibition of oxidative phosphorylation; (f) lowering of oxygen concentration (see [38]); and (g) inhibition of the tricarboxylic acid cycle [86]. Such stimulation of AOX activity, as deduced from the increasing amounts of AOX transcripts or AOX protein and from activation of cyanide-resistant hydroxamate-sensitive oxidative pathway, was demonstrated not only with the yeast D. magnusii [83, 87], C. albicans [88, 89], H. anomala [90], P. pastoris [16] and the fungi N. crassa [85, 91], P. anserine [92, 93], M. grisea [49], A. niger [94], Phycomyces blakesleeanus [50], and U. maydis [42], the insect pathogenic fungus M. anisopliae [53], but also with H. capsulatum [57], vascular plants (see [95-97] and references therein), and Drosophila [71].
The appearance of cyanide- and antimycin A-insensitive respiration in mitochondria may also arise from changes in the physiological condition of the tissue, organ, or organism. The first classical example is the manifold avalanche-like enhancement of AOX activity in thermogenic tissues of aroid plants over a few days ([10, 98, 99] and references therein). In this case AOX becomes the only terminal oxidase, and oxidation of the substrate in the respiratory chain releases heat sufficient to produce volatiles that attract pollinating insects. Another example is the different degrees of AOX expression in trypanosomes at various stages of the organism’s development. Mitochondria of the bloodstream form are devoid of cytochromes, and respiration is carried out exclusively by AOX [100]. Respiration of the parasitic ciliate Philasterides dicentrarchi causing a disease in turbot is sensitive to antimycin A under normal oxygenation, while cyanide- and antimycin A-insensitive respiratory pathway is activated under hypoxic conditions [68]. In the yeasts Y. lipolytica (formerly C. lipolytica and Schizosaccharomycopsis lipolytica) and P. membranifaciens grown on glucose, AOX is induced upon the transition of culture to the phase of stationary growth; in this case, AOX operates simultaneously with the cytochrome part of the respiratory chain [37, 79, 101, 102]. In the phytopathogenic fungus U. maydis, AOX activity depended on the growth stage of the culture, on the carbon source, and temperature [103]. In the yeast P. pastoris the AOX activity increased monotonically during the culture growth and fell sharply upon depletion of glucose in the growth medium [16]. In mitochondria of the fungus Metarhizium anisopliae, the highest AOX activity was observed at the beginning and at the end of the fungal developmental cycle, during germination of aerial conidia and upon the formation of submerged conidia [53]. In the dimorphic fungus P. brasiliensis causing paracoccidioidomycosis in humans, the expression of AOX-encoding gene increased significantly during conidia germination and formation of the yeast form [104, 105]. In the hemibiotrophic fungus M. perniciosa, a tropical pathogen causing the disease of cocoa, which first colonizes the living host tissues (biotrophic phase) and then grows on dead plant (necrotic phase), the largest number of AOX transcripts was observed in the biotrophic phase [56]. In Philasterides dicentrarchi, the highest activity of AOX occurred in the phase of stationary growth [68]. Aging of potato slices [106] and fruit ripening were also shown to be accompanied by a significant activation of AOX [107, 108].
In the yeast P. anomala (Hansenula anomala), the level of AOX expression depended on the carbon source. It was low on glucose-containing medium but increased dramatically in media containing glycerol, lactate, or raffinose [109]. Similar data were obtained for the yeast C. albicans, where expression of the AOX-encoding gene increased in media containing glycerol or ethanol [110].
In fungi and yeasts, the AOX activity and the content of AOX transcript increased substantially upon mild heat shock [44, 101], as well as under conditions of oxidative [19, 44, 49, 89, 90, 105, 111] and osmotic [44] stresses. Nevertheless, there are some exceptions: the amount of AOX transcript in P. anserine diminished under the action of reactive oxygen species [112]. In the fungus Ph. blakesleeanus, the AOX activity increased under temporary hypoxia or anoxia [50]. In the pathogenic fungi, AOX activity increased sharply under the action of antifungal drugs ([113] and references therein).
The expression of AOX in plants depended on availability of nutrients, such as phosphate [114-116], nitrate, or ammonium [117-119]. Depending on the type of tissue or organ, developmental phase, and metabolic status [120-123], the expression of AOX was significantly increased in response to a wide range of stresses and adverse environmental conditions, such as changes in temperature and light intensity [122, 124-131], osmotic stress [132-134], drought ([132, 135] and references therein), oxidative stress [126, 136-138], attack by incompatible strains of bacterial pathogens, phytopathogens or their elicitors ([139-144] and references therein), treatment with ethylene [145] and NO ([144] and references therein), and the addition of salicylic acid [10, 124, 126, 146, 147] and citrate [124]. In Arabidopsis, the gene AOX1a (one of the genes encoding AOX) is used as a model gene for studying the responses to various kinds of stress [95, 148].
In some cases, an interrelation was found between the action of certain stress factors and AOX induction in plants. For example, drought and osmotic stress are known to be accompanied by oxidative stress. In photosynthesizing organisms, the chloroplasts transform light energy into reducing equivalents. Since CO2 fixation consumes only about 50% of the absorbed light energy, it is obvious that photosynthesis produces an excess of reducing equivalents. In the absence of energy dissipation, the excess of reducing equivalents would cause oxidative stress and damage to the photosynthetic apparatus. External stress factors inhibit CO2 fixation, thus promoting the accumulation of excess reactive oxygen species (ROS) in chloroplasts. Obviously, mechanisms are needed to prevent the overreduction of the photosynthetic electron-transport chain components. This purpose is accomplished by virtue of AOX induction, as well as by export of malate from chloroplasts to mitochondria via the malate-oxaloacetate shunting pathway and through the export of glycolate into peroxisomes, where glycolate is converted to glycine with the subsequent export of glycine to mitochondria, where it is oxidized to serine ([128] and references therein). The elicitors and toxins of plant pathogens facilitate mitochondrial ROS formation [149]. Infection by pathogens and viruses leads to the accumulation of hydrogen peroxide, NO, ethylene, salicylic acid, and the methyl ester of jasmonic acids, which act as signaling molecules for the induction of a protective response of plants and induce the expression of the gene coding AOX or elevate the content of the plant protein ([150] and references therein). These compounds inhibit the cytochrome-dependent electron transport in the respiratory chain. Recent studies revealed that the promoter of gene AOX1a is sensitive to hydrogen peroxide [148]. Infection by pathogens activates also the pentose phosphate pathway and NADP-dependent malic enzyme, which in turn increases the pools of NADPH and pyruvate acting as AOX activator in plants (see below).
In other cases, relationships between the action of the stress factors and AOX activation remain to be elucidated. But it should be clear that the induction of AOX elevates the metabolic plasticity of cells, which is beneficial under variable temperature, light intensity, and availability of nutrients, as well as under the action of biotic and abiotic factors restricting the activity of the main cytochrome oxidation pathway.
GENE STRUCTURES CODING FOR AOX
The AOX gene has nuclear origin. The gene structures encoding AOX have been investigated most thoroughly in the case of plants. In plant genomes, AOX is represented by two different gene subfamilies, AOX1 and AOX2 [122, 151]. The first gene group is expressed in monocots and dicots, whereas the latter family is characteristic only for dicotyledonous plants [152]. The genes of the AOX1 subfamily are inducibly expressed in response to some types of stress including oxidative stress and parasite attacks, whereas the genes of AOX2 subfamily are expressed constitutively or their expression is controlled by other processes [151, 153]. The expression of AOX1 genes is believed to be associated with the involvement of AOX in preventing ROS-induced apoptosis [137]. The genes of the AOX1 family in A. thaliana plants represent a classic example of genes that are activated in response to inhibition of the electron transport chain [15, 95, 120, 151, 154-156]. The genome of this plant species contains four members of the AOX1 family and only one gene of the AOX2 family. Depending on the ROS level in mitochondria, a different number of AOX1 genes are expressed simultaneously [95, 120, 151], and expression of the Aox1a and Aox1d genes is enhanced to the highest extent under stress conditions [127].
The genome of Vitis vinifera has two genes of the AOX1 family (Aox1a and Aox1b); these tissue-specific genes are induced in response to oxidative stress. The gene Aox1a is expressed only in roots and leaves, whereas the gene Aox1b is characteristic to flowers. The plant has also one gene of the AOX2 subfamily that is expressed constitutively in all tissues [157].
There are some exceptions to the latter rule. For example, in genomes of Vigna unguiculata (the genome contains Aox1, Aox2a, and Aox2b genes [126, 158]), Medicago sativa, and Medicago truncatula, the gene Aox2b of the AOX2 subfamily is inducible, and its expression increases in response to oxidative stress [158].
Carrot (Daucus carota L.) is characterized by a unique set of AOX genes [159]. It has two genes of each family, specifically DcAOX1a, DcAOX1b, DcAOX2a, and DcAOX2b. The expression of these genes varies depending on the stage of plant development [160].
Bioinformatics approaches demonstrated that the genes of the AOX1 family occur in lower plants such as green and brown algae [24].
The genes encoding AOX in fungi were found to be less diverse than the plant AOX genes; these organisms usually contain the gene of only one of the two subfamilies [1]. The saprophytic fungus A. fumigatus has a single AOX gene, EFax, composed of 1172 bp, which encodes a ~40-kDa protein. Expression of the gene is activated by the addition into the medium of the prooxidants menadione and paraquat, which confirms the involvement of AOX in antioxidant defense. The filamentous fungus N. crassa also has only one AOX gene, whose expression depends on the functional state of mitochondria [85].
In the yeast P. anomala, AOX is encoded by a single nuclear gene, while in the yeast C. albicans AOX is encoded by two genes of the AOX1 family [1, 37, 89]. In the latter organism AOX1a gene is constitutively expressed, while the gene AOX1b is induced by stress conditions [110, 161].
In the fungus A. niger under elevated production of citric acid, the AOX activity is regulated through the transcriptional regulation of expression of the AOX1a gene [44, 162].
In pathogenic fungi, e.g. P. brasiliensis and H. capsulatum, increased expression of AOX genes was found to depend on the stage of the life cycle [104]. The AOX activity in another pathogenic fungus, Metarhizium anisopliae, peaked at the period of sporulation [53], although AOX was expressed intensively throughout the life cycle.
It has been recognized recently that the yeasts contain normally one or, more often, several AOX isoforms, whereas the absence of AOX is an apparent exception related to the transition to anoxia in facultative anaerobes [35, 163].
In trypanosomes the AOX genes are confined into large polycistronic structures controlled by a single promoter. Consequently, the level of AOX activity in these organisms is regulated primarily at the transcriptional level, as well as at the level of mRNA stability [1].
The genome of the amoeba A. castellanii contains two copies of an AOX gene measuring 1113 and 1125 bp. Each gene encodes a ~42-kDa protein and has an N-terminal leader sequence containing information on mitochondrial localization [62]. Transformation of E. coli by any of these genes results in the appearance of cyanide-resistant respiration that is sensitive to salicyl hydroxamate.
Investigation of A. thaliana AOX gene promoters has shown that the promoter of the AOX1a gene consists of 93 bp located upstream of the gene. Many nucleotide pairs are quite conserved and indispensable for activation of transcription by factors of mitochondrial retrograde regulation in response to suppression of the electron transport chain with antimycin A or inhibition of the tricarboxylic acid cycle by monofluoroacetate [95]. Mutations in the promoter sequence led to partial or complete inhibition of AOX1a gene expression [95]. Studies with the fungus N. crassa revealed the presence of an induction motif consisting of two CCG repeats separated by a 7-bp region that is also characteristic of transcription factors having zinc clusters. This motif is assumed to be common to all the genes encoding fungal AOX proteins [164].
Typically, AOX genes have four exons separated by thee gene-specific introns of different lengths [126], which may provide a mechanism for regulation of gene expression. The popular notion that genes exhibiting high constitutive expression have short introns and are small in number is not justified for the plant AOXs, where the constitutive gene AOX2 has an intron whose length is more than 60% of the gene length (up to 62% in Vitis vinifera). On the other hand, the introns of inducible AOX1 genes expressed in response to oxidative stress have total length of less than 15% of the gene length [157]. In the process of evolution, extra introns could be acquired or lost; therefore, some organisms possess AOX-encoding genes with two or four introns [165].
The AOX encoded by the nuclear genome and synthesized by cytoplasmic ribosomes must be transported into the mitochondrial inner membrane, the eventual destination of AOX. To cross the membranes, the proteins should exist in import-competent state, which is achieved by means of interaction with cytoplasmic chaperones in the presence of ATP. To be imported into mitochondria, the proteins must have a specific “address” targeting their transport into these organelles. These proteins are usually synthesized as precursors whose molecular weight is higher than that of functionally competent mature proteins. Despite substantial differences in lengths of leader sequences among different mitochondria-targeted proteins, a common feature of all these precursors is that they are devoid of negatively charged amino acids; enriched in positively charged amino acid residues, alanine, leucine, and serine; and are able to form amphiphilic α-helix. The mitochondrial leader sequences contain N- and C-termini. The N-terminus is presumably responsible for the “recognition” of the organelle, and the C-terminus contains information ensuring the “correct” cleavage of the leader sequence [166, 167]. The protein import into the inner mitochondrial membrane requires the presence of specific receptors on the mitochondrial surface and of specific translocase complexes, TOM and TIM (translocases of the outer mitochondrial membrane and of the inner mitochondrial membrane). The main driving force for the import of most proteins into the inner mitochondrial membrane (with a few exceptions) is the membrane potential generated during oxidation of energy substrates by the respiratory chain components. The precursor proteins imported into mitochondria are processed by specific peptidases to be converted to the mature forms. The information required for recognition of the proper site for the cleavage of the leader peptide is usually localized inside the leader sequence [166, 167].
Since information on AOX transport into mitochondria of various organisms is fragmentary, we present here all relevant data now available. Import of AOX in soybean cotyledons and roots depends on the membrane potential and ATP; it is accompanied by processing of 34- and 36-kDa proteins synthesized on cytoplasmic ribosomes, during which the precursors are converted into the mature 32-kDa protein. According to data of site-directed mutagenesis, the replacement of arginine in the second position with glutamine completely inhibited the precursor processing [168]. The import of AOX precursor into spinach mitochondria depended not only on the total positive charge of the leader sequence, but also on the position of positively charged amino acids in the C-terminus [166]. The expression of AOX from S. guttatum in the yeast S. pombe led to appearance in the yeast of cyanide-resistant respiration that was inhibited by octyl gallate. In this system, the precursor protein was effectively imported and processed to the mature form. These data indicate similarities and interchangeability of the systems that are responsible for translocation of proteins in plants and yeasts [169].
In the pathogenic form of T. brucei where AOX acts as the only terminal oxidase, the signal targeting AOX import into mitochondria is localized at the N-terminus of the molecule, presumably between the 115th and 146th amino acid residues [170]. The import of AOX into mitochondria of the two forms of T. brucei is carried out by two different mechanisms. In the procyclic form of the parasite, the AOX import depends on the membrane potential and requires the presence of external cytosolic factors and ATP. However, in the bloodstream form, the AOX import depends on ATP but is unaffected by the membrane potential [171].
STRUCTURE OF AOX
The early studies focused primarily on establishing the membrane localization of AOX in plant mitochondria. It was originally thought that AOX has two transmembrane helices and a short loop located in the matrix [172, 173]. Later research established that AOX is a semi-integral protein located in the inner mitochondrial membrane, with the active site localized in the matrix [76, 174, 175]. It was also found that the composition of the AOX active site includes a nonheme iron [176, 177], whereas the catalytically active AOX contacts also with quinones, oxygen atoms, and the membrane.
Sequencing of the genomes of well-studied organisms revealed conserved glutamate and histidine residues in the composition of AOX [173]. It turned out later that these residues were the ligands in the diiron active site; therefore, the protein was assigned to the family of diiron carboxylate proteins [173, 174, 178-180]. The nonheme diiron enzymes belong to a diverse and widespread family of metalloproteins. All of them react with oxygen and participate in redox reactions [1, 37]. They use various substrates and are divided into different subfamilies with different sets of catalytic functions, such as oxidation, hydroxylation, and desaturation. Despite the functional diversity, most of the proteins in this family have similar structural elements [174]. These include a four-helix bundle domain where the diiron active site is coordinated by carboxylate groups. In addition, this domain accounts for the activation of molecular oxygen [1, 2, 22, 37]. The presence in AOX of the diiron cluster was proved by the ESR method [179, 181]. The diiron active site of AOX is represented by two ExxH motifs, where the conserved histidine and glutamate residues participate in the coordination of the two iron atoms. Only one of the iron atoms is involved in binding of oxygen during the catalytic cycle [182].
The AOX of plants exists as a dimer and is posttranslationally regulated by α-keto acids and succinate. Unlike the AOX in plants, the AOX of fungi and yeasts is a monomer; it is not regulated by α-keto acids and succinate but is controlled by nucleotide mono- and diphosphates (see chapter “Regulation of AOX” for details).
In 2000, Joseph-Horne et al. were the first to imitate the structure of diiron cluster of fungal AOX. They also found that the conserved cysteine residues required for the dimerization of AOX protein in plants are missing in fungal AOX [43].
The first attempt to crystallize and investigate the AOX structure by X-ray analysis was reported in 2010. A highly purified AOX from T. brucei was used as the material [20]. As a result, the structure was obtained with resolution of 2.9 Å and R-factor value of 9.5%. The AOX of T. brucei was found to be a monomer, and the structure of the diiron cluster was determined; however, attempts to reproduce the complete model of AOX structure did not succeed.
Apart from the diiron active site that reduces oxygen to water, AOXs of all organisms possess a site where ubiquinone is attached as a reducing agent. Based on the resolved architecture of ubiquinone-binding proteins, the amino acid residues characteristic of the ubiquinone-binding site in AOXs were determined [183]. The composition and location of the pocket accommodating the ubiquinone molecule were determined by point mutations of relevant amino acid residues. It was found by this approach that mutations of residues Gln242, Tyr253, Ser256, His261, or Arg262 significantly reduced the activity of AOX of T. brucei, while the replacement of Trp206 with phenylalanine or tyrosine completely blocked the operation of the enzyme [1].
In the AOX from S. guttatum, the ubiquinone molecule kept in the pocket is coordinated by the hydrophobic environment formed by amino acid residues of conserved helices submerged into the membrane (Val155, Arg159, Arg173, Leu177, Val180, Leu267, Glu270, Ala271, and Ser274). Positively charged amino acid residues of arginine and serine coordinate the position of oxygen atoms by electrostatic interactions [22]. Point mutations of these amino acid residues greatly reduced both the Km value for oxygen and the maximal AOX activity. Thus, it was found that the ubiquinone molecule is located at a distance of 4-5 Å from the diiron center, while the isoprenoid tail of the molecule might be in contact with the membrane [22].
The first model of the AOX spatial structure (AOX from T. brucei), resolved by X-ray analysis was reported in 2013 [76]. The structure of the AOX active centers and the mechanism of AOX inhibition were clarified [22, 76]. The ubiquinone binding site of T. brucei is a recessed pocket formed between the helices submerged into the membrane; it is formed by the following amino acid residues: Arg96, Asp100, Arg118, Leu122, Glu123, Ala126, Asp162, His165, Leu212, Asp213, Asp215, Ala216, Thr219, and Asp266. The aromatic ring of the ubiquinone molecule is located at a distance of 4.3 Å from the diiron center, which is entirely consistent with the data obtained for the plant AOX [76]. The exact mechanism of electron transfer during oxygen reduction to water is unknown; however, it is supposed that the four-electron reduction of oxygen to water involves ubiquinone and the nearby tyrosine residue (Tyr220 in the case of T. brucei).
The structure of the yeast AOX has not yet been established. Therefore, we carried out the search and subsequent construction of the three-dimensional model of AOX from the yeast Y. lipolytica. The genome of Y. lipolytica is sequenced [40], and the library of cDNA for this organism is available. However, the genome is not fully annotated; furthermore, the majority of the genes were annotated automatically. Therefore, we had to solve the following problems.
1. Search for the AOX sequence in the genome of Y. lipolytica (UniProt ID – Q6C9M5_YARLI).
2. Determination the secondary structure of the studied protein; search for homologous proteins with resolved spatial structure.
3. Structural simulation of the yeast AOX based on sequence homology and gaining information on the active centers and structural features of the protein.
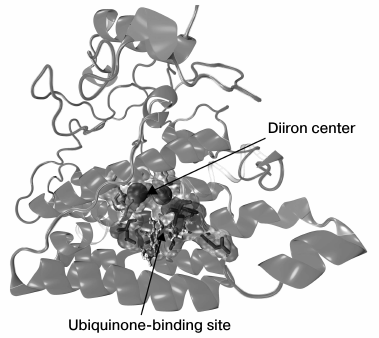
Our work resulted in a model of AOX spatial structure in Y. lipolytica; the model is schematically depicted in Fig. 3.
Fig. 3. Model of spatial structure for AOX of the yeast Y. lipolytica.
Using multiple alignments of known structures, we found the numbers and positions of residues in the active diiron center. In the AOX of Y. lipolytica, two iron atoms are coordinated by six amino acid residues: four glutamate residues (Glu151, Glu190, Glu241, and Glu297) and two histidine residues (His193 and His300). These amino acid residues are highly conserved, whereas the remaining residues participating in the domain formation are more variable and are presumably involved in maintaining the correct positioning of amino acid residues in the active center. The iron atoms were inserted using the VMD software [184].
We performed the search for the ubiquinone-binding site that is necessary for the functioning of AOX. In this way, we identified the amino acid residues that coordinate the ubiquinone molecule: Arg146, Phe147, Leu150, Ile153, Leu240, Thr247, and the residues involved in the formation of a hydrophobic “pocket” accommodating the ubiquinone molecule. The mutual arrangement of the ubiquinone molecule and the diiron active site in Y. lipolytica was found similar to that in the AOX structure of T. brucei [76]; this arrangement is convenient for electron transfer between the active center and ubiquinone. We note that were we the first to obtain a three-dimensional model of the yeast AOX.
BIOGENESIS OF AOX
Analysis of biogenesis of the cyanide-resistant alternative oxidative pathway showed that it comprises two stages. The first is believed to involve synthesis of the protein component on cytoplasmic ribosomes (this process is blocked by cycloheximide, an inhibitor of cytoplasmic protein synthesis). This step is relatively slow and depends on temperature. The second stage is fast, comparatively independent of temperature, and insensitive to cycloheximide. This stage requires the presence of Fe (III) ions and probably includes the activation of the precursor protein synthesized in the first step as well as the incorporation of iron [38].
REGULATION OF AOX
Regulation of expression of AOX genes is a highly complex and, unfortunately, insufficiently characterized process. Since AOX operates in mitochondria whereas its genes are encoded by the nuclear genome, AOX is a good example of a retrogradely regulated protein [85]. Nevertheless, the exact nature of the retrograde regulatory pathways for AOX is still unknown. By studying the coexpression of the AOX gene and various other factors in N. crassa, 62 strains were obtained that displayed reduced AOX activity during its induction. The AOX activity was affected by transcription factors, kinases, monothiol glutaredoxin, the mitochondrial receptor Tom70 (a translocase component of the outer mitochondrial membrane), and other factors [85].
The AOX activity is known to be regulated by transcriptional and posttranslational mechanisms. The first mechanism consists of the control of AOX gene expression and mRNA functional condition. The second includes all the mechanisms controlling AOX activity by virtue of changes in structure, activity, and properties of the mature protein.
A series of direct repressors and activators of AOX are known for A. thaliana. These include the ABA-insensitive transcription factor 4 (ABI4) [154], No-Apical-Meristem (NAC) factor, ATAF1-2 (Arabidopsis thaliana Activating Transcription Factor 1-2), CUC2 (Cup-Shaped Cotyledon2), ANAC013 (Arabidopsis NAC domain-containing protein13) [185], and ANAC017 (Arabidopsis NAC domain-containing protein17) as regulators of AOX1a2 [186], and the transcription factors WRKY40 and WRKY63 (factors containing the motif Trp-Arg-Lys-Thr) that are linked to the promoter region of the Aox1a gene by means of specific cis-elements [187].
It was shown that AOX expression in plants is regulated by interaction of the AOX gene promoter with auxin transport proteins such as rao3p, rao4p, rao5p, and rao6p [188], whose expression depends on the functional state of mitochondria. Consequently, this type of regulation can be considered a reciprocal response to stress conditions.
The regulated expression of AOX in fungi is controlled by several known genes. Transcriptional regulation of AOX in C. albicans involves histidine kinase [110]. Early studies identified the aod-2 gene in the genome of the fungus N. crassa, which is responsible for AOX activity (at that time the AOX-encoding gene was called aod-1) [9, 189]. Later studies revealed that the regulation of AOX gene expression in N. crassa and many other fungi involves up to five different transcription factors having also other functions [164, 190]. The genes were provisionally designated aod-4, aod-5, aod-6, and aod-7; the knockout of any of these genes led to the decrease in AOX expression. The most significant effect was observed upon the knockout of aod-2 and aod-5 genes [164, 191]. These transcription factors comprise two zinc clusters in their composition [192]. The orthologs of aod-2 and aod-5 were found in many organisms including P. anserine [193] and A. nidulans [194].
Expression of AOX genes in fungi and plants may depend on intracellular levels of cAMP and calcium ions. The addition of cyanide to a culture of Y. lipolytica featuring the aerobic type metabolism resulted in a twofold increase in intracellular ADP content and a fivefold increase in the AMP content, which activated cyanide-resistant respiration [27, 195, 196].
At the posttranslational level, AOX activity is regulated by availability of the substrate, i.e. by the concentration of ubiquinone, its reduction level, and oxygen concentration in the cell [81]. Such regulation is typical of many plant species [1] and that of other organisms [197].
In higher plants, where AOX exists in the form of dimers connected by cysteine “bridges” at sites CysI and CysII, the activity of the protein is regulated through changes in the type of binding between the monomers. AOX molecules interconnected by noncovalent bonds are active, while the formation of covalent bonds between regulatory cysteine residues after their oxidation [198] leads to inactivation of the enzyme [199].
The active protein (in its reduced form) in plants can be regulated by α-keto acids [200]. This is a complex process dependent on the kind of activator, as well as on the locations of cysteine residues. In isoenzymes where CysI is replaced with serine, the AOX activity was not stimulated by pyruvate but was enhanced by succinate [1, 201]. The thermogenic plant A. maculatum contains a variety of AOXs that are insensitive to regulation by pyruvate [99, 202]. The explanation of this fact takes into account the presence in plant AOXs of a conserved region, the ENV-element. In the gene AmAOX1e that is constitutively expressed in several tissues of A. maculatum [99] and codes for one form of AOX, the ENV-regulatory element is replaced with a so-called QNT-element characteristic of other kingdoms of life. Furthermore, it was shown that A. maculatum also has an AOX isoform that is activated by pyruvate but insensitive to succinate. Pyruvate transforms AOX into the functionally active state irrespective of the level of ubiquinone reduction [202].
In fungi and yeasts, the situation is principally different. The yeast AOX exists as a monomer and does not have the regulatory cysteine residues (see section “Structure of AOX” for details) and, consequently, is not controlled by α-keto acids [1, 172]. However, it is activated upon attachment of purine mono- and diphosphates, i.e. AMP, ADP, dAMP, and GMP [27, 31, 32, 41-44, 172]. The AOX from the fungus U. maydis grown in the presence of succinate can be activated by the addition of either GMP or pyruvate [42].
In the parasitic protist A. castellanii, ATP, unlike other nucleotides, inhibits AOX. Remarkably, ATP competitively binds to the protein at the GMP-binding site. Thus, the AOX activity is regulated by the proportion of intercellular purine nucleotides [62-64]. A similar mechanism of regulation is observed in the amoebae D. discoideum [62, 64] and the yeasts C. maltosa [62, 64], Y. lipolytica [27], and C. parapsilosis [29]. However, the mechanism of interactions of small molecules with the AOX protein remains poorly understood.
Interestingly, the impact of ROS on the fungus P. anserina lowered the amount of mRNA and, accordingly, the AOX activity [112]. This example is an exception from the general rule that implies the universal involvement of AOX in the response to oxidative stress.
PHYSIOLOGICAL ROLE OF AOX
The role of the alternative respiratory pathway, unrelated to energy storage, has attracted attention of researchers for a few decades. Several hypotheses have been proposed as outlined below.
The physiological role of AOX in thermogenesis of thermogenic plants is quite obvious. In thermogenic flowering organs, mitochondria undergo fundamental rearrangements of the respiratory chain: AOX becomes the only terminal oxidase with very high activity. The energy of substrate oxidation by this respiratory chain is converted into heat, which promotes evaporation of volatile substances, thus attracting pollinating insects ([1, 99] and references therein).
The role of AOX in inhibiting the terminal components of the main respiratory chain or in losing the ability to synthesize these components (see section “Induction of AOX”) is also quite conceivable. Operation of the alternative pathway under these conditions enables the reoxidation of cytoplasmic NADH, thus increasing the chances of cell survival. In addition, the phosphorylating activity at the first coupling site and the substrate phosphorylation are retained, while AOX ensures high oxidative activity required for biosynthetic processes in mitochondria. This situation occurs also in T. brucei, the causative agent of sleeping sickness. At a certain stage of parasite development (the bloodstream form), AOX is the only terminal oxidase ensuring respiration and survival of the pathogen in host cells [100]. Similar events were observed in Philasterides dicentrarchi, causing the disease in turbot. Under normal oxygenation, respiration of the pathogen was sensitive to inhibition of the cytochrome chain by antimycin A, whereas under hypoxic conditions cyanide- and antimycin A-resistant respiration was induced [203].
The fundamental significance of AOX in alleviating or preventing oxidative stress is also evident. The decrease in reduction state of coenzyme Q, the donor of reducing equivalents for AOX, diminishes the production of superoxide anion radical and, eventually, production of hydrogen peroxide, the most stable reactive oxygen species. Such data have been obtained for the yeasts Y. lipolytica [27, 79], C. krusei [80], C. neoformans [36], and P. pastoris [16, 153], the fungi U. maydis [42], S. sclerotiorum [204], P. anserina [48], M. grisea [205], and H. capsulatum [53], the alga E. gracilis [153], and the higher plants A. thaliana [206], N. tabacum [131, 141, 207], and O. sativa [208]. It was also shown that increased expression of AOX decreases the production of reactive nitrogen species [209]. Owing to its involvement in suppression of oxidative stress, AOX may determine the threshold for the induction of programmed cell death [153]. Overexpression of the AtAox1a gene in A. thaliana prevented aluminum-induced cell death by reducing ROS levels at early stages of the stress response [210].
In this connection, the role of AOX in the response to light stress is becoming clear [125]. During germination of A. thaliana seeds under continuous strong light, higher rates of ROS production and less efficient use of reducing equivalents in chloroplasts were characteristic of the mutants defective in the Aox1a gene, as compared to the wild-type seeds [125, 206]. The inhibition of AOX was accompanied by fast accumulation of NADPH in illuminated chloroplasts, and consequently, by overreduction of the acceptor side of photosystem I. Thus, oxidation of reducing equivalents by AOX protects plants against light stress [141, 207] and prevents the destruction of the photosynthetic apparatus.
Apart from the aforementioned roles of AOX in optimizing respiratory metabolism, protection against excess ROS, and occasional cell survival, AOX performs other functions according to the stage of development or the metabolic and physiological status of cells, organs, and tissues. It is commonly accepted that the induction of AOX increases metabolic plasticity of cells, which could be useful for rapid adaptation to variable nutrient sources or to biotic and abiotic stress factors [1, 211-213]. Different AOX functions may complement each other, rather than be mutually exclusive [2, 153]. Moreover, AOX is presently considered as a marker of stress conditions and as a candidate player involved in cell reprogramming under these conditions [1, 211, 212].
Salt-stressed A. thaliana plants were marked by an increase in intracellular Na+ content and elevated ROS production. In these plants, the expression of the AtAox1a gene was sharply increased [133]. The high AOX activity in roots and shoots provides A. thaliana plants with the ability to grow on media with high concentrations of NaCl and to maintain intracellular Na+ at low levels [133]. The involvement of AOX in response to salt stress was also shown for M. truncatula [214]. In experiments with the alga Ch. reinhardtii grown in nitrate-rich media, participation of AOX in nitrate and nitrite reduction was demonstrated [60].
Leaves of N. tabacum plants lacking AOX had elevated levels of H2O2 and superoxide O2– [215-217]. In addition, N. tabacum mutants devoid of AOX were less tolerant to stresses caused by attacks of pathogenic bacteria, fungi, and sucking insects. Protective metabolites were less abundant in mutant plants than in the wild type plants, whereas the ROS content and the percentage of cell death were higher than in the wild type [141, 218]. Thus, AOX in plants might represent a universal mechanism of response to oxidative stress [219] and stress conditions in general [141].
In the pathogenic fungi A. fumigatus [220] and H. capsulatum, AOX is involved in protection under stresses and ensures the survival of the pathogens during their existence inside the host [57]. In the pathogenic fungus S. sclerotiorum, AOX participates in regulation of growth, development, and responses to oxidative stress [204].
In the pathogenic yeast C. neoformans, AOX is expressed in response to changes in temperature of the host organism. The mutant of C. neoformans lacking the AOX gene featured low virulence and low tolerance of oxidative stress [36]. Similar data were obtained for the pathogenic fungus P. brasiliensis [54], the causative agent of the paracoccidioidomycosis in humans. One of the most important stages of P. brasiliensis development is the transition of the fungus from the mycelial into the yeast form. This transformation was decelerated after inhibition of AOX or respiratory chain complexes III and IV, and it was completely stopped upon the inhibition of both branches of the respiratory chain, which suggests the possible involvement of AOX in metabolic rearrangements of this pathogenic organism [105].
AOX may participate in antiviral defense. This was shown for tomato and petunia plants [221].
In embryogenesis of carrot (Daucus carota L.), the AOX genes DcAOX1a and DcAOX2a are expressed differentially. During somatic embryogenesis, the gene DcAOX1a encoding one AOX isoform is not expressed, whereas the gene DcAOX2a encoding the other AOX isoform is expressed very actively. At later stages of embryogenesis, expression of DcAOX2a is lowered. The addition of salicylhydroxamic acid, an AOX inhibitor at the stage of somatic embryogenesis, impaired the development of embryonic structures and retarded the growth of the embryos. This process was reversible and depended on the concentration of the inhibitor added. These results imply the participation of AOX in metabolic rearrangements in plants during embryogenesis and cell differentiation [159].
AOX has recently attracted attention of researchers as a possible therapeutic agent for treating disorders in mitochondrial oxidative phosphorylation. The genes encoding AOX in the ascidian Ciona intestinalis and representative members of the Araceae family were functionally expressed in cultures of human cells [222-225]. The expression was found to eliminate lactate accumulation and excessive ROS formation [222, 223], the two commonly manifested symptoms of damage to the oxidative phosphorylation system. The AOX expression prevented also the inhibition of growth and removed the elevated sensitivity to prooxidants in cell lines deficient in cytochrome oxidase [224]. Coexpression of NADH-dehydrogenase (Ndi1) from S. cerevisiae and AOX from Emericella nidulans fully restored the NADH DH/CoQ-reductase CoQ-oxidase activity in nonviable mice lacking the mitochondrial DNA [45]. Expression of AOX of C. intestinalis in D. melanogaster prevented largely or completely the mortality caused by toxins and by global or partial tissue-specific knockout of COX CoVb and cIV subunits of cytochrome oxidase in the respiratory chain [71], as well as mortality caused by deficiency of Surf1 factor responsible for assembling cytochrome oxidase [226]. The expression also prevented the locomotive defect and excess ROS production in drosophila flies with mutated gene dj-1beta, the homolog of human gene DJ1 involved in Parkinson’s disease [226], and promoted restoration of dopamine-dependent neuroregulation [61]. Expression of AOX in mice had a positive influence on cyanide-resistant respiration in intact organs and facilitated long-term protection against lethal concentrations of cyanide in animals. Furthermore, the enzyme properties, activity of the main respiratory chain components, and the efficiency of the oxidative phosphorylation system in isolated mitochondria remained unchanged [227].
These data indicate that AOX might be an important tool to overcome the adverse aftereffects of restricted activity of the main respiratory chain in cells and in animals. The presence of active AOX provides not only the bypass of a defective cytochrome pathway, but also ensures the sustained activity of the tricarboxylic acid cycle under constrained activity of the main cytochrome chain [228] and alleviates the cell damage caused by mitochondrial ROS. It should be remembered that mitochondrial dysfunctions and overproduction of superoxide in mitochondria is an important factor in many human diseases, ranging from systemic pathology in children to cardiomyopathy, ischemia, cancer, and neurodegenerative disorders.
This study was supported by the Russian Foundation for Basic Research (grant No. 13-04-01530), by the Russian Science Foundation (grant No. 14-24-00107), and by the Russian Academy of Sciences (program “Molecular and Cell Biology”).
REFERENCES
1.Albury, M. S., Elliott, C., and Moore, A. L. (2009)
Towards a structural elucidation of the alternative oxidase in plants,
Physiol. Plant., 137, 316-327.
2.Vanlerberghe, G. C., Cvetkovska, M., and Wang, J.
(2009) Is the maintenance of homeostatic mitochondrial signaling during
stress a physiological role for alternative oxidase? Physiol.
Plant., 137, 392-406.
3.Lloyd, D., and Edwards, S. W. (1978) Electron
transport pathways alternative to the main phosphorylating respiratory
chain, in Functions of Alternative Terminal Oxidases (Degn, H.,
Lloyd, D., and Hill, G. C., eds.) Pergamon Press, pp. 1-10.
4.Schonbaum, G. R., Bonner, W. D., Jr., Storey, B.
T., and Bahr, J. T. (1971) Specific inhibition of the
cyanide-insensitive respiratory pathway in plant mitochondria by
hydroxamic acids, Plant Physiol., 47, 124-128.
5.Huq, S., and Palmer, J. M. (1978) Isolation of a
cyanide-resistant duroquinol oxidase from Arum maculatum
mitochondria, FEBS Lett., 95, 217-220.
6.Elthon, T. E., and McIntosh, L. (1986)
Characterization and solubilization of the alternative oxidase of
Sauromatum guttatum mitochondria, Plant Physiol.,
82, 1-6.
7.Elthon, T. E., and McIntosh, L. (1987)
Identification of the alternative terminal oxidase of higher plant
mitochondria, Proc. Natl. Acad. Sci. USA, 84,
8399-8403.
8.Elthon, T. E., Nickels, R. L., and McIntosh, L.
(1989) Monoclonal antibodies to the alternative oxidase of higher plant
mitochondria, Plant Physiol., 89, 1311-1317.
9.Lambowitz, A. M., Sabourin, J. R., Bertrand, H.,
Nickels, R., and McIntosh, L. (1989) Immunological identification of
the alternative oxidase of Neurospora crassa mitochondria,
Mol. Cell. Biol., 9, 1362-1364.
10.Rhoads, D. M., and McIntosh, L. (1992) Salicylic
acid regulation of respiration in higher plants: alternative oxidase
expression, Plant Cell, 4, 1131-1139.
11.Eriksson, M., Gardestrom, P., and Samuelsson, G.
(1995) Isolation, purification, and characterization of mitochondria
from Chlamydomonas reinhardtii, Plant Physiol.,
107, 479-483.
12.Chaudhuri, M., Ajayi, W., Temple, S., and Hill,
G. C. (1995) Identification and partial purification of a
stage-specific 33 kDa mitochondrial protein as the alternative oxidase
of the Trypanosoma brucei brucei bloodstream trypomastigotes,
J. Eukaryot. Microbiol., 42, 467-472.
13.Affourtit, C., and Moore, A. L. (2004)
Purification of the plant alternative oxidase from Arum
maculatum: measurement, stability and metal requirement,
Biochim. Biophys. Acta, 1608, 181-189.
14.Whelan, J., McIntosh, L., and Day, D. A. (1993)
Sequencing of a soybean alternative oxidase cDNA clone, Plant
Physiol., 103, 1481.
15.Vanlerberghe, G. C., and McIntosh, L. (1994)
Mitochondrial electron transport regulation of nuclear gene expression.
Studies with the alternative oxidase gene of tobacco, Plant
Physiol., 105, 867-874.
16.Kern, A., Hartner, F. S., Freigassner, M.,
Spielhofer, J., Rumpf, C., Leitner, L., Frohlich, K. U., and Glieder,
A. (2007) Pichia pastoris “just in time” alternative
respiration, Microbiology, 153, 1250-1260.
17.Ito-Inaba, Y., Hida, Y., and Inaba, T. (2009)
What is critical for plant thermogenesis? Differences in mitochondrial
activity and protein expression between thermogenic and non-thermogenic
skunk cabbages, Planta, 231, 121-130.
18.Nihei, C., Fukai, Y., Kawai, K., Osanai, A.,
Yabu, Y., Suzuki, T., Ohta, N., Minagawa, N., Nagai, K., and Kita, K.
(2003) Purification of active recombinant trypanosome alternative
oxidase, FEBS Lett., 538, 35-40.
19.Magnani, T., Soriani, F. M., Martins, V. P.,
Nascimento, A. M., Tudella, V. G., Curti, C., and Uyemura, S. A. (2007)
Cloning and functional expression of the mitochondrial alternative
oxidase of Aspergillus fumigatus and its induction by oxidative
stress, FEMS Microbiol. Lett., 271, 230-238.
20.Kido, Y., Sakamoto, K., Nakamura, K., Harada, M.,
Suzuki, T., Yabu, Y., Saimoto, H., Yamakura, F., Ohmori, D., Moore, A.,
Harada, S., and Kita, K. (2010) Purification and kinetic
characterization of recombinant alternative oxidase from Trypanosoma
brucei brucei, Biochim. Biophys. Acta, 1797,
443-450.
21.Elliott, C., Young, L., May, B., Shearman, J.,
Albury, M. S., Kido, Y., Kita, K., and Moore, A. L. (2014) Purification
and characterization of recombinant DNA encoding the alternative
oxidase from Sauromatum guttatum, Mitochondrion,
S1567-7249(14)00028-2.
22.Young, L., May, B., Pendlebury-Watt, A.,
Shearman, J., Elliott, C., Albury, M. S., Shiba, T., Inaoka, D. K.,
Harada, S., Kita, K., and Moore, A. L. (2014) Probing the
ubiquinol-binding site of recombinant Sauromatum guttatum
alternative oxidase expressed in E. coli membranes through
site-directed mutagenesis, Biochim. Biophys. Acta, 1837,
1219-1225.
23.McDonald, A. E. (2009) Alternative oxidase: what
information can protein sequence comparisons give us? Physiol.
Plant., 137, 328-341.
24.Neimanis, K., Staples, J. F., Huner, N. P., and
McDonald, A. E. (2013) Identification, expression, and taxonomic
distribution of alternative oxidases in non-angiosperm plants,
Gene, 526, 275-286.
25.Matsunaka, S., Morita, S., and Conti, S. F.
(1966) Respiratory system of Rhodotorula glutinis. I.
Inhibitor tolerance and cytochrome components, Plant Physiol.,
41, 1364-1369.
26.Nyns, E. J., and Hamaide-Deplus, M. C. (1972)
Cyanide-insensitive respiration of Candida lipolytica, Arch.
Int. Physiol. Biochim., 80, 978-980.
27.Medentsev, A. G., Arinbasarova, A. Y., and
Akimenko, V. K. (2004) Reactivation of the alternative oxidase of
Yarrowia lipolytica by nucleoside monophosphates, FEMS Yeast
Res., 5, 231-236.
28.Guerin, M., and Camougrand, N. (1986) The
alternative oxidase of Candida parapsilosis, Europ. J.
Biochem./FEBS, 159, 519-524.
29.Milani, G., Jarmuszkiewicz, W., Sluse-Goffart, C.
M., Schreiber, A. Z., Vercesi, A. E., and Sluse, F. E. (2001)
Respiratory chain network in mitochondria of Candida
parapsilosis: ADP/O appraisal of the multiple electron pathways,
FEBS Lett., 508, 231-235.
30.Ruy, F., Vercesi, A. E., and Kowaltowski, A. J.
(2006) Inhibition of specific electron transport pathways leads to
oxidative stress and decreased Candida albicans proliferation,
J. Bioenerg. Biomembr., 38, 129-135.
31.Costa-de-Oliveira, S., Sampaio-Marques, B.,
Barbosa, M., Ricardo, E., Pina-Vaz, C., Ludovico, P., and Rodrigues, A.
G. (2012) An alternative respiratory pathway on Candida krusei:
implications on susceptibility profile and oxidative stress, FEMS
Yeast Res., 12, 423-429.
32.Sakajo, S., Minagawa, N., and Yoshimoto, A.
(1997) Effects of nucleotides on cyanide-resistant respiratory activity
in mitochondria isolated from antimycin A-treated yeast Hansenula
anomala, Bioscience Biotechnol. Biochem., 61,
396-399.
33.Shi, N. Q., Cruz, J., Sherman, F., and Jeffries,
T. W. (2002) SHAM-sensitive alternative respiration in the
xylose-metabolizing yeast Pichia stipites, Yeast,
19, 1203-1220.
34.Cabrera-Orefice, A., Guerrero-Castillo, S.,
Luevano-Martinez, L. A., Pena, A., and Uribe-Carvajal, S. (2010)
Mitochondria from the salt-tolerant yeast Debaryomyces hansenii
(halophilic organelles?), J. Bioenerg. Biomembr., 42,
11-19.
35.Cabrera-Orefice, A., Chiquete-Felix, N.,
Espinasa-Jaramillo, J., Rosas-Lemus M., Guerrero-Castillo, S., Pena,
A., and Uribe-Carvajal, S. (2014) The branched mitochondrial
respiratory chain from Debaryomyces hansenii: components and
supramolecular organization, Biochim. Biophys. Acta,
1837, 73-84.
36.Akhter, S., McDade, H. C., Gorlach, J. M.,
Heinrich, G., Cox, G. M., and Perfect, J. R. (2003) Role of alternative
oxidase gene in pathogenesis of Cryptococcus neoformans,
Infect. Immun., 71, 5794-5802.
37.Veiga, A., Arrabaca, J. D., and Loureiro-Dias, M.
C. (2003) Cyanide-resistant respiration, a very frequent metabolic
pathway in yeasts, FEMS Yeast Res., 3, 239-245.
38.Zvyagilskaya, R. A., and Kotelnikova, A. V.
(1991) Structure and Functional Activity of Yeast Mitochondria
(Monograph), Series Biological Chemistry [in Russian],
Vol. 36, VINITI, Moscow.
39.Joseph-Horne, T., Hollomon, D. W., and Wood, P.
M. (2001) Fungal respiration: a fusion of standard and alternative
components, Biochim. Biophys. Acta, 1504, 179-195.
40.Kerscher, S., Durstewitz, G., Casaregola, S.,
Gaillardin, C., and Brandt, U. (2001) The complete mitochondrial genome
of Yarrowia lipolytica, Compar. Funct. Genom., 2,
80-90.
41.Juarez, O., Guerra, G., Martinez, F., and Pardo,
J. P. (2004) The mitochondrial respiratory chain of Ustilago
maydis, Biochim. Biophys. Acta, 1658, 244-251.
42.Sierra-Campos, E., Velazquez, I., Matuz-Mares,
D., Villavicencio-Queijeiro, A., and Pardo, J. P. (2009) Functional
properties of the Ustilago maydis alternative oxidase under
oxidative stress conditions, Mitochondrion, 9,
96-102.
43.Joseph-Horne, T., Babij, J., Wood, P. M.,
Hollomon, D., and Sessions, R. B. (2000) New sequence data enable
modeling of the fungal alternative oxidase and explain an absence of
regulation by pyruvate, FEBS Lett., 481, 141-146.
44.Honda, Y., Hattori, T., and Kirimura, K. (2012)
Visual expression analysis of the responses of the alternative oxidase
gene (aox1) to heat shock, oxidative, and osmotic stresses in
conidia of citric acid-producing Aspergillus niger, J.
Biosci. Bioeng., 113, 338-342.
45.Perales-Clemente, E., Bayona-Bafaluy, M. P.,
Perez-Martos, A., Barrientos, A., Fernandez-Silva, P., and Enriquez, J.
A. (2008) Restoration of electron transport without proton pumping in
mammalian mitochondria, Proc. Natl. Acad. Sci. USA, 105,
18735-18739.
46.Dinant, M., Baurain, D., Coosemans, N., Joris,
B., and Matagne, R. F. (2001) Characterization of two genes encoding
the mitochondrial alternative oxidase in Chlamydomonas
reinhardtii, Curr. Genet., 39, 101-108.
47.El-Khoury, R., and Sainsard-Chanet, A. (2010)
Deletion of the mitochondrial NADH kinase increases mitochondrial DNA
stability and life span in the filamentous fungus Podospora
anserina, Exp. Gerontol., 45, 543-549.
48.Scheckhuber, C. Q., Houthoofd, K., Weil, A. C.,
Werner, A., De Vreese, A., Vanfleteren, J. R., and Osiewacz, H. D.
(2011) Alternative oxidase dependent respiration leads to an increased
mitochondrial content in two long-lived mutants of the aging model
Podospora anserina, PLoS One, 6, e16620.
49.Yukioka, H., Inagaki, S., Tanaka, R., Katoh, K.,
Miki, N., Mizutani, A., and Masuko, M. (1998) Transcriptional
activation of the alternative oxidase gene of the fungus Magnaporthe
grisea by a respiratory-inhibiting fungicide and hydrogen peroxide,
Biochim. Biophys. Acta, 1442, 161-169.
50.Stanic, M., Zakrzewska, J., Hadzibrahimovic, M.,
Zizic, M., Markovic, Z., Vucinic, Z., and Zivic, M. (2013) Oxygen
regulation of alternative respiration in fungus Phycomyces
blakesleeanus: connection with phosphate metabolism, Res.
Microbiol., 164, 770-778.
51.Vanderleyden, J., Kurth, J., and Verachtert, H.
(1979) Characterization of cyanide-insensitive respiration in
mitochondria and submitochondrial particles of Moniliella
tomentosa, Biochem. J., 182, 437-443.
52.Xu, T., Wang, Y. T., Liang, W. S., Yao, F., Li,
Y. H., Li, D. R., Wang, H., and Wang, Z. Y. (2013) Involvement of
alternative oxidase in the regulation of sensitivity of Sclerotinia
sclerotiorum to the fungicides azoxystrobin and procymidone, J.
Microbiol. (Seoul, Korea), 51, 352-358.
53.Uribe, D., and Khachatourians, G. G. (2008)
Identification and characterization of an alternative oxidase in the
entomopathogenic fungus Metarhizium anisopliae, Canad. J.
Microbiol., 54, 119-127.
54.Ruiz, O. H., Gonzalez, A., Almeida, A. J.,
Tamayo, D., Garcia, A. M., and Restrepo, A. (2011) Alternative oxidase
mediates pathogen resistance in Paracoccidioides brasiliensis
infection, PLoS Neglect. Tropic. Dis., 5, e1353.
55.Dolgikh, V. V., Senderskiy, I. V., Pavlova, O.
A., Naumov, A. M., and Beznoussenko, G. V. (2011) Immunolocalization of
an alternative respiratory chain in Antonospora
(Paranosema) locustae spores: mitosomes retain their role
in microsporidial energy metabolism, Eukaryot. Cell, 10,
588-593.
56.Thomazella, D. P., Teixeira, P. J., Oliveira, H.
C., Saviani, E. E., Rincones, J., Toni, I. M., Reis, O., Garcia, O.,
Meinhardt, L. W., Salgado, I., and Pereira, G. A. (2012) The
hemibiotrophic cacao pathogen Moniliophthora perniciosa depends
on a mitochondrial alternative oxidase for biotrophic development,
New Phytol., 194, 1025-1034.
57.Johnson, C. H., Prigge, J. T., Warren, A. D., and
McEwen, J. E. (2003) Characterization of an alternative oxidase
activity of Histoplasma capsulatum, Yeast, 20,
381-388.
58.Sharpless, T. K., and Butow, R. A. (1970) An
inducible alternate terminal oxidase in Euglena gracilis
mitochondria, J. Biol. Chem., 245, 58-70.
59.Mathy, G., Cardol, P., Dinant, M., Blomme, A.,
Gerin, S., Cloes, M., Ghysels, B., DePauw, E., Leprince, P., Remacle,
C., Sluse-Goffart, C., Franck, F., Matagne, R. F., and Sluse, F. E.
(2010) Proteomic and functional characterization of a Chlamydomonas
reinhardtii mutant lacking the mitochondrial alternative oxidase 1,
J. Proteome Res., 9, 2825-2838.
60.Gerin, S., Mathy, G., Blomme, A., Franck, F., and
Sluse, F. E. (2010) Plasticity of the mitoproteome to nitrogen sources
(nitrate and ammonium) in Chlamydomonas reinhardtii: the logic
of Aox1 gene localization, Biochim. Biophys. Acta,
1797, 994-1003.
61.Humphrey, D. M., Parsons, R. B., Ludlow, Z. N.,
Riemensperger, T., Esposito, G., Verstreken, P., Jacobs, H. T., Birman,
S., and Hirth, F. (2012) Alternative oxidase rescues
mitochondria-mediated dopaminergic cell loss in Drosophila,
Hum. Mol. Genet., 21, 2698-2712.
62.Henriquez, F. L., McBride, J., Campbell, S. J.,
Ramos, T., Ingram, P. R., Roberts, F., Tinney, S., and Roberts, C. W.
(2009) Acanthamoeba alternative oxidase genes: identification,
characterization and potential as antimicrobial targets, Int. J.
Parasitol., 39, 1417-1424.
63.Antos-Krzeminska, N., and Jarmuszkiewicz, W.
(2014) Functional expression of the Acanthamoeba castellanii
alternative oxidase in Escherichia coli; regulation of the
activity and evidence for AcAox gene function, Biochem. Cell
Biol., 92, 235-241.
64.Woyda-Ploszczyca, A. M., Sluse, F. E., and
Jarmuszkiewicz, W. (2009) Regulation of Acanthamoeba castellanii
alternative oxidase activity by mutual exclusion of purine nucleotides;
ATP’s inhibitory effect, Biochim. Biophys. Acta,
1787, 264-271.
65.Kimura, K., Kuwayama, H., Amagai, A., and Maeda,
Y. (2010) Developmental significance of cyanide-resistant respiration
under stressed conditions: experiments in Dictyostelium cells,
Development Growth Different., 52, 645-656.
66.Chaudhuri, M., Ott, R. D., and Hill, G. C. (2006)
Trypanosome alternative oxidase: from molecule to function, Trends
Parasitol., 22, 484-491.
67.Murphy, A. D., and Lang-Unnasch, N. (1999)
Alternative oxidase inhibitors potentiate the activity of atovaquone
against Plasmodium falciparum, Antimicrob. Agents
Chemother., 43, 651-654.
68.Mallo, N., Lamas, J., and Leiro, J. M. (2013)
Evidence of an alternative oxidase pathway for mitochondrial
respiration in the scuticociliate Philasterides dicentrarchi,
Protist, 164, 824-836.
69.Suzuki, T., Hashimoto, T., Yabu, Y., Majiwa, P.
A., Ohshima, S., Suzuki, M., Lu, S., Hato, M., Kido, Y., Sakamoto, K.,
Nakamura, K., Kita, K., and Ohta, N. (2005) Alternative oxidase (AOX)
genes of African trypanosomes: phylogeny and evolution of AOX and
plastid terminal oxidase families, J. Eukaryot. Microbiol.,
52, 374-381.
70.McDonald, A. E., Vanlerberghe, G. C., and
Staples, J. F. (2009) Alternative oxidase in animals: unique
characteristics and taxonomic distribution, J. Exp. Biol.,
212, 2627-2634.
71.Kemppainen, K. K., Rinne, J., Sriram, A.,
Lakanmaa, M., Zeb, A., Tuomela, T., Popplestone, A., Singh, S., Sanz,
A., Rustin, P., and Jacobs, H. T. (2014) Expression of alternative
oxidase in Drosophila ameliorates diverse phenotypes due to
cytochrome oxidase deficiency, Hum. Mol. Genet., 23,
2078-2093.
72.Stenmark, P., and Nordlund, P. (2003) A
prokaryotic alternative oxidase present in the bacterium
Novosphingobium aromaticivorans, FEBS Lett., 552,
189-192.
73.Fu, A., Aluru, M., and Rodermel, S. R. (2009)
Conserved active site sequences in Arabidopsis plastid terminal
oxidase (PTOX): in vitro and in planta mutagenesis studies,
J. Biol. Chem., 284, 22625-25632.
74.McDonald, A. E., and Vanlerberghe, G. C. (2006)
Origins, evolutionary history, and taxonomic distribution of
alternative oxidase and plastoquinol terminal oxidase, Comparat.
Biochem. Physiol. Pt. D, Genom. Proteom., 1,
357-364.
75.Madden, T. L., Tatusov, R. L., and Zhang, J.
(1996) Applications of network BLAST server, Methods Enzymol.,
266, 131-141.
76.Shiba, T., Kido, Y., Sakamoto, K., Inaoka, D. K.,
Tsuge, C., Tatsumi, R., Takahashi, G., Balogun, E. O., Nara, T., Aoki,
T., Honma, T., Tanaka, A., Inoue, M., Matsuoka, S., Saimoto, H., Moore,
A. L., Harada, S., and Kita, K. (2013) Structure of the trypanosome
cyanide-insensitive alternative oxidase, Proc. Natl. Acad. Sci.
USA, 110, 4580-4585.
77.McWilliam, H., Li, W., Uludag, M., Squizzato, S.,
Park, Y. M., Buso, N., Cowley, A. P., and Lopez, R. (2013) Analysis
tool web services from the EMBL-EBI, Nucleic Acids Res.,
41, 597-600.
78.Boc, A., Diallo, A. B., and Makarenkov, V. (2012)
T-REX: a web server for inferring, validating and visualizing
phylogenetic trees and networks, Nucleic Acids Res., 40,
573-579.
79.Medentsev, A. G., Arinbasarova, A. Y.,
Golovchenko, N. P., and Akimenko, V. K. (2002) Involvement of the
alternative oxidase in respiration of Yarrowia lipolytica
mitochondria is controlled by the activity of the cytochrome pathway,
FEMS Yeast Res., 2, 519-524.
80.Juszczuk, I. M., and Rychter, A. M. (2003)
Alternative oxidase in higher plants, Acta Biochim. Polon.,
50, 1257-1271.
81.Sluse, F. E., and Jarmuszkiewicz, W. (1998)
Alternative oxidase in the branched mitochondrial respiratory network:
an overview on structure, function, regulation, and role, Brazil.
Med. Biol. Res., 31, 733-747.
82.Crichton, P. G., Albury, M. S., Affourtit, C.,
and Moore, A. L. (2010) Mutagenesis of the Sauromatum guttatum
alternative oxidase reveals features important for oxygen binding and
catalysis, Biochim. Biophys. Acta, 1797, 732-737.
83.Zvyagilskaya, R. A., Korosteleva, N. L., and
Kotelnikova, A. V. (1977) Study of Endomyces magnusii
respiratory system. Properties of mitochondria grown in the
presence of antimycin A, Biokhimiya, 42, 1888-1895.
84.Veiga, A., Arrabaca, J. D., and Loureiro-Dias, M.
C. (2000) Cyanide-resistant respiration is frequent, but confined to
yeasts incapable of aerobic fermentation, FEMS Microbiol. Lett.,
190, 93-97.
85.Nargang, F. E., Adames, K., Rub, C., Cheung, S.,
Easton, N., Nargang, C. E., and Chae, M. S. (2012) Identification of
genes required for alternative oxidase production in the Neurospora
crassa gene knockout library, G3 (Bethesda, Md.),
2, 1345-1356.
86.Gupta, K. J., Shah, J. K., Brotman, Y., Jahnke,
K., Willmitzer, L., Kaiser, W. M., Bauwe, H., and Igamberdiev, A. U.
(2012) Inhibition of aconitase by nitric oxide leads to induction of
the alternative oxidase and to a shift of metabolism towards
biosynthesis of amino acids, J. Exp. Bot., 63,
1773-1784.
87.Zvjagilskaya, R. A., Korosteleva, N. L., and
Kotelnikova, A. V. (1978) An antimycin A- and cyanide-insensitive
variant of Endomyces magnusii, in Functions of Alternative
Terminal Oxidases (Degn, H., et al., eds.) Pergamon Press,
Oxford-New-York, pp. 179-185.
88.Helmerhorst, E. J., Stan, M., Murphy, M. P.,
Sherman, F., and Oppenheim, F. G. (2005) The concomitant expression and
availability of conventional and alternative, cyanide-insensitive,
respiratory pathways in Candida albicans, Mitochondrion,
5, 200-211.
89.Yan, L., Li, M., Cao, Y., Gao, P., Cao, Y., Wang,
Y., and Jiang, Y. (2009) The alternative oxidase of Candida
albicans causes reduced fluconazole susceptibility, J.
Antimicrob. Chemother., 64, 764-773.
90.Minagawa, N., Koga, S., Nakano, M., Sakajo, S.,
and Yoshimoto, A. (1992) Possible involvement of superoxide anion in
the induction of cyanide-resistant respiration in Hansenula
anomala, FEBS Lett., 302, 217-219.
91.Tanton, L. L., Nargang, C. E., Kessler, K. E.,
Li, Q., and Nargang, F. E. (2003) Alternative oxidase expression in
Neurospora crassa, Fungal Genet. Biol., 39,
176-190.
92.Osiewacz, H. D., and Stumpfer, S. W. (2001)
Metabolism and aging in the filamentous fungus Podospora
anserina, Arch. Gerontol. Geriatr., 32, 185-197.
93.Stumpfer, S. W., Stephan, O., and Osiewacz, H. D.
(2004) Impact of a disruption of a pathway delivering copper to
mitochondria on Podospora anserine metabolism and life span,
Eukaryot. Cell, 3, 200-211.
94.Kirimura, K., Matsui, T., Sugano, S., and Usami,
S. (1996) Enhancement and repression of cyanide-insensitive respiration
in Aspergillus niger, FEMS Microbiol. Lett., 141,
251-254.
95.Dojcinovic, D., Krosting, J., Harris, A. J.,
Wagner, D. J., and Rhoads, D. M. (2005) Identification of a region of
the Arabidopsis AtAOX1a promoter necessary for
mitochondrial retrograde regulation of expression, Plant Mol.
Biol., 58, 159-175.
96.Strodtkotter, I., Padmasree, K., Dinakar, C.,
Speth, B., Niazi, P. S., Wojtera, J., Voss, I., Do, P. T., Nunes-Nesi,
A., Fernie, A. R., Linke, V., Raghavendra, A. S., and Scheibe, R.
(2009) Induction of the AOX1D isoform of alternative oxidase in A.
thaliana T-DNA insertion lines lacking isoform AOX1A is
insufficient to optimize photosynthesis when treated with antimycin
A, Mol. Plant, 2, 284-297.
97.Zubo, Y. O., Potapova, T. V., Tarasenko, V. I.,
Borner, T., and Konstantinov, Y. M. (2014) The rate of transcription in
Arabidopsis chloroplasts depends on activity of alternative
electron transfer pathway in mitochondria, Doklady Biochem.
Biophys., 455, 76-79.
98.Zhu, Y., Lu, J., Wang, J., Chen, F., Leng, F.,
and Li, H. (2011) Regulation of thermogenesis in plants: the
interaction of alternative oxidase and plant uncoupling mitochondrial
protein, J. Integr. Plant Biol., 53, 7-13.
99.Ito, K., Ogata, T., Kakizaki, Y., Elliott, C.,
Albury, M. S., and Moore, A. L. (2011) Identification of a gene for
pyruvate-insensitive mitochondrial alternative oxidase expressed in the
thermogenic appendices in Arum maculatum, Plant Physiol.,
157, 1721-1732.
100.Walker, R., Jr., Saha, L., Hill, G. C., and
Chaudhuri, M. (2005) The effect of over-expression of the alternative
oxidase in the procyclic forms of Trypanosoma brucei, Mol.
Biochem. Parasitol., 139, 153-162.
101.Biryukova, E. N., Arinbasarova, A. Yu., and
Medentsev, A. G. (2008) Adaptation of Yarrowia lipolytica yeast
to ethanol, Mikrobiologiya, 78, 186-191.
102.Guerrero-Castillo, S., Cabrera-Orefice, A.,
Vazquez-Acevedo, M., Gonzalez-Halphen, D., and Uribe-Carvajal, S.
(2012) During the stationary growth phase, Yarrowia lipolytica
prevents the overproduction of reactive oxygen species by activating an
uncoupled mitochondrial respiratory pathway, Biochim. Biophys.
Acta, 1817, 353-362.
103.Juarez, O., Guerra, G., Velazquez, I.,
Flores-Herrera, O., Rivera-Perez, R. E., and Pardo, J. P. (2006) The
physiologic role of alternative oxidase in Ustilago maydis,
FEBS J., 273, 4603-4615.
104.Hernandez, O., Garcia, A. M., Almeida, A. J.,
Tamayo, D., Gonzalez, A., Restrepo, A., and McEwen, J. G. (2011) Gene
expression during activation of Paracoccidioides brasiliensis
conidia, Yeast, 28, 771-781.
105.Martins, V. P., Dinamarco, T. M., Soriani, F.
M., Tudella, V. G., Oliveira, S. C., Goldman, G. H., Curti, C., and
Uyemura, S. A. (2011) Involvement of an alternative oxidase in
oxidative stress and mycelium-to-yeast differentiation in
Paracoccidioides brasiliensis, Eukaryot. Cell, 10,
237-248.
106.Hiser, C., and McIntosh, L. (1990) Alternative
oxidase of potato is an integral membrane protein synthesized de
novo during aging of tuber slices, Plant Physiol.,
93, 312-318.
107.Almeida, A. M., Jarmuszkiewicz, W., Khomsi, H.,
Arruda, P., Vercesi, A. E., and Sluse, F. E. (1999) Cyanide-resistant,
ATP-synthesis-sustained, and uncoupling-protein-sustained respiration
during postharvest ripening of tomato fruit, Plant Physiol.,
119, 1323-1330.
108.Considine, M. J., Daley, D. O., and Whelan, J.
(2001) The expression of alternative oxidase and uncoupling protein
during fruit ripening in mango, Plant Physiol., 126,
1619-1629.
109.Sakajo, S., Minagawa, N., and Yoshimoto, A.
(1999). Structure and regulatory expression of a single copy
alternative oxidase gene from the yeast Pichia anomala,
Biosci. Biotechnol. Biochem., 63, 1889-1894.
110.Huh, W. K., and Kang, S. O. (2001)
Characterization of the gene family encoding alternative oxidase from
Candida albicans, Biochem. J., 356, 595-604.
111.Angelova, M. B., Pashova, S. B., Spasova, B.
K., Vassilev, S. V., and Slokoska, L. S. (2005) Oxidative stress
response of filamentous fungi induced by hydrogen peroxide and
paraquat, Mycol. Res., 109, 150-158.
112.Borghouts, C., Scheckhuber, C. Q., Stephan, O.,
and Osiewacz, H. D. (2002) Copper homeostasis and aging in the fungal
model system Podospora anserina: differential expression of
PaCtr3 encoding a copper transporter, Int. J. Biochem. Cell
Biol., 34, 1355-1371.
113.Kunova, A., Pizzatti, C., and Cortesi, P.
(2013) Impact of tricyclazole and azoxystrobin on growth, sporulation
and secondary infection of the rice blast fungus, Magnaporthe
oryzae, Pest Manag. Sci., 69, 278-284.
114.Parsons, H. L., Yip, J. Y., and Vanlerberghe,
G. C. (1999) Increased respiratory restriction during phosphate-limited
growth in transgenic tobacco cells lacking alternative oxidase,
Plant Physiol., 121, 1309-1320.
115.Sieger, S. M., Kristensen, B. K., Robson, C.
A., Amirsadeghi, S., Eng, E. W., Abdel-Mesih, A., Moller, I. M., and
Vanlerberghe, G. C. (2005) The role of alternative oxidase in
modulating carbon use efficiency and growth during macronutrient stress
in tobacco cells, J. Exp. Bot., 56, 1499-1515.
116.Florez-Sarasa, I., Lambers, H., Wang, X.,
Finnegan, P. M., and Ribas-Carbo, M. (2014) The alternative respiratory
pathway mediates carboxylate synthesis in white lupine cluster roots
under phosphorus deprivation, Plant Cell Environ., 37,
922-928.
117.Escobar, M. A., Geisler, D. A., and Rasmusson,
A. G. (2006) Reorganization of the alternative pathways of the
Arabidopsis respiratory chain by nitrogen supply: opposing
effects of ammonium and nitrate, Plant J., 45,
775-788.
118.Watanabe, C. K., Hachiya, T., Takahara, K.,
Kawai-Yamada, M., Uchimiya, H., Uesono, Y., Terashima, I., and Noguchi,
K. (2010) Effects of AOX1a deficiency on plant growth, gene
expression of respiratory components and metabolic profile under
low-nitrogen stress in Arabidopsis thaliana, Plant Cell
Physiol., 51, 810-822.
119.Hachiya, T., and Noguchi, K. (2011) Integrative
response of plant mitochondrial electron transport chain to nitrogen
source, Plant Cell Rep., 30, 195-204.
120.Clifton, R., Lister, R., Parker, K. L., Sappl,
P. G., Elhafez, D., Millar, A. H., Day, D. A., and Whelan, J. (2005)
Stress-induced co-expression of alternative respiratory chain
components in Arabidopsis thaliana, Plant Mol. Biol.,
58, 193-212.
121.Figueira, T. R., and Arruda, P. (2011)
Differential expression of uncoupling mitochondrial protein and
alternative oxidase in the plant response to stress, J. Bioenerg.
Biomembr., 43, 67-70.
122.Millar, A. H., Whelan, J., Soole, K. L., and
Day, D. A. (2011) Organization and regulation of mitochondrial
respiration in plants, Ann. Rev. Plant Biol., 62,
79-104.
123.Vanlerberghe, G. C. (2013) Alternative oxidase:
a mitochondrial respiratory pathway to maintain metabolic and signaling
homeostasis during abiotic and biotic stress in plants, Int. J. Mol.
Sci., 14, 6805-6847.
124.Djajanegara, I., Finnegan, P. M., Mathieu, C.,
McCabe, T., Whelan, J., and Day, D. A. (2002) Regulation of alternative
oxidase gene expression in soybean, Plant Mol. Biol., 50,
735-742.
125.Zhang, D. W., Xu, F., Zhang, Z. W., Chen, Y.
E., Du, J. B., Jia, S. D., Yuan, S., and Lin, H. H. (2010) Effects of
light. on cyanide-resistant respiration and alternative oxidase
function in Arabidopsis seedlings, Plant Cell Environ.,
33, 2121-2131.
126.Costa, J. H., Mota, E. F., Cambursano, M. V.,
Lauxmann, M. A., de Oliveira, L. M., Silva Lima Mda, G., Orellano, E.
G., and Fernandes de Melo, D. (2010) Stress-induced co-expression of
two alternative oxidase (VuAox1 and 2b) genes in Vigna
unguiculata, J. Plant Physiol., 167, 561-570.
127.Xu, F., Yuan, S., and Lin, H. H. (2011)
Response of mitochondrial alternative oxidase (AOX) to light signals,
Plant Signal. Behav., 6, 55-58.
128.Yoshida, K., Watanabe, C. K., Hachiya, T.,
Tholen, D., Shibata, M., Terashima, I., and Noguchi, K. (2011) Distinct
responses of the mitochondrial respiratory chain to long- and
short-term high-light environments in Arabidopsis thaliana,
Plant Cell Environ., 34, 618-628.
129.Searle, S. Y., Thomas, S., Griffin, K. L.,
Horton, T., Kornfeld, A., Yakir, D., Hurry, V., and Turnbull, M. H.
(2011) Leaf respiration and alternative oxidase in field-grown alpine
grasses respond to natural changes in temperature and light, New
Phytol., 189, 1027-1039.
130.Searle, S. Y., and Turnbull, M. H. (2011)
Seasonal variation of leaf respiration and the alternative pathway in
field-grown Populus × canadensis, Physiol. Plant.,
141, 332-342.
131.Wang, J., Rajakulendran, N., Amirsadeghi, S.,
and Vanlerberghe, G. C. (2011) Impact of mitochondrial alternative
oxidase expression on the response of Nicotiana tabacum to cold
temperature, Physiol. Plant., 142, 339-351.
132.Costa, J. H., Jolivet, Y., Hasenfratz-Sauder,
M. P., Orellano, E. G., da Guia Silva Lima, M., Dizengremel, P., and
Fernandes de Melo, D. (2007) Alternative oxidase regulation in roots of
Vigna unguiculata cultivars differing in drought/salt tolerance,
J. Plant Physiol., 164, 718-727.
133.Smith, C. A., Melino, V. J., Sweetman, C., and
Soole, K. L. (2009) Manipulation of alternative oxidase can influence
salt tolerance in Arabidopsis thaliana, Physiol. Plant.,
137, 459-472.
134.Skirycz, A., De Bodt, S., Obata, T., De Clercq,
I., Claeys, H., De Rycke, R., Andriankaja, M., Van Aken, O., Van
Breusegem, F., Fernie, A. R., and Inze, D. (2010) Developmental stage
specificity and the role of mitochondrial metabolism in the response of
Arabidopsis leaves to prolonged mild osmotic stress, Plant
Physiol., 152, 226-244.
135.Wang, J., and Vanlerberghe, G. C. (2013) A lack
of mitochondrial alternative oxidase compromises capacity to recover
from severe drought stress, Physiol. Plant., 149,
461-473.
136.Xiao, M., Ma, J., Li, H., Jin, H., and Feng, H.
(2010) Effects of hydrogen sulfide on alternative pathway respiration
and induction of alternative oxidase gene expression in rice suspension
cells, Zeitschrift Naturforschung. Sec. C: Biosci., 65,
463-471.
137.Mlejnek, P. (2013) Cytokinin-induced cell death
is associated with elevated expression of alternative oxidase in
tobacco BY-2 cells, Protoplasma, 250, 1195-1202.
138.Andronis, E. A., Moschou, P. N., Toumi, I., and
Roubelakis-Angelakis, K. A. (2014) Peroxisomal polyamine oxidase and
NADPH-oxidase cross-talk for ROS homeostasis which affects respiration
rate in Arabidopsis thaliana, Front. Plant Sci.,
5, 132.
139.Simons, B. H., Millenaar, F. F., Mulder, L.,
Van Loon, L. C., and Lambers, H. (1999) Enhanced expression and
activation of the alternative oxidase during infection of
Arabidopsis with Pseudomonas syringae pv. tomato,
Plant Physiol., 120, 529-538.
140.Fu, L. J., Shi, K., Gu, M., Zhou, Y. H., Dong,
D. K., Liang, W. S., Song, F. M., and Yu, J. Q. (2010) Systemic
induction and role of mitochondrial alternative oxidase and nitric
oxide in a compatible tomato–tobacco mosaic virus interaction,
Mol. Plant–Microbe Interact., 23, 39-48.
141.Zhang, L., Oh, Y., Li, H., Baldwin, I. T., and
Galis, I. (2012) Alternative oxidase in resistance to biotic stresses:
Nicotiana attenuata AOX contributes to resistance to a pathogen
and a piercing-sucking insect but not Manducasexta larvae,
Plant Physiol., 160, 1453-1467.
142.Liao, Y. W., Shi, K., Fu, L. J., Zhang, S., Li,
X., Dong, D. K., Jiang, Y. P., Zhou, Y. H., Xia, X. J., Liang, W. S.,
and Yu, J. Q. (2012) The reduction of reactive oxygen species formation
by mitochondrial alternative respiration in tomato basal defense
against TMV infection, Planta, 235, 225-238.
143.Colombatti, F., Gonzalez, D. H., and Welchen,
E. (2014) Plant mitochondria under pathogen attack: a sigh of relief or
a last breath? Mitochondrion, S1567-7249(14)00032-4.
144.Li, Z., Liang, W. S., and Carr, J. P. (2014)
Effects of modifying alternative respiration on nitric oxide-induced
virus resistance and PR1 protein accumulation, J. Gen. Virol.,
95, 2075-2081.
145.Ederli, L., Morettini, R., Borgogni, A.,
Wasternack, C., Miersch, O., Reale, L., Ferranti, F., Tosti, N., and
Pasqualin, S. (2006) Interaction between nitric oxide and ethylene in
the induction of alternative oxidase in ozone-treated tobacco plants,
Plant Physiol., 142, 595-608.
146.Lei, T., Yan, Y. C., Xi, D. H., Feng, H., Sun,
X., Zhang, F., Xu, W. L., Liang, H. G., and Lin, H. H. (2008) Effects
of salicylic acid on alternative pathway respiration and alternative
oxidase expression in tobacco calli, Zeitschrift Naturforschung.
Sec. C: Biosci., 63, 706-712.
147.Matos, A. R., Mendes, A. T., Scotti-Campos, P.,
and Arrabaca, J. D. (2009) Study of the effects of salicylic acid on
soybean mitochondrial lipids and respiratory properties using the
alternative oxidase as a stress-reporter protein, Physiol.
Plant., 137, 485-497.
148.Ho, L. H., Giraud, E., Uggalla, V., Lister, R.,
Clifton, R., Glen, A., Thirkettle-Watts, D., Van Aken, O., and Whelan,
J. (2008) Identification of regulatory pathways controlling gene
expression of stress-responsive mitochondrial proteins in
Arabidopsis, Plant Physiol., 147, 1858-1873.
149.Rhoads, D. M., Umbach, A. L., Subbaiah, C. C.,
and Siedow, J. N. (2006) Mitochondrial reactive oxygen species.
Contribution to oxidative stress and interorganellar signaling,
Plant Physiol., 141, 357-366.
150.Koornneef, A., and Pieterse, C. M. (2008) Cross
talk in defense signaling, Plant Physiol., 146,
839-844.
151.Clifton, R., Millar, A. H., and Whelan, J.
(2006) Alternative oxidases in Arabidopsis: a comparative
analysis of differential expression in the gene family provides new
insights into function of non-phosphorylating bypasses, Biochim.
Biophys. Acta, 1757, 730-741.
152.Considine, M. J., Holtzapffel, R. C., Day, D.
A., Whelan, J., and Millar, A. H. (2002) Molecular distinction between
alternative oxidase from monocots and dicots, Plant Physiol.,
129, 949-953.
153.Van Aken, O. V., Giraud, E., Clifton, R., and
Whelan, J. (2009) Alternative oxidase: a target and regulator of stress
responses, Physiol. Plant., 137, 354-361.
154.Giraud, E., Van Aken, O., Ho, L. H., and
Whelan, J. (2009) The transcription factor ABI4 is a regulator of
mitochondrial retrograde expression of ALTERNATIVE OXIDASE1a, Plant
Physiol., 150, 1286-1296.
155.Gray, G. R., Villarimo, A. R., Whitehead, C.
L., and McIntosh, L. (2004) Transgenic tobacco (Nicotiana tabacum
L.) plants with increased expression levels of mitochondrial
NADP+-dependent isocitrate dehydrogenase: evidence
implicating this enzyme in the redox activation of the alternative
oxidase, Plant Cell Physiol., 45, 1413-1425.
156.Vanlerberghe, G. C., and McIntosh, L. (1996)
Signals regulating the expression of the nuclear gene encoding
alternative oxidase of plant mitochondria, Plant Physiol.,
111, 589-595.
157.Costa, J. H., de Melo, D. F., Gouveia, Z.,
Cardoso, H. G., Peixe, A., and Arnholdt-Schmitt, B. (2009) The
alternative oxidase family of Vitis vinifera reveals an
attractive model to study the importance of genomic design, Physiol.
Plant., 137, 553-565.
158.Cavalcanti, J. H., Oliveira, G. M., Saraiva, K.
D., Torquato, J. P., Maia, I. G., de Melo, D. F., and Costa, J. H.
(2013) Identification of duplicated and stress-inducible Aox2b
gene co-expressed with Aox1 in species of the Medicago
genus reveals a regulation linked to gene rearrangement in
leguminous genomes, J. Plant Physiol., 170,
1609-1619.
159.Frederico, A. M., Campos, M. D., Cardoso, H.
G., Imani, J., and Arnholdt-Schmitt, B. (2009) Alternative oxidase
involvement in Daucus carota somatic embryogenesis, Physiol.
Plant., 137, 498-508.
160.Campos, M. D., Cardoso, H. G., Linke, B.,
Costa, J. H., de Melo, D. F., Justo, L., Frederico, A. M., and
Arnholdt-Schmitt, B. (2009) Differential expression and co-regulation
of carrot AOX genes (Daucus carota), Physiol. Plant.,
137, 578-591.
161.Huh, W. K., and Kang, S. O. (1999) Molecular
cloning and functional expression of alternative oxidase from
Candida albicans, J. Bacteriol., 181,
4098-4102.
162.Hattori, T., Kino, K., and Kirimura, K. (2009)
Regulation of alternative oxidase at the transcription stage in
Aspergillus niger under the conditions of citric acid
production, Curr. Microbiol., 58, 321-325.
163.Gonzalez-Barroso, M. M., Ledesma, A., Lepper,
S., Perez-Magan, E., Zaragoza, P., and Rial, E. (2006) Isolation and
bioenergetic characterization of mitochondria from Pichia
pastoris, Yeast (Chichester, England), 23,
307-313.
164.Chae, M. S., Lin, C. C., Kessler, K. E.,
Nargang, C. E., Tanton, L. L., Hahn, L. B., and Nargang, F. E. (2007)
Identification of an alternative oxidase induction motif in the
promoter region of the aod-1 gene in Neurospora crassa,
Genetics, 175, 1597-1606.
165.Polidoros, A. N., Mylona, P. V., and
Arnholdt-Schmittm, B. (2009) Aox gene structure, transcript
variation and expression in plants, Physiol. Plant., 137,
342-353.
166.Tanudji, M., Sjoling, S., Glaser, E., and
Whelan, J. (1999) Signals required for the import and processing of the
alternative oxidase into mitochondria, J. Biol. Chem.,
274, 1286-1293.
167.Mokranjac, D., and Neupert, W. (2009) Thirty
years of protein translocation into mitochondria: unexpectedly complex
and still puzzling, Biochim. Biophys. Acta, 1793,
33-41.
168.Whelan, J., Hugosson, M., Glaser, E., and Day,
D. A. (1995) Studies on the import and processing of the alternative
oxidase precursor by isolated soybean mitochondria, Plant Mol.
Biol., 27, 769-778.
169.Albury, M. S., Dudley, P., Watts, F. Z., and
Moore, A. L. (1996) Targeting the plant alternative oxidase protein to
Schizosaccharomyces pombe mitochondria confers
cyanide-insensitive respiration, J. Biol. Chem., 271,
17062-17066.
170.Hamilton, V., Singha, U. K., Smith, J. T.,
Weems, E., and Chaudhuri, M. (2014) Trypanosome alternative oxidase
possesses both an N-terminal and internal mitochondrial targeting
signal, Eukaryot. Cell, 13, 539-547.
171.Williams, S., Saha, L., Singha, U. K., and
Chaudhuri, M. (2008) Trypanosoma brucei: differential
requirement of membrane potential for import of proteins into
mitochondria in two developmental stages, Exp. Parasitol.,
118, 420-433.
172.Umbach, A. L., and Siedow, J. N. (2000)
Covalent and noncovalent dimers of the cyanide-resistant alternative
oxidase protein in higher plant mitochondria and their relationship to
enzyme activity, Plant Physiol., 103, 845-854.
173.Siedow, J. N., Umbach, A. L., and Moore, A. L.
(1995) The active site of the cyanide-resistant oxidase from plant
mitochondria contains a binuclear iron center, FEBS Lett.,
362, 10-14.
174.Berthold, D. A., and Stenmark, P. (2003)
Membrane-bound diiron carboxylate proteins, Ann. Rev. Plant
Biol., 54, 497-517.
175.Moore, A. L., Shiba, T., Young, L., Harada, S.,
Kita, K., and Ito, K. (2013) Unraveling the heater: new insights into
the structure of the alternative oxidase, Ann. Rev. Plant Biol.,
64, 637-663.
176.Bendall, D. S., and Bonner, W. D. (1971)
Cyanide-insensitive respiration in plant mitochondria, Plant
Physiol., 47, 236-245.
177.Minagawa, N., Sakajo, S., Komiyama, T., and
Yoshimoto, A. (1990) Essential role of ferrous iron in
cyanide-resistant respiration in Hansenula anomala, FEBS
Lett., 267, 114-116.
178.Moore, A. L., Umbach, A. L., and Siedow, J. N.
(1995) Structure-function relationships of the alternative oxidase of
plant mitochondria: a model of the active site, J. Bioenerg.
Biomembr., 27, 367-377.
179.Berthold, D. A., Voevodskaya, N., Stenmark, P.,
Graslund, A., and Nordlund, P. (2002) EPR studies of the mitochondrial
alternative oxidase. Evidence for a diiron carboxylate center, J.
Biol. Chem., 277, 43608-43614.
180.Marechal, A., Kido, Y., Kita, K., Moore, A. L.,
and Rich, P. R. (2009) Three redox states of Trypanosoma brucei
alternative oxidase identified by infrared spectroscopy and
electrochemistry, J. Biol. Chem., 284, 31827-31833.
181.Moore, A. L., Carre, J. E., Affourtit, C.,
Albury, M. S., Crichton, P. G., Kita, K., and Heathcote, P. (2008)
Compelling EPR evidence that the alternative oxidase is a diiron
carboxylate protein, Biochim. Biophys. Acta, 1777,
327-330.
182.Young, L., Shiba, T., Harada, S., Kita, K.,
Albury, M. S., and Moore, A. L. (2013) The alternative oxidases: simple
oxidoreductase proteins with complex functions, Biochem. Soc.
Transact., 41, 1305-1311.
183.Albury, M. S., Elliott, C., and Moore, A. L.
(2010) Ubiquinol-binding site in the alternative oxidase: mutagenesis
reveals features important for substrate binding and inhibition,
Biochim. Biophys. Acta, 1797, 1933-1939.
184.Humphrey, W., Dalke, A., and Schulten, K.
(1996) VMD – visual molecular dynamics, J. Mol. Graph.,
14, 33-38.
185.De Clercq, I., Vermeirssen, V., Van Aken, O.,
Vandepoele, K., Murcha, M. W., Law, S. R., Inze, A., Ng, S., Ivanova,
A., Rombaut, D., van de Cotte, B., Jaspers, P., Van de Peer, Y.,
Kangasjarvi, J., Whelan, J., and Van Breusegem, F. (2013) The
membrane-bound NAC transcription factor ANAC013 functions in
mitochondrial retrograde regulation of the oxidative stress response in
Arabidopsis, Plant Cell, 25, 3472-3490.
186.Ng, S., Ivanova, A., Duncan, O., Law, S. R.,
Van Aken, O., De Clercq, I., Wang, Y., Carrie, C., Xu, L., Kmiec, B.,
Walker, H., Van Breusegem, F., Whelan, J., and Giraud, E. (2013) A
membrane-bound NAC transcription factor, ANAC017, mediates
mitochondrial retrograde signaling in Arabidopsis, Plant
Cell, 25, 3450-3471.
187.Van Aken, O., Zhang, B., Law, S., Narsai, R.,
and Whelan, J. (2013) AtWRKY40 and AtWRKY63 modulate the expression of
stress-responsive nuclear genes encoding mitochondrial and chloroplast
proteins, Plant Physiol., 162, 254-271.
188.Ivanova, A., Law, S. R., Narsai, R., Duncan,
O., Lee, J. H., Zhang, B., Van Aken, O., Radomiljac, J. D., van der
Merwe, M., Yi, K., and Whelan, J. (2014) A functional antagonistic
relationship between auxin and mitochondrial retrograde signaling
regulates alternative oxidase1a expression in Arabidopsis,
Plant Physiol., 165, 1233-1254.
189.Li, Q., Ritzel, R. G., McLean, L. L., McIntosh,
L., Ko, T., Bertrand, H., and Nargang, F. E. (1996) Cloning and
analysis of the alternative oxidase gene of Neurospora crassa,
Genetics, 142, 129-140.
190.Chae, M. S., and Nargang, F. E. (2009)
Investigation of regulatory factors required for alternative oxidase
production in Neurospora crassa, Physiol. Plant.,
137, 407-418.
191.Descheneau, A. T., Cleary, I. A., and Nargang,
F. E. (2005) Genetic evidence for a regulatory pathway controlling
alternative oxidase production in Neurospora crassa,
Genetics, 169, 123-135.
192.MacPherson, S., Larochelle, M., and Turcotte,
B. (2006) A fungal family of transcriptional regulators: the zinc
cluster proteins, Microbiol. Mol. Biol. Rev. MMBR, 70,
583-604.
193.Sellem, C. H., Bovier, E., Lorin, S., and
Sainsard-Chanet, A. (2009) Mutations in two zinc-cluster proteins
activate alternative respiratory and gluconeogenic pathways and restore
senescence in long-lived respiratory mutants of Podospora
anserina, Genetics, 182, 69-78.
194.Suzuki, Y., Murray, S. L., Wong, K. H., Davis,
M. A., and Hynes, M. J. (2012) Reprogramming of carbon metabolism by
the transcriptional activators AcuK and AcuM in Aspergillus
nidulans, Mol. Microbiol., 84, 942-964.
195.Medentsev, A. G., Arinbasarova, A. Y., and
Akimenko, V. K. (1999) Regulation and physiological role of
cyanide-resistant oxidases in fungi and plants, Biochemistry
(Moscow), 64, 1230-1243.
196.Medentsev, A. G., Arinbasarova, A. Iu., and
Akimenko, V. K. (2001) Level of cyclic AMP during induction of
alternative oxidase in Yarrowia lipolytica yeast cells,
Mikrobiologiya, 70, 29-33.
197.Jarmuszkiewicz, W., Czarna, M., and Sluse, F.
E. (2005) Substrate kinetics of the Acanthamoeba castellanii
alternative oxidase and effects of GMP, Biochim. Biophys. Acta,
1708, 71-78.
198.Rhoads, D. M., Umbach, A. L., Sweet, C. R.,
Lennon, A. M., Rauch, G. S., and Siedow, J. N. (1998) Regulation of the
cyanide-resistant alternative oxidase from plant
mitochondria – identification of the cysteine residue
involved in α-keto acid stimulation and intersubunit disulfide
bond formation, J. Biol. Chem., 273, 30750-30756.
199.Day, D. A., and Wiskich, J. T. (1995)
Regulation of alternative oxidase activity in higher plants, J.
Bioenerg. Biomembr., 27, 379-385.
200.Vanlerberghe, G. C., Yip, J. Y., and Parsons,
H. L. (1999) In organello and in vivo evidence of the importance
of the regulatory sulfhydryl/disulfide system and pyruvate for
alternative oxidase activity in tobacco, Plant Physiol.,
121, 793-803.
201.Grant, N., Onda, Y., Kakizaki, Y., Ito, K.,
Watling, J., and Robinson, S. (2009) Two cys or not two cys? That is
the question; alternative oxidase in the thermogenic plant sacred
lotus, Physiol. Plant., 150, 987-995.
202.Carre, J. E., Affourtit, C., and Moore, A. L.
(2011) Interaction of purified alternative oxidase from thermogenic
Arum maculatum with pyruvate, FEBS Lett., 585,
397-401.
203.Mallo, N., Lamas, J., and Leiro, J. M. (2014)
Alternative oxidase inhibitors as antiparasitic agents against
scuticociliatosis, Parasitology, 14, 1-11.
204.Xu, T., Yao, F., Liang, W. S., Li, Y. H., Li,
D. R., Wang, H., and Wang, Z. Y. (2012) Involvement of alternative
oxidase in the regulation of growth, development, and resistance to
oxidative stress of Sclerotinia sclerotiorum, J. Microbiol.
(Seoul, Korea), 50, 594-602.
205.Avila-Adame, C., and Koller, W. (2002)
Disruption of the alternative oxidase gene in Magnaporthe grisea
and its impact on host infection, Mol. Plant–Microbe Interact.
MPMI, 15, 493-500.
206.Giraud, E., Ho, L. H., Clifton, R., Carroll,
A., Estavillo, G., Tan, Y. F., Howell, K. A., Ivanova, A., Pogson, B.
J., Millar, A. H., and Whelan, J. (2008) The absence of ALTERNATIVE
OXIDASE1a in Arabidopsis results in acute sensitivity to
combined light and drought stress, Plant Physiol., 147,
595-610.
207.Zhang, Y., Xi, D., Wang, J., Zhu, D., and Guo,
X. (2009) Functional analysis reveals effects of tobacco alternative
oxidase gene (NtAOX1a) on regulation of defense responses
against abiotic and biotic stresses, Biosci. Rep., 29,
375-383.
208.Feng, H., Hou, X., Li, X., Sun, K., Wang, R.,
Zhang, T., and Ding, Y. (2013) Cell death of rice roots under salt
stress may be mediated by cyanide-resistant respiration, Zeitschrift
Naturforschung. Sec. C: Biosci., 68, 39-46.
209.Cvetkovska, M., Dahal, K., Alber, N. A., Jin,
C., Cheung, M., and Vanlerberghe, G. C. (2014) Knockdown of
mitochondrial alternative oxidase induces the “stress
state” of signaling molecule pools in Nicotiana tabacum,
with implications for stomatal function, New Phytol.,
203, 449-461.
210.Liu, J., Li, Z., Wang, Y., and Xing, D. (2014)
Overexpression of ALTERNATIVE OXIDASE1a alleviates
mitochondria-dependent programmed cell death induced by aluminium
phytotoxicity in Arabidopsis, J. Exp. Bot., 65,
4465-4478.
211.Rasmusson, A. G., Fernie, A. R., and van
Dongen, J. T. (2009) Alternative oxidase: a defense against metabolic
fluctuations? Physiol. Plant., 137, 371-382.
212.Chai, T. T., Colmer, T. D., and Finnegan, P. M.
(2010) Alternative oxidase, a determinant of plant gametophyte fitness
and fecundity, Plant Signal. Behav., 5, 604-606.
213.Hanqing, F., Kun, S., Mingquan, L., Hongyu, L.,
Xin, L., Yan L., and Yifeng, W. (2010) The expression, function and
regulation of mitochondrial alternative oxidase under biotic stresses,
Mol. Plant Pathol., 11, 429-440.
214.Mhadhbi, H., Fotopoulos, V., Mylona, P. V.,
Jebara, M., Aouani, M. E., and Polidoros, A. N. (2013) Alternative
oxidase 1 (Aox1) gene expression in roots of Medicago
truncatula is a genotype-specific component of salt stress
tolerance, J. Plant Physiol., 170, 111-114.
215.Maxwell, D. P., Wang, Y., and McIntosh, L.
(1999) The alternative oxidase lowers mitochondrial reactive oxygen
production in plant cells, Proc. Natl. Acad. Sci. USA,
96, 8271-8276.
216.Yip, J. Y., and Vanlerberghe, G. C. (2001)
Mitochondrial alternative oxidase acts to dampen the generation of
active oxygen species during a period of rapid respiration induced to
support a high rate of nutrient uptake, Physiol. Plant.,
112, 327-333.
217.Amirsadeghi, S., McDonald, A. E., and
Vanlerberghe, G. C. (2007) A glucocorticoid-inducible gene expression
system can cause growth defects in tobacco, Planta, 226,
453-463.
218.Robson, C. A., and Vanlerberghe, G. C. (2002)
Transgenic plant cells lacking mitochondrial alternative oxidase have
increased susceptibility to mitochondria-dependent and -independent
pathways of programmed cell death, Plant Physiol., 129,
1908-1920.
219.Cvetkovska, M., and Vanlerberghe, G. C. (2012)
Alternative oxidase modulates leaf mitochondrial concentrations of
superoxide and nitric oxide, New Phytol., 195, 32-39.
220.Tudella, V. G., Curti, C., Soriani, F. M.,
Santos, A. C., and Uyemura, S. A. (2004) In situ evidence of an
alternative oxidase and an uncoupling protein in the respiratory chain
of Aspergillus fumigatus, Int. J. Biochem. Cell Biol.,
36, 162-172.
221.Ma, H., Song, C., Borth, W., Sether, D.,
Melzer, M., and Hu, J. (2011) Modified expression of alternative
oxidase in transgenic tomato and petunia affects the level of tomato
spotted wilt virus resistance, BMC Biotechnol., 11,
96.
222.Hakkaart, G. A., Dassa, E. P., Jacobs, H. T.,
and Rustin, P. (2006) Allotopic expression of a mitochondrial
alternative oxidase confers cyanide resistance to human cell
respiration, EMBO Rep., 7, 341-345.
223.Matsukawa, K., Kamata, T., and Ito, K. (2009)
Functional expression of plant alternative oxidase decreases antimycin
A-induced reactive oxygen species production in human cells, FEBS
Lett., 583, 148-152.
224.Dassa, E. P., Dufour, E., Goncalves, S.,
Jacobs, H. T., and Rustin, P. (2009) The alternative oxidase, a tool
for compensating cytochrome c oxidase deficiency in human cells,
Physiol. Plant., 137, 427-434.
225.Kakizaki, Y., Seymour, R. S., and Ito, K.
(2010) A novel functional element in the N-terminal region of Arum
concinnatum alternative oxidase is indispensable for catalytic
activity of the enzyme in HeLa cells, Biochim. Biophys. Acta,
1797, 20-28.
226.Fernandez-Ayala, D. J., Sanz, A., Vartiainen,
S., Kemppainen, K. K., Babusiak, M., Mustalahti, E., Costa, R.,
Tuomela, T., Zeviani, M., Chung, J., O’Dell, K. M., Rustin, P.,
and Jacobs, H. T. (2009) Expression of the Ciona intestinalis
alternative oxidase (AOX) in Drosophila complements defects in
mitochondrial oxidative phosphorylation, Cell Metab., 9,
449-460.
227.El-Khoury, R., Dufour, E., Rak, M.,
Ramanantsoa, N., Grandchamp, N., Csaba, Z., Duvillie, B., Benit, P.,
Gallego, J., Gressens, P., Sarkis, C., Jacobs, H. T., and Rustin, P.
(2013) Alternative oxidase expression in the mouse enables bypassing
cytochrome c oxidase blockade and limits mitochondrial ROS
overproduction, PLoS Genet., 9, e1003182.
228.Vanlerberghe, G. C., Vanlerberghe, A. E., and
McIntosh, L. (2011) Molecular genetic evidence of the ability of
alternative oxidase to support respiratory carbon metabolism, Plant
Physiol., 113, 657-661.