REVIEW: Diversity of Phosphorus Reserves in Microorganisms
T. V. Kulakovskaya*, L. P. Lichko, and L. P. Ryazanova
Skryabin Institute of Biochemistry and Physiology of Microorganisms, Russian Academy of Sciences, pr. Nauki 5, 142290 Pushchino, Moscow Region, Russia; E-mail: alla@ibpm.pushchino.ru* To whom correspondence should be addressed.
Received June 10, 2014
Phosphorus compounds are indispensable components of the Earth’s biomass metabolized by all living organisms. Under excess of phosphorus compounds in the environment, microorganisms accumulate reserve phosphorus compounds that are used under phosphorus limitation. These compounds vary in their structure and also perform structural and regulatory functions in microbial cells. The most common phosphorus reserve in microorganism is inorganic polyphosphates, but in some archae and bacteria insoluble magnesium phosphate plays this role. Some yeasts produce phosphomannan as a phosphorus reserve. This review covers also other topics, i.e. accumulation of phosphorus reserves under nutrient limitation, phosphorus reserves in activated sludge, mycorrhiza, and the role of mineral phosphorus compounds in mammals.
KEY WORDS: microorganism, phosphorus, inorganic polyphosphate, magnesium phosphate, phosphomannan, EBPR, limited growth, phosphorus reserveDOI: 10.1134/S0006297914130100
Abbreviations: Pi, orthophosphate; polyP, inorganic polyphosphates.
Phosphorus compounds are indispensable components of the Earth’s
biomass metabolized by all living organisms. Phosphorus is part of the
most important organic compounds including nucleic acids, ATP, and
other nucleoside phosphates, phospholipids, and phosphorylated proteins
and carbohydrates. The long-term research in the field of experimental
and theoretical modeling of prebiological stages of the origin of the
biosphere suggested a hypothesis about abiogenic origin of ATP, RNA,
phosphorylated sugars, and inorganic polyphosphates and their
involvement in progenote metabolic pathways [1-15].
Insufficiency of phosphorus sources in the environment limits the growth and development of microorganisms, while their excess has a negative effect on regulation of phosphate metabolism. The intracellular content of Pi in living cells is strictly regulated. This regulation is provided by variations in the activity of specific transport systems of the cytoplasmic membrane and, in eukaryotes, the activity of transport systems of organelles. Pi homeostasis also involves the enzymes providing phosphate conversion into osmotically inert forms, which are specifically compartmentalized in cells. Microorganisms living in constantly varying environments have various mechanisms of adaptation to phosphate deficiency and excess. One such mechanism is Pi transport systems with different affinity and mechanisms of action.
Some species of microorganisms possess transporting systems with high affinity to Pi and can survive and grow at very low Pi concentrations in the medium. Such properties are typical of the bacterium GFAJ-1 inhabiting waters with enhanced content of arsenates and low content of phosphates [16]. When this microorganism was found, it was supposed to utilize arsenic instead of phosphorus [16]. However, later it was shown that the adaptive mechanism providing its existence under these unfavorable conditions is the presence of a Pi transporting system with extremely high affinity, which allows the cells of this bacterium to take up Pi from the concentration of 1.7 µmole/liter [17].
Most bacteria have two phosphate transporting systems: Pit and Pst [18-21]. The Pit transporter is constitutive, has a low affinity to Pi, and is capable of Pi absorption and export together with a bivalent metal cation in the form of MeHPO4, with consumption of energy of proton motive force [19, 20]. The Pst system is induced at Pi concentrations in the medium below 20 µM and has high affinity to Pi [18, 19].
Yeasts also have several phosphate transporting systems with different affinity to Pi [22, 23]. The presence of numerous transporting systems allows microbial cells to take up phosphate from media with either low or high phosphate content.
This review is devoted to another pathway of microbial adaptation to changes in phosphorus accessibility in the environment, namely, the formation of reserve phosphorus compounds that are accumulated or utilized under excess or deficiency of phosphorus sources in the medium, respectively. These compounds are of diverse chemical nature and not only play the role of relatively inert phosphorus reserves in the microbial cell but also perform structural, bypassing, and regulatory functions.
DIVERSITY OF PHOSPHORUS RESERVES IN MICROORGANISMS
Reserve phosphorus compounds in microorganisms include substances accumulated under phosphate excess in the medium and utilized under its deficiency. These are both mineral and organic phosphorus compounds localized inside the cell, in different cell compartments, and outside the cells. Extracellular phosphorus reserve compounds either are adsorbed onto the cell surface or are present in the medium. The accumulation of phosphorus reserve compounds in microorganisms is characterized by quantitative and qualitative diversity.
Orthophosphate. The simplest reserve phosphorus compounds of microorganisms are low-solubility phosphates: MgPO4OH·4H2O formed in the halophilic archaea Halobacterium salinarium and Halorubrum distributum [24-26] and NH4MgPO4·6H2O formed in bacteria of the Brevibacterium genus [26] and Acetobacter xylinum [27].
The archaea H. salinarium and H. distributum concentrate phosphate from aqueous solutions during their growth [24-26]. Pi consumption is suppressed by FCCP, an uncoupler dissipating the transmembrane proton gradient on the membranes [25, 26]. At excess concentration of Pi, a considerable part is not used in biosynthetic processes but accumulates in biomass (Table 1).
Table 1. Content of Pi and
inorganic polyphosphate (polyP) in biomass (% of Pi consumed
from the medium) during cultivation on media with excess Pi
(8-11 mM) [25]
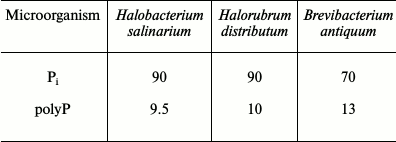
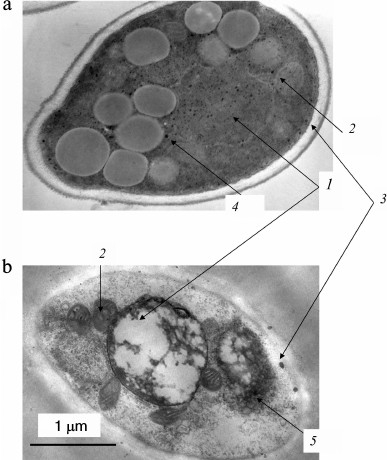
The Pi content in the biomass of both archaea increases with an increase in the initial Pi concentration in the medium and considerably exceeds the content of inorganic polyphosphates. The accumulation of Pi from the medium leads to changes in the morphology of archaeal cells [24-26]. Only some of the cells in the population remain intact under Pi excess. Thin sections show compression of the cytoplasm, accumulation of electron-dense material in the cells (lead citrate staining), an appreciable number of damaged cells, and extracellular crystalline material (Fig. 1). The biomass of these archaea grown under Pi excess is treated with distilled water, followed by cell lysis, and a water-insoluble precipitate (orthophosphate) is obtained by repeated washing with water and centrifugation.
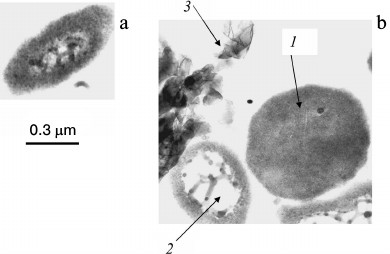
Fig. 1. Ultrathin sections of Halorubrum distributum cells [25]: a) cells grown on medium with 0.05 mM Pi; b) cells grown on medium with 11.5 mM Pi: 1) cells with thickened cytoplasm; 2) enlarged nucleoid zone; 3) extracellular crystals of magnesium phosphate.
This compound was identified by X-ray structure analysis as Mg2PO4OH·4H2O (International Center for Diffraction Data, 1999, No. 44-0774), and the content of H2O was determined by thermogravimetric analysis [26]. The chemical composition of the precipitate is in agreement with the fact that the cells of halophilic archaea need magnesium ions to scavenge Pi from the medium [26]. Excess production of this poorly soluble salt leads to destructives changes in some of the cells in the populations of H. salinarium and H. distributum. During cultivation of H. salinarium and H. distributum in Pi-deficient medium, the content of magnesium phosphate in the biomass decreased fourfold [25]. After reinoculation into Pi-deficient medium, the biomass increment was greater with the inoculum pregrown in medium with higher Pi concentration [25]. This fact confirms the hypothesis that both intracellular and extracellular Pi as a poorly soluble salt performs the function of phosphate reserve for the entire population.
Reserves of phosphate as poorly soluble salts was also revealed in several species of brevibacteria, which during their growth almost completely consumed Pi from the medium at its concentration of about 11 mM (Table 1) [26]. Analysis of phosphorus compounds of the biomass showed also the accumulation of mainly orthophosphate (Table 1). In contrast to the archaea, brevibacteria demonstrated intracellular accumulation of Pi. The reserve phosphorus compound was extracted from B. antiquum cells by high-pressure extrusion. It was identified by X-ray structure analysis as NH4MgPO4·6H2O (International Center for Diffraction Data, 1999, No. 15-0762) [26]. The presence of NH4+ ions was confirmed by infrared spectroscopy, and the content of H2O and NH4 was determined by thermogravimetry [26].
The accumulation of Pi in brevibacterial cells was accompanied by cell shape changes, the appearance of electron-dense zones in the cytoplasm and cell wall, and cell wall thickening (Fig. 2). It seems that cell wall thickening allows these bacteria, in contrast to halophilic archaea, to remain intact in spite of the high degree of mineralization.
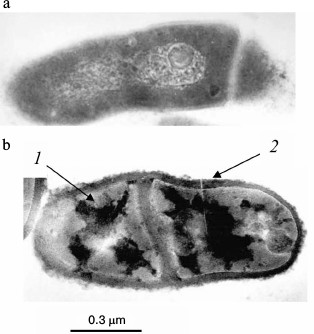
Fig. 2. Ultrathin sections of Brevibacterium antiquum cells [25]: a) cells grown in medium with 0.05 mM Pi; b) cells grown in medium with 11.5 mM Pi: 1) electron dense regions of the cytoplasm; 2) thickened cell wall with electron dense regions.
The cyanobacterium Microcoleus chthonoplastes accumulated polyP in cells up to 1.4% P/g dry biomass when Pi concentration was increased to 0.55 mM; its increase to 1.2 mM resulted in Pi precipitation on the mucous sheaths of the cells and their mineralization [28]. The mineral sheaths of cyanobacteria contain phosphorus and calcium [28]. Increase in Pi concentration to 2.5 mM resulted in trichomes mineralization and cell death. Degradation of the natural cyanobacterial mat is accompanied by destruction of these structures, and Pi released into the medium is sufficient for surviving cyanobacteria [29]. This process is generally similar to mineralization in the culture of halophilic archaea described above. The accumulation of orthophosphate (including extracellular orthophosphate) is also typical of Acetobacter xylinum under carbon deficiency [27].
Inorganic polyphosphates (polyP). In most microorganisms, the role of phosphate reserve is performed by inorganic polyphosphates (polyP), the linear polymers of orthophosphoric acid, containing from three to several hundred of phosphate residues (Fig. 3a) [5]. PolyP, being polymers, have no effect on osmotic pressure and simultaneously are an energy reserve, because the energy of their phosphodiester bond is the same as in the ATP molecule. According to the modern concepts of the role of polyP in microbial cells, phosphate reservation is not the only function; they are involved in the regulation of enzyme activity, the level of expression of many genes, and stress adaptation processes [13, 30-32].
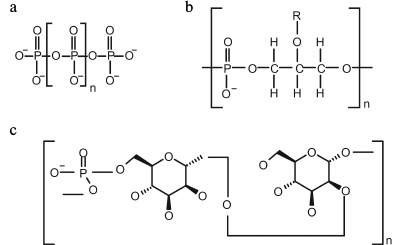
Fig. 3. Structure of inorganic polyphosphate (a), glycerol teichoic acid (b), and phosphomannan of Kuraishia capsulata (c).
The role of polyP as a phosphate reserve has been proven for many microorganisms belonging to different taxa from archaea to fungi [5, 33-35]. The amount of these polymers is lower under phosphate starvation and higher with sufficient phosphate content in the medium. PolyP is rapidly consumed under phosphate starvation even in E. coli characterized by low polyP reserve [36].
Some bacteria are champions in polyphosphate accumulation. For example, polyP was up to 30% of dry biomass in the bacterium A. johnsonii under Pi excess [37]. Corynebacterium glutamicum accumulates up to 600 mM Pi in the cytosol as polyP, and polyP granules can make up to 37% of the cell volume [38]. Representatives of the genera Mycobacteria and Corynebacteria accumulate a large amount of polyP as cytoplasmic granules [5, 13, 38]. It seems that the high ability to accumulate polyP is associated with the fact that the bacteria of this systematic group show a close relationship between the function of polyP as a phosphate reserve and the energy function of these polymers. In addition to polyphosphate kinase, which is the key enzyme of polyP synthesis in prokaryotes [30], representatives of this group of bacteria possess enzymes providing the direct consumption of polyP energy for substrate phosphorylation, such as polyphosphate glucokinase [39, 40], NAD kinase [41, 42], and fructose and mannose kinases [43]. Considerable amounts of polyP are accumulated by bacterial associates from activated sludge of wastewater treatment plants when wastewaters contain excess Pi [44-46].
In most yeast species studied in this respect, the basic reserve phosphorus compound is inorganic polyphosphates [13]. In the typical case of cultivation in complete medium with excess Pi (20 mM), S. cerevisiae cells accumulate little Pi (~94 µmole P/g dry biomass) and much polyP (~658 µmol P/g dry biomass) [47]. PolyP with chain lengths of 3-8 to 200-260 phosphate residues were obtained from yeasts [47]. PolyP has been found in yeasts in most cell compartments [48]. Pi deficiency in the medium causes a decrease in polyP level in S. cerevisiae cells [5, 34, 49]. However, even phosphate-starved cells maintain a low but quite reliable level of polyP [49]. It seems that some polyP in yeast cells performs the function of phosphate reserve, while another smaller fraction of these polymers is responsible for various regulatory functions, e.g. regulation of glucose transferase activity in the cell wall [50], maintenance of negative charge of the cell wall [51], or other hypothetical functions associated with regulation of gene expression [13].
Pi-prestarved S. cerevisiae cells transferred into complete medium accumulate more polyP than cells growing normally in complete medium, i.e. there is a phenomenon of hypercompensation, or “phosphate overplus” [49], which is also known for bacteria [5, 13].
The following yeasts accumulating considerable amounts of polyP were isolated from wastewaters containing excess phosphate: Candida humicola [52, 53], Hansenula fabianii, and Hansenula anomala [54].
Organic phosphorus compounds. Teichoic acids (polymeric compounds of cell walls of Gram-positive bacteria) consist of repeating polyol or glycosyl polyol residues linked by phosphodiester bonds (Fig. 3b). The structures of these polymers are diverse and taxonomically interesting [55]. These polymers can contain up to 30% of the total phosphorus of the cells and are consumed in Pi-deficient medium [56]. The addition of teichoic acid into a phosphate-limited cultivation medium stimulated the growth of Bacillus subtilis [56]. Hence, it was supposed that one of the functions of teichoic acids is phosphate reservation. It has been shown that B. subtilis strains with point mutations in the genes coding for the enzymes of teichoic acid biosynthesis are not viable under phosphate-limiting conditions [57].
This function of teichoic acids is now rarely discussed in the literature and is considered secondary, since it has been shown that these polymers are involved in bacterial cell morphogenesis, regulate activity of autolysins, and participate in processes of adhesion and regulation of ionic composition of the cell wall [58]. However, it should be borne in mind that polyfunctionality is a characteristic feature of the majority of biological macromolecules. It should be remembered that inorganic polyphosphates, being largely a phosphorus reserve, also perform other regulatory functions not always associated with phosphate metabolism.
The yeast Kuraishia (Hansenula) capsulata on medium with excess phosphate accumulates extracellular phosphomannan (Fig. 3c) [59]. Its amount decreases at lower Pi concentrations in the medium [60]. Further evidence of the reserve role of this polymer is the ability of this yeast to utilize phosphomannan from the medium under phosphate starvation [61].
FORMATION OF PHOSPHORUS RESERVES BY MICROORGANISMS UNDER MODEL
LIMITING CONDITIONS
Formation of reserve compounds in microbial cells is usually associated with growth limitation in unbalanced media, when there is an excess of some nutrients but not enough of other nutrients to provide growth and reproduction. It is also typical of the accumulation of reserve phosphorus compounds. Escherichia coli cells accumulate inorganic polyphosphates under amino acid deficiency. The level of guanosine penta- and tetraphosphate, (p)ppGpp, one of the substrates of the gppA polyphosphatase, increases in response to amino acid starvation, resulting in competitive inhibition of polyphosphate hydrolysis by this enzyme [62]. An increase in the level of ATP and polyphosphate synthesis by polyphosphate kinase results from the inhibition of cell growth and division [30].
The diversity of phosphorus reserve formation by microorganisms belonging to different taxonomic groups was comparatively studied in an experimental model where the cells were placed into unified media containing no carbon or nitrogen sources and not maintaining growth (Table 2).
Table 2. Phosphate uptake by microorganisms
under limited conditions (% of initial content in the medium)
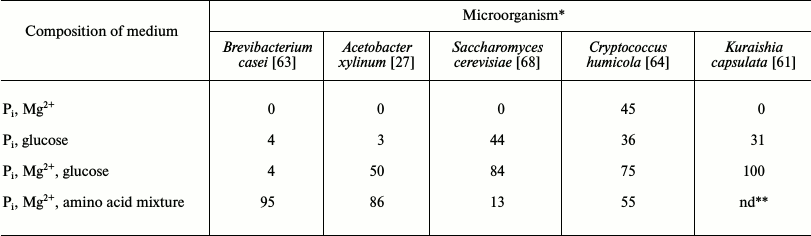
* Incubation time 15 h for B. casei and A. xylinum,
5 h for S. cerevisiae and Cr. humicola, and
24 h for K. capsulata. Concentrations of medium components:
KH2PO4, 5 mM; MgSO4, 5 mM;
glucose, 30 mM; Difco amino acid mixture, 5 g/liter.
** nd, no data.
Phosphate uptake ability was analyzed in this system for the three bacterial species – E. coli, Brevibacterium casei [63], and A. xylinum [27] – and for several species of ascomycetous and basidiomycetous yeasts [64]. This ability proved to be different. E coli cells consumed a small amount of Pi, while B. casei and A. xylinum cells almost completely scavenged Pi from the medium at its initial concentration of 5 mM. Brevibacterium casei and A. xylinum accumulated up to 0.3-0.5 mmol P/g wet biomass. This was close to the accumulation of mineral phosphorus compounds by bacteria isolated from activated sludge of phosphate-contaminated wastewaters: Acinetobacter johnsonii [65], Microlunatus phosphovorus [66], and Rhodocyclus sp. [67].
The analyzed yeasts were shown to contain species with efficient phosphate uptake and species taking up small amounts of Pi [64]. There was no relationship between the taxonomic position of and phosphate uptake by a species. Some species of the Cryptococcus genus (order Sporidiales) consumed little Pi (Cr. terreus), while other species consumed nearly all of it at a concentration of 5 mM (Cr. humicola). Pseudozyma fusiformata, a representative of the order Ustilaginales, also related to basidiomycetes, consumed Pi only twice as poorly as Cr. humicola. Ascomycetes also demonstrated considerable differences in the ability to take up Pi. At the same time, uptake ability comparable to that of Cr. humicola was characteristic of the taxonomically distant S. cerevisiae. This yeast consumed 0.7 and 0.4 mmol Pi/g dry biomass in the complete medium but 2.1 and 1.9-mmol Pi/g dry biomass in the nitrogen-limited medium with Pi and Mg excess.
The compositions of reserve phosphorus compounds were also compared in some microorganisms showing high level of Pi removal from the medium (Table 3). Comparison of the conditions favoring phosphorus reserve formation led to the following conclusions. The bacteria B. casei and A. xylinum form poorly soluble orthophosphate salts under carbon source limitation and nitrogen excess [27, 63]. The indispensable component for such form of phosphorus mineralization is magnesium ions. This is in agreement with features of the functioning of the Pit transport system, which transports phosphate in the form of MeHPO4 under Pi excess. The requirement for magnesium ions is also associated with the chemical structure of the poorly soluble salt formed. Excess of nitrogen as a mixture of amino acids evidently results in the activation of their catabolism and release of ammonium ions. Histidine, arginine, glutamine, or α-ketoglutarate and ammonium sulfate can be added as a nitrogen source instead of amino acid mixture [27, 63]. Ammonium sulfate and magnesium per se did not stimulate the formation of these phosphate reserves, probably because the Pit system depends on the proton gradient across the cytoplasmic membrane.
Table 3. Composition of reserve phosphorus
compounds in representatives of bacteria and yeasts under
nutrient-limited conditions [27, 61, 63, 64,
68]
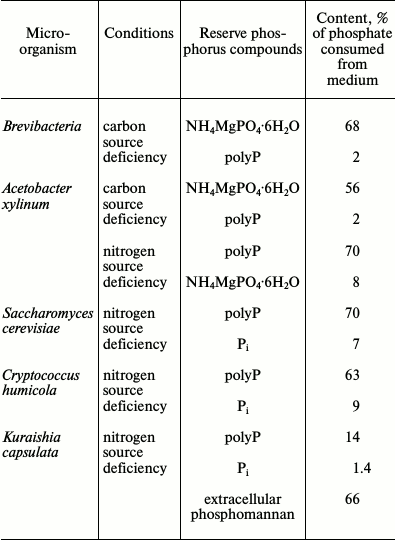
In contrast to brevibacteria, A. xylinum demonstrated the accumulation of phosphate reserves not only under nitrogen limitation, but also under nitrogen deficiency in the presence of glucose. In the latter case, the main phosphate reserve was polyP [27] (Table 3). Electron microscopy shows many electron-dense materials, both bound with the cell surface of these bacteria and lying free in the biomass, which emerged as a result of incubation with the amino acid mixture (Fig. 4). Data obtained during the extraction of phosphorus compounds from the biomass suggest that this material is a poorly soluble orthophosphate. It seems that acetobacteria not only consume Pi but also alkalize the medium by catabolizing some amino acids under carbon deficiency. At the same time, magnesium phosphate forms a precipitate that is adsorbed on the cell surface. The following two ways of phosphate reservation are implemented in A. xylinum depending on environmental conditions: as poorly soluble Pi salts under energy deficiency and nitrogen excess, and as high molecular weight polyP in the presence of an energy source but under nitrogen deficiency.
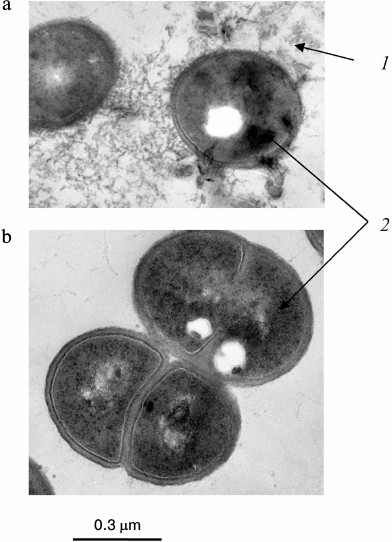
Fig. 4. Ultrathin sections of Acetobacter xylinum cells [27]: a) cells were incubated in medium containing 5 mM KH2PO4, 5 mM MgSO4, and 5 g/liter of amino acid mixture (Casamino acid; Difco) for 15 h; b) cells were incubated in medium containing 5 mM KH2PO4, 5 mM MgSO4, and 30 mM glucose for 15 h: 1) extracellular electron dense material, probably poorly soluble salt of Pi; 2) intracellular electron dense material, probably polyP.
Most of the yeast species used in our model system accumulated polyP, and the necessary condition for accumulation was the presence of glucose as a carbon source. No orthophosphate accumulation was observed (Table 3). Magnesium ions were not an obligatory component for such accumulation; however, they stimulated polyP accumulation (twofold for S. cerevisiae and Cr. humicola). In the presence of magnesium ions, both yeast species demonstrated higher content of polyphosphates with chain lengths of 70 and more phosphate residues and lower content of short-chain polyP with chain lengths of about 15-45 phosphate residues [64].
In yeast cells grown in Pi-depleted medium, the content of polyP was low and the typical electron-dense inclusions were not observed [68]. The S. cerevisiae and Cr. humicola cells that had accumulated polyP in the presence of Mg2+ contained numerous small electron-dense inclusions in the cytoplasm, vacuoles, and mitochondria (Fig. 5). The peculiar feature of these inclusions in S. cerevisiae was their localization close to the cytoplasmic membrane, as well as in association with large electron-transparent inclusions, probably of lipid nature (Fig. 5a). The Cr. humicola cells, which had accumulated polyP in the presence of Mg2+, also contained many small electron-dense inclusions in the cytoplasm (Fig. 5b), vacuoles, especially close to the vacuolar membrane, and mitochondria. The peculiarity of polyP localization in the cells of this yeast species was the presence of large aggregated polyP granules in the cytoplasm and close to the vacuolar membrane and single large granules close to the cell wall. Cr. humicola accumulated longer-chain polyP compared to S. cerevisiae [64].
Fig. 5. Ultrathin sections of the cells of S. cerevisiae (a) and Cr. humicola (b) after 5-h incubation in minimal medium containing 5 mM KH2PO4, 5 mM MgSO4, and 30 mM glucose: 1) vacuole; 2) mitochondria; 3) polyP granules associated with cell envelope; 4) polyP granules associated with lipid inclusions; 5) polyP aggregates in cytoplasm near vacuolar membrane.
The only exception in the studied sample was the yeast not accumulating polyP under carbon excess and nitrogen deficiency (Table 3). It should be noted that K. capsulata completely utilizes Pi at the initial concentration of 5 mM in about 24 h, while most of the yeast species utilize it during 5 h. At the same time, the content of polyP remained low. The incubation medium contained organic compounds, where phosphorus could be detected only after chemical mineralization. When the phosphomannan preparation was obtained from the incubation medium by precipitation with Cetavlon [61], it was shown to contain the major portion of Pi consumed from the medium. The resultant phosphomannan fraction contained 3.35 µmole of total phosphorus per g of the preparation (~57% of total phosphorus in the incubation medium) and contained neither Pi nor labile phosphorus. These data suggest that the extracellular phosphomannan of K. capsulata is a secondary metabolite formed under nitrogen deficiency as well as phosphorus and carbon excess.
The experiments with simulated limiting conditions made it possible to estimate the diversity of structure, localization, and peculiarities of formation of reserve phosphorus compounds in microorganisms from different taxa.
ROLE OF PHOSPHATE-RESERVING MICROORGANISMS IN NATURAL AND
TECHNOGENIC ECOLOGICAL NICHES
Water bodies and bottom sediments. The concentration of Pi in natural water reservoirs, including the ocean, is usually too low to provide the primary formation of calcium phosphates from solution (nucleation) [69]. However, the formation of such minerals does occur in some water bodies. Many data demonstrate that microorganisms are primarily responsible for assimilation and remineralization of phosphorus in the ocean [69-71]. Many microorganisms inhabiting the ocean are able to concentrate Pi as intracellular polyP under conditions when oxygen is available (in surface water layers). This is followed by utilization of the polyP as an energy source under anaerobic conditions (in bottom sediments), release of Pi, increase in its local concentration, and precipitation of apatite in calcium-rich seawater [69-71]. Such release and hydrolysis of polyP can occur after cell death in the bottom sediments [69-71]. This process is supposed to involve marine bacteria belonging to the genera Pseudomonas and Acinetobacter [69], as well as the sulfide-oxidizing bacteria Beggiatoa and Thiomargarita [72-74] that form bacterial mats. It has been shown that Pi concentration increases to 300 µM in oceanic bottom sediments containing up to 25% hydroxyapatite and inhabited with Thiomargarita, but is usually below 1 µM in ocean water [71]. These bacteria were shown to accumulate phosphates under aerobic conditions, while phosphate release from the cells and apatite formation were observed under anaerobic conditions [73]. Beggiatoa and Thiomargarita accumulated polyP in the presence of sulfur and nitrate [74]. Under laboratory conditions, polyP in Beggiatoa cells was depolymerized at higher sulfide concentrations and under oxygen deficiency, while Pi was released into the medium [74].
Diatoms are also capable of polyP accumulation [75]. PolyP granules found in bottom sediments are similar in size to those found in diatoms. It is supposed that the accumulated polyP enters bottom sediments after the death of diatom cells and destruction of their silicate cell walls; then Pi is released by alkaline phosphatase localized on the cell surface [75].
Novel genetic and bioinformatics approaches have made it possible to ascertain the broad distribution of the ppk1 and ppk2 genes coding for polyphosphate kinases and the ppx gene coding for polyphosphatase among marine oligotrophic microorganisms living under Pi deficiency [76]. These data are evidence in favor of the global spreading of phosphorus concentration as polyP by microorganisms in the World Ocean.
Activated sludge of wastewater treatment plants. Recently, polyP has been used worldwide as a component of detergents. The hydrolysis of polyP by microbial enzymes results in excessive release of Pi into wastewater and hence causes eutrophication. The use of polyP in detergents is now banned in Europe and the USA; however, according to the literature data, wastewaters contain 4-12 mg P/liter, most often as Pi, which is easily utilized by microorganisms including cyanobacteria [77]. Therefore, the problem of wastewater purification from excess Pi is still relevant. The basic microbiological approach to wastewater purification from excess Pi is so-called Enhanced Biological Phosphorus Removal (EBPR). Numerous studies describing the microbiota of activated sludge and the mechanisms of Pi uptake and accumulation of reserve phosphorous compounds by the microorganisms involved in EBPR are generalized in a great number of reviews. Hence, let us give but a few references to relatively recent reviews [77-80]. The role of polyP accumulation by sludge bacteria during wastewater purification from excessive phosphates was demonstrated relatively long ago [81-86]. The engineering design process was created still earlier. In treatment plants successively used in some countries, the content of Pi in wastewaters is minimized by activated sludge microorganisms. Technologies have been developed for using the phosphates accumulated in sludge as phosphate fertilizers [78]. The main problem in this respect is the high content of heavy metals and xenobiotics in sludge of wastewater treatment plants.
Hence, the microbiota of activated sludge of treatment plants is complex and the process of phosphate absorption depends on many factors including the composition of microbial associations and wastewater composition, and thus investigation of the mechanisms of Pi removal from the dissolved phase remains an urgent problem. Wastewater purification from phosphate needs alternation of anaerobic and aerobic conditions, which is achieved most often via the serial arrangement of anaerobic and aerobic zones in a series of flow-through systems, with sludge returning into the cycle. In the anaerobic stage, the activated sludge bacteria take up the organic substrates of wastewaters. Intracellular polyP is used as an energy source, while Pi is released into the medium. Such conditions favor the accumulation of polyhydroxybutyrate (PHB) and other polyhydroxyalkanoates (PHA). It is considered that the bacteria accumulating large amounts of polyP have a selective advantage in the anaerobic zone. In the aerobic zone, PHA is hydrolyzed, ATP is synthesized, and the sludge consumes more Pi than has been released in the previous aerobic stage. The Pi scavenged from wastewaters accumulates in bacterial cells as a large amount of polyP. A certain amount of phosphate is retained also by extracellular polymers associated with the agglomerations of microbial cells [87]. Phosphorus-enriched sludge is then removed from the system.
The first pure cultures isolated from EBPR systems and accumulating large amounts of polyP were different strains of Acinetobacter sp. [81]. Later, bacteria of numerous systematic groups were isolated from activated sludge; novel species and genera were described. They include Microlunatus phosphovorus [88], Rhodocyclus sp. [46], corynebacteria [89], Microthrix parvicella [90], Tetracoccus cechii [91], Tetrasphaera [92, 93], Gemmatimonas aurantiaca [94], and Accumulibacter phosphatis [95]. Several yeast species were also found in activated sludge [52-54]. Sludge was also shown to contain many uncultured species, which are identified by molecular biological and fluorescence methods [80]. Specific dyes for polyP and PHA make it possible to evaluate the ability to accumulate these biopolymers directly in sludge preparations.
By modeling EBPR in laboratory reactors, it has been concluded that the process cannot be performed completely by any individual microbial species. It is now believed that the composition of the microbial communities performing EBPR is determined by wastewater chemical composition, temperature, pH, and other factors. EBPR water treatment plants are unique technogenic ecological niches whose peculiarities are determined just by the presence of anaerobic and aerobic zones, with different bacterial species or associations gaining selective advantage in each zone [77-80]. Regarding biochemical peculiarities, they contain PAO (Polyphosphate-Accumulating Microorganisms): bacteria accumulating large amounts of polyP. They were believed to inhibit phosphate transport under nitrate excess. Later, sludge was shown to contain DPAO (Denitrifying Polyphosphate Accumulating Organisms), which can simultaneously take up phosphate and reduce nitrate in the absence of oxygen, utilizing nitrate as an electron acceptor. The next group is the so-called GAO (Glycogen-Accumulating Microorganisms), which accumulate glycogen under aerobic conditions and compete with PAO for the carbon source [77-80]. The competition between PAO and GAO and its dependence on temperature, pH, carbon source accessibility, and the Pi and acetate ratio in wastewaters have been described in detail in a review [80]. However, the problem of stable EBPR is still far from solved.
Among the fundamental problems associated with the application of polyP-accumulating microorganisms in wastewater bioremediation, we should mention, first, the problem of directed development of the most productive microbial consortia for phosphate scavenge from wastewaters; second, substantial dependence of phosphate uptake efficiency on wastewater composition; and, third, the problem of further application of activated sludge. It is obvious that polyP-accumulating microorganisms are the key element of excess phosphate uptake from wastewaters, and further studies of polyP metabolism are important for the development of improved variants of biotechnologies for wastewater purification.
Mycorrhiza. Some observations demonstrate that mycorrhiza contains large amounts of Pi and polyP. A substantial amount of polyP was detected by X-ray microassay in vacuoles of the fungus Pisolithus tinctorius in the ectomycorrhiza formed with the roots of Eucalyptus pilularis [96]. Microsclerotia of the fungus Phialocephala fortinii accumulated polyP at an early stage of interaction with the roots of Asparagus officinalis [97]. Studies in obligate mycorrhizal fungi have shown that polyP is accumulated in fungal cells and then locally hydrolyzed to supply phosphate to symbiotic plants [98]. The content of polyP in the fungus varies during mycorrhiza development and can be used as an activity indicator of the fungus as a phosphate supplier for the plant [98]. The obligate mycorrhizal fungi have recently been shown to have a polyP-synthetase activity in the presence of ATP [99]. Mycorrhizal fungi play a key role in phosphorus supply to symbiotic plants. It is associated with the ability of fungal cells to concentrate Pi from soil, to dissolve poorly soluble mineral phosphorus compounds due to organic acid excretion into the medium, and to accumulate polyP [100].
CONCLUSION
The data on the diversity of phosphate reserves in microorganisms suggest that most often they are present as mineral compounds. Organic phosphorus reserve compounds occur rarely. The formation of mineral phosphorus compounds is related to the phenomenon termed biological mineralization, or biomineralization. Biological molecules are matrices or catalysts for the formation of mineral compounds in living cells; therefore, these compounds are characterized by structural peculiarities different from minerals of abiotic origin. The term “phosphate mineral nucleation”, which is also accepted in the literature, denotes the case when the local increase in the concentrations of phosphate and metal cations (especially calcium) results in the initial formation of apatite crystals, while further formation of this biomineral is controlled by specific proteins [70].
Some pathways of phosphorus biomineralization have been maintained during the evolution from prokaryotes to the higher eukaryotes. They are observed primarily in mitochondria, which, according to the endosymbiotic theory, originated from ancient bacteria. Electron-dense granules (so-called “dense granules”) with high Ca and P concentrations were found in rat liver mitochondria as early as in 1964 [70]. It was unclear why crystalline apatite was not formed in these granules. However, later it was shown that such granules contained not Pi, but polyP [70]. They have been found in protozoa and in mammals: in special cell organelles (acidocalcisomes) [101] and in the platelets [102] and mitochondria of bone tissue cells [103], respectively. PolyP was also shown to participate in calcium homeostasis and transport across the membrane in mitochondria of other mammalian tissues [104].
To date, ideas of the role of polyP in bone tissues are in brief as follows [103, 105-107]. Mitochondria accumulate calcium and polyP in osteoclasts, forming dense granules. Because of exocytosis, these granules are released into the extracellular space in the place of bone growth or repair. Here, the granules are destroyed and alkaline phosphatase hydrolyzes polyP and releases Pi. With the involvement of osteoblast-specific proteins, structured bone apatite is formed from the released Pi and calcium. There are still many unclear aspects in this process. It is not known what enzymes are responsible for polyP synthesis in mitochondria, because the gene of the typical polyphosphate kinase responsible for polyP synthesis in bacteria has not been found in mammals [32]. Also, it is not known what signals cause the release of polyP granules from osteoclasts.
PolyP-rich granules have also been found in platelets [102]. On destruction of platelets, polyP is released into blood, where it is involved in the coagulation cascade, being bound by factor XII and activating it, and then polyP and calcium ions enter the thrombus to increase its stability [108-112].
There is an evolutionarily significant analogy between phosphorus mineralization in microorganisms and bone apatite formation and individual stages of clotting in mammals. The most pronounced similarity is observed between the formation of sedimentary apatites in water bodies with the involvement of microorganisms and the formation of bone tissue apatite in mammals:
– individual stages of these processes are characterized by predominance of either uptake of phosphorus mineral compounds from the medium or their release from cells (and/or release from cells in case of death);
– phosphate concentration from the medium is accompanied by local accumulation of inorganic polyphosphates in the cells;
– under varying environmental conditions or cell death, polyP is released into the extracellular medium and hydrolyzed by phosphatases; apatite is formed from the released Pi in the presence of calcium ions.
The study of phosphate reserves in microorganisms, their structure, and conditions of formation and destruction is significant not only for understanding phosphorus turnover in the biosphere, but also for modeling normal and pathological processes in the human organism associated with phosphate metabolism.
The authors are grateful to Dr. N. E. Suzina (Skryabin Institute of Biochemistry and Physiology of Microorganisms, Russian Academy of Sciences) for the electron micrographs.
This work was supported by the Program of Presidium of the Russian Academy of Sciences “Problems of Biosphere Origin and Evolution”.
REFERENCES
1.Miller, S. L., and Parris, M. (1964) Synthesis of
pyrophosphate under primitive earth conditions, Nature,
204, 1248-1249.
2.Beck, A., and Orgel, L. E. (1965) The formation of
condensed phosphate in aqueous solution, Proc. Natl. Acad. Sci.
USA, 54, 664-669.
3.Lohmann, R., and Orgel, L. E. (1968) Prebiotic
synthesis: phosphorylation in aqueous solution, Science,
161, 64-66.
4.Osterberg, R., and Orgel, L. E. (1972)
Polyphosphate and trimetaphosphate formation under potentially
perbiotic conditions, J. Mol. Evol., 1, 241-252.
5.Kulaev, I. S. (1975) Biochemistry of Inorganic
Polyphosphates [in Russian], MGU Publishers, Moscow.
6.Miller, S. L. (1986) Current status of the
prebiotic synthesis of small molecules, Chem. Scripta,
26B, 5-11.
7.Oro, J., Miller, S. L., and Lazcano, A. (1990) The
origin and early evolution of life on Earth, Ann. Rev. Earth Planet
Sci., 18, 317-356.
8.Kornberg, A. (1995) Inorganic polyphosphate: toward
making a forgotten polymer unforgettable, J. Bacteriol.,
177, 491-496.
9.Arrhenius, G., Sales, B., Mojzsis, S., and Lee, T.
(1997) Entropy and charge in molecular evolution: the case of
phosphate, J. Theor. Biol., 187, 503-522.
10.Baltscheffsky, H. (1997) Major
“Anastrophes” in the origin and early evolution of
biological energy conversion, J. Theor. Biol.,
187, 495-501.
11.De Graaf, R. M., and Schwartz, A. W. (2000)
Reduction and activation of phosphate on the primitive earth, Origin
Life Evol. Biospheres, 30, 405-410.
12.Spirin, A. S. (2001) Protein biosynthesis, the
world of RNA, and life origin, Vestnik RAN, 71,
146-153.
13.Kulaev, I. S., Vagabov, V. M., and Kulakovskaya,
T. V. (2005) High Molecular Weight Inorganic Polyphosphates:
Biochemistry, Cell Biology, Biotechnology [in Russian], Nauchnyi
Mir, Moscow.
14.Galimov, E. M. (2006) Phenomenon of Life.
Between Equilibrium and Nonlinearity. Origin and Principles of
Evolution [in Russian], URSS Publisher, Moscow.
15.Cavalier-Smith, T. (2006) Cell evolution and
Earth history: stasis and revolution, Philos. Trans. R. Soc. Lond.
B. Biol. Sci., 361, 969-1006.
16.Wolfe-Simon, F., Switzer Blum, J., Kulp, T. R.,
Gordon, G. W., Hoeft, S. E., Pett-Ridge, J., Stolz, J. F., Webb, S. M.,
Weber, P. K., Davies, P. C., Anbar, A. D., and Oremland, R. S. (2011) A
bacterium that can grow by using arsenic instead of phosphorus,
Science, 332, 1163-1166.
17.Erb, T. J., Kiefer, P., Hattendorf, B., Gunther,
D., and Vorholt, J. A. (2012) GFAJ-1 is an arsenate-resistant,
phosphate-dependent organism, Science, 337, 467-470.
18.Rao, N. N., and Torriani, A. (1990) Molecular
aspects of phosphate transport in Escherichia coli, Mol.
Microbiol., 4, 1083-1090.
19.Van Veen, H. W., Abee, T., Kortstee, G. J. J.,
Konings, W. N., and Zehnder, A. J. B. (1994) Translocation of
metal phosphate via the phosphate inorganic transport system of
Escherichia coli, Biochemistry, 33, 1766-1770.
20.Harris, R. M., Webb, D. C., Howitt, S. M., and
Cox, G. B. (2001) Characterization of PitA and PitB from Escherichia
coli, J. Bacteriol., 183, 5008-5014.
21.Spira, B., Aguena, M., de Castro Oliveira, J. V.,
and Yagil, E. (2010) Alternative promoters in the pst operon of
Escherichia coli, Mol. Genet. Genom., 284,
489-498.
22.Persson, B. L., Lagerstedt, J. O., Pratt, J. R.,
Pattison-Granberg, J., Lundh, K., Shokrollahzadeh, S., and Lundh, F.
(2003) Regulation of phosphate acquisition in Saccharomyces
cerevisiae, Curr. Genet., 43, 225-244.
23.Dick, C. F., Dos-Santos, A. L., and
Meyer-Fernandes, J. R. (2014) Inorganic phosphate uptake in
unicellular eukaryotes, Biochim. Biophys. Acta, 1840,
2123-2127.
24.Smirnov, A. V., Suzina, N. E., Kulakovskaya, T.
V., and Kulaev, I. S. (2002) Magnesium orthophosphate – the novel
form of phosphate reservation in the halophilic archaeon
Halobacterium salinarium, Mikrobiologiya, 71,
786-793.
25.Smirnov, A. V. (2003) Phosphate Uptake and
Reservation by Some Archaea and Bacteria: Candidate’s
dissertation [in Russian], Pushchino.
26.Smirnov, A., Suzina, N., Chudinova, N.,
Kulakovskaya, T., and Kulaev, I. (2005) Formation of insoluble
phosphate during growth of the archae Halorubrum distributum and
Halobacterium salinarium and the bacterium Brevibacterium
antiquum, FEMS Microbiol. Ecol., 52, 129-137.
27.Ryazanova, L. P., Suzina, N. E., Kulakovskaya, T.
V., and Kulaev, I. S. (2009) Phosphate accumulation of Acetobacter
xylinum, Arch. Microbiol., 191, 467-471.
28.Gerasimenko, L. M., Goncharova, I. V., and
Zaytseva, L. V. (1998) The influence of phosphorus content in the
medium on cyanobacterial growth and mineralization,
Mikrobiologiya, 67, 249-254.
29.Goncharova, I. V., and Gerasimenko, L. M. (1993)
The dynamics of inorganic phosphorus uptake by the cells of
Microcoleus chthonoplastes, Mikrobiologiya,
62, 1048-1055.
30.Kornberg, A., Rao, N. N., and Ault-Riche, D.
(1999) Inorganic polyphosphate: a molecule with many functions, Ann.
Rev. Biochem., 68, 89-125.
31.Reusch, R. N. (2000) Transmembrane ion transport
by polyphosphate/poly-(R)-3-hydroxybutyrate complexes, Biochemistry
(Moscow), 65, 280-295.
32.Rao, N. N., Gomez-Garcia, M. R., and Kornberg, A.
(2009) Inorganic polyphosphate: essential for growth and survival,
Ann. Rev. Biochem., 78, 605-647.
33.Harold, F. M. (1966) Inorganic polyphosphates in
biology: structure, metabolism, and functions, Bacteriol. Rev.,
30, 772-794.
34.Kulaev, I. S., and Vagabov, V. M. (1983)
Polyphosphate metabolism in microorganisms, Adv. Microbiol.
Physiol., 24, 83-171.
35.Wood, H. G., and Clark, J. E. (1988) Biological
aspects of inorganic polyphosphates, Ann. Rev. Biochem.,
57, 235-260.
36.Nesmeyanova, M. A. (2000) Polyphosphates and
enzymes of polyphosphate metabolism in Escherichia coli,
Biochemistry (Moscow), 65, 309-314.
37.Deinema, M. H., Habers, L. H. A., Scholten, J.,
Turkstra, E., and Webers, H. A. A. M. (1980) The accumulation of
polyphosphate in Acinetobacter spp., FEMS Microbiol.
Lett., 9, 275-279.
38.Lindner, S. N., Knebel, S., Pallerla, S. R.,
Schoberth, S. M., and Wendisch, V. F. (2010) Cg2091 encodes a
polyphosphate/ATP-dependent glucokinase of Corynebacterium
glutamicum, Appl. Microbiol. Biotechnol., 87,
703-713.
39.Szymona, M. (1957) Utilization of inorganic
polyphosphates for phosphorylation of glucose in Micobacterium
phlei, Bull. Acad. Pol. Sci. Ser. Sci. Biol.,
5, 379-381.
40.Hsieh, P. C., Shenoy, B. C., Samols, D., and
Phillips, N. F. B. (1996) Cloning, expression and characterization of
polyphosphate glucokinase from Mycobacterium tuberculosis, J.
Biol. Chem., 271, 4909-4915.
41.Kawai, S., Mori, S., Mukai, T., Suzuki, S.,
Yamada, T., Hashimoto, W., and Murata, K. (2000) Inorganic
polyphosphate/ATP-NAD kinase of Micrococcus flavus and
Mycobacterium tuberculosis H37Rv, Biochem. Biophys. Res.
Commun., 276, 57-63.
42.Mori, S., Yamasaki, M., Maruyama, Y., Momma, K.,
Kawai, S., Hashimoto, W., Mikami, B., and Murata, K. (2004)
Crystallographic studies of Mycobacterium tuberculosis
polyphosphate/ATP-NAD kinase complexed with NAD, J. Biosci.
Bioeng., 98, 391-393.
43.Mukai, T., Kawai, S., Matsukawa, H., Matuo, Y.,
and Murata, K. (2003) Characterization and molecular cloning of a novel
enzyme, inorganic polyphosphate/ATP-glucomannokinase, of
Arthrobacter sp. strain KM, Appl. Environ. Microbiol.,
69, 3849-3857.
44.Kortstee, G. J. J., Appeldoorn, K. J., Bonting,
C. F. C., van Niel, E. W. J., and van Veen, H. W. (2000) Recent
developments in the biochemistry and ecology of enhanced biological
phosphorus removal, Biochemistry (Moscow), 65,
332-340.
45.Mino, T. (2000) Microbial selection of
polyphosphate-accumulating bacteria in activated sludge wastewater
treatment processes for enhanced biological phosphate removal,
Biochemistry (Moscow), 65, 341-348.
46.Keasling, J. D., van Dien, S. J., Trelstad, P.,
Renninger, N., and McMahon, K. (2000) Application of polyphosphate
metabolism to environmental and biotechnological problems,
Biochemistry (Moscow), 65, 324-331.
47.Vagabov, V. M., Trilisenko, L. V., Shchipanova,
I. N., Sibeldina, L. A., and Kulaev, I. S. (1998) Variation of
inorganic polyphosphate chain length depending on Saccharomyces
cerevisiae growth stage, Mikrobiologiya, 67,
193-198.
48.Lichko, L., Kulakovskaya, T., Pestov, N., and
Kulaev, I. (2006) Inorganic polyphosphates and exopolyphosphatases in
cell compartments of the yeast Saccharomyces cerevisiae under
inactivation of PPX1 and PPN1 genes, Biosci. Rep., 26,
45-54.
49.Vagabov, V. M., Trilisenko, L. V., and Kulaev, I.
S. (2000) Dependence of inorganic polyphosphate chain length on the
orthophosphate content in the culture medium of the yeast
Saccharomyces cerevisiae, Biochemistry (Moscow),
65, 349-354.
50.Kalebina, T. S., Egorov, S. N., Arbatsky, N. P.,
Bezsonov, E. E., Gorkovsky, A. A., and Kulaev, I. S. (2008) On the role
of high molecular polyphosphates in the activation of glucan
transferase Bgl2p from the cell wall of the yeast Saccharomyces
cerevisiae, Dokl. Akad. Nauk, 420, 695-698.
51.Ivanov, A. Yu., Vagabov, V. M., Fomchenkov, V.
M., and Kulaev, I. S. (1996) Investigation of the influence of cell
wall polyphosphates on sensitivity of the yeast Saccharomyces
carlsbergensis to the damage by cetyl trimethyl ammonium bromide,
Mikrobiologiya, 65, 611-616.
52.McGrath, J. W., and Quinn, J. P. (2000)
Intracellular accumulation of polyphosphate by the yeast Candida
humicola G-1 in response to acid pH, Appl. Environ.
Microbiol., 66, 4068-4073.
53.McGrath, J. W., Kulakova, A. N., Kulakov, L. A.,
and Quinn, J. P. (2005) In vitro detection and characterization
of a polyphosphate synthesizing activity in the yeast Candida
humicola G-1, Res. Microbiol., 156, 485-491.
54.Watanabe, T., Ozaki, N., Iwashita, K., Fujii, T.,
and Lefuji, H. (2008) Breeding of wastewater treatment yeasts that
accumulate high concentration of phosphorus, Appl. Microbiol.
Biotechnol., 80, 331-338.
55.Potekhina, N. V., Streshinskaya, G. M.,
Tul’skaya, E. M., Kozlova, Yu. I., Senchenkova, S. N., and
Shashkov, A. S. (2011) Phosphate-containing cell wall polymers of
bacilli, Biochemistry (Moscow), 76, 745-754.
56.Grant, W. D. (1979) Cell wall teichoic acid as a
reserve phosphate source in Bacillus subtilis, J.
Bacteriol., 137, 35-43.
57.Bhavsar, A. P., Erdman, L. K., Schertzer, J. W.,
and Brown, E. D. (2004) Teichoic acid is an essential polymer in
Bacillus subtilis that is functionally distinct from teichuronic
acid, J. Bacteriol., 186, 7865-7873.
58.Brown, S., Santa Maria, J. P., Jr., and Walker,
S. (2013) Wall teichoic acids of gram-positive bacteria, Ann. Rev.
Microbiol., 67, 313-336.
59.Slodki, M. E. (1963) Structure of Hansenula
capsulata NRRL Y-1842 phosphomannan, Biochim. Biophys. Acta,
69, 96-102.
60.Avigad, G., and Kalina, M. (1979) Effect of
orthophosphate limitation on the production of phosphomannan by
Hansenula capsulata, FEMS Microbiol. Lett., 6,
111-114.
61.Lichko, L. P., Kulakovskaya, T. V., and Kulaev,
I. S. (2013) Extracellular phosphomannan as a phosphate reserve in the
yeast Kuraishia capsulata, Biochemistry (Moscow),
78, 674-677.
62.Kuroda, A. (2006) A polyphosphate–Lon
protease complex in the adaptation of Escherichia coli to amino
acid starvation, Biosci. Biotechnol. Biochem., 70,
325-331.
63.Ryazanova, L. P., Smirnov, A. V., Kulakovskaya,
T. V., and Kulaev, I. S. (2007) Reduction of phosphate concentration in
the medium by the cells of Brevibacterium casei,
Mikrobiologiya, 76, 752-758.
64.Breus, N. A., Ryazanova, L. P., Dmitriev, V. V.,
Kulakovskaya, T. V., and Kulaev, I. S. (2012) Accumulation of phosphate
and polyphosphate by Cryptococcus humicola and Saccharomyces
cerevisiae in the absence of nitrogen, FEMS Yeast Res.,
12, 617-624.
65.Van Niel, E. W. J., De Best, J. H., Kets, E. P.
W., Bonting, C. F. C., and Kortstee, G. J. J. (1999) Polyphosphate
formation by Acinetobacter johnsonii 210A: effect of cellular
energy status and phosphate-specific transport system, Appl.
Microbiol. Biotechnol., 51, 639-646.
66.Santos, M. M., Lemos, P. C., Reis, M. A. M., and
Santos, H. (1999) Glucose metabolism and kinetics of phosphorus removal
by the fermentative bacterium Microlunatus phosphovorus,
Appl. Environ. Microbiol., 65, 3920-3928.
67.Zilles, J. L., Peccia, J., Kim, M. W., Hung, C.
H., and Noguera, D. R. (2002) Involvement of Rhodocyclus-related
organisms in phosphorus removal in full scale wastewater treatment
plants, Appl. Environ. Microbiol., 68,
2763-2769.
68.Breus, N. A., Ryazanova, L. P., Suzina, N. E.,
Kulakovskaya, T. V., Valiakhmetov, A. Ya., Yashin, V. A., Sorokin, V.
V., and Kulaev, I. S. (2010) Accumulation of inorganic polyphosphates
in Saccharomyces cerevisiae under nitrogen deficiency:
stimulation by magnesium ions and peculiarities of localization,
Mikrobiologiya, 80, 612-618.
69.Diaz, J., Ingall, E., Benitez-Nelson, C.,
Paterson, D., de Jonge, M. D., McNulty, I., and Brandes, J. A. (2008)
Marine polyphosphate: a key player in geologic phosphorus
sequestration, Science, 320, 652-655.
70.Omelon, S., Ariganello, M., Bonucci, E., Grynpas,
M., and Nanci, A. (2013) A review of phosphate mineral nucleation in
biology and geobiology, Calcif. Tissue Int., 93,
382-396.
71.Karl, D. M. (2014) Microbially mediated
transformations of phosphorus in the sea: new views of an old cycle,
Ann. Rev. Marine Sci., 6, 279-337.
72.Schulz, H. N., and Schulz, H. D. (2005) Large
sulfur bacteria and the formation of phosphorite, Science,
307, 416-418.
73.Goldhammer, T., Bruchert, V., Ferdelman, T. G.,
and Zabel, M. (2010) Microbial sequestration of phosphorus in anoxic
upwelling sediments, Nat. Geosci., 3, 557-561.
74.Brock, J., and Schulz-Vogt, H. N. (2011) Sulfide
induced phosphate release from polyphosphate in cultures of marine
Beggitoa strain, ISME J., 5, 497-506.
75.Dyhrman, S. T., Jenkins, B. D., Rynearson, T. A.,
Saito, M. A., Mercier, M. L., Alexander, H., Whitney, L. P.,
Drzewianowski, A., Bulygin, V. V., Bertrand, E. M., Wu, Z.,
Benitez-Nelson, C., and Heithoff, A. (2012) The transcriptome and
proteome of the diatom Thalassiosira pseudonana reveal a diverse
phosphorus stress response, PLoS One, 7, e33768; DOI:
0.1371/journal.pone.0033768.
76.Temperton, B., Gilbertm, J. A., Quinn, J. P., and
McGrath, J. W. (2011) Novel analysis of oceanic surface water
metagenomes suggests importance of polyphosphate metabolism in
oligotrophic environments, PLoS One, 6, e16499; DOI:
10.1371/journal.pone.0016499.
77.McMahon, K. D., and Read, E. K. (2013) Microbial
contribution to phosphorus cycling in eutrophic lakes and wastewater,
Ann. Rev. Microbiol., 67, 199-219.
78.Hirota, R., Kuroda, A., Kato, J., and Ohtake, H.
(2010) Bacterial phosphate metabolism and its application to phosphorus
recovery and industrial bioprocesses, J. Biosci. Bioeng.,
109, 423-432.
79.Yuan, Z., Pratt, S., and Batstone, D. J. (2012)
Phosphorus recovery from wastewater through microbial processes,
Curr. Opin. Biotechnol., 23, 878-883.
80.Gebremariam, S. Y., Beutel, M. W., Christian, D.,
and Hess, T. F. (2011) Research advances and challenges in the
microbiology of enhanced biological phosphorus removal – a
critical review, Water Environ. Res., 83, 195-219.
81.Fuhs, G. W., and Chen, M. (1975) Microbiological
basis of phosphorus removal in the activated sludge process for the
treatment of wastewaters, Microb. Ecol., 2, 119-138.
82.Mino, T., Kawakami, T., and Matsuo, T. (1985)
Location of phosphorus in activated sludge and function of
intracellular polyphosphates in biological phosphorus removal process,
Water Sci. Technol., 17, 93-106.
83.Toerien, D. F., Gerber, A., Lotter, L. H., and
Cloete, T. E. (1990) Enhanced phosphorus removal systems in activated
sludge systems, Adv. Microb. Ecol., 11, 173-230.
84.Seviour, R. J., and Blackall, L. L. (eds.) (1999)
The Microbiology of Activated Sludge, Kluwer Academic
Publishing, Boston.
85.Blackall, L. L., Crocetti, G. R., Saunders, A.
M., and Bond, P. L. (2002) A review and update of the microbiology of
enhanced biological phosphorus removal in wastewater treatment plants,
Antonie Van Leeuwenhoek, 81, 681-691.
86.Kortstee, G. J. J., Appeldoorn, K. J., Bonting,
C. F. C., van Niel, E. W. J., and van Veen, H. W. (1994) Biology of
polyphosphate accumulating bacteria, involved in enhanced biological
phosphorus removal, FEMS Microbiol. Rev., 15,
137-153.
87.Cloete, T. E., and Oosthuizen, D. J. (2001) The
role of extracellular exopolymers in the removal of phosphorus from
activated sludge, Water Res., 35, 3595-3598.
88.Nakamura, K., Hiraishi, A., Yoshimi, Y.,
Kawaharasaki, M., Masuda, K., and Kamagata, Y. (1995). Microlunatus
phosphovorus gen. nov. sp. nov., a new gram-positive
polyphosphate-accumulating bacterium isolated from activated sludge,
Int. J. System. Bacteriol., 45, 17-22.
89.Bark, K., Kampfer, P., Sponner, A., and Dott, W.
(1993) Polyphosphate-dependent enzymes in some coryneform bacteria
isolated from sewage sludge, FEMS Microbiol. Lett., 107,
133-138.
90.Erhart, R., Bradford, D., Sevior, R. J., Amann,
R., and Blackall, L. L. (1997) Development and use of fluorescent in
situ hybridization probes for the detection and identification of
Microtrix parvicella in activated sludge, Syst. Appl.
Microbiol., 20, 310-318.
91.Blackall, L. L., Crocetti, G. R., Saunders, A.
M., and Bond, P. L. (2002) A review and update of the microbiology of
enhanced biological phosphorus removal in wastewater treatment plants,
Antonie Van Leeuwenhoek, 81, 681-691.
92.Maszenan, A. M., Seviour, R. J., Patel, B. K.,
Schumann, P., Burghardt, J., Tokiwa, Y., and Stratton, H. M. (2000)
Three isolates of novel polyphosphate-accumulating gram-positive cocci,
obtained from activated sludge, belong to a new genus,
Tetrasphaera gen. nov., and description of two new species,
Tetrasphaera japonica sp. nov. and Tetrasphaera
australiensis sp. nov., Int. J. Syst. Evol. Microbiol.,
50, 593-603.
93.Hanada, S., Liu, W. T., Shintani, T., Kamagata,
Y., and Nakamura, K. (2002) Tetrasphaera elongata sp. nov., a
polyphosphate-accumulating bacterium isolated from activated sludge,
Int. J. Syst. Evol. Microbiol., 52, 883-887.
94.Zhang, H., Sekiguchi, Y., Hanada, S., Hugenholtz,
P., Kim, H., Kamagata, Y., and Nakamura, K. (2003) Gemmatimonas
aurantiaca gen. nov., sp. nov., a gram-negative, aerobic,
polyphosphate-accumulating microorganism, the first cultured
representative of the new bacterial phylum Gemmatimonadetes phyl. nov.,
Int. J. Syst. Evol. Microbiol., 53, 1155-1163.
95.Liu, W. T., Nielsen, A. T., Wu, J. H., Tsai, C.
S., Matsuo, Y., and Molin, S. (2001) In situ identification of
polyphosphate- and polyhydroxyalkanoate-accumulating traits for
microbial populations in a biological phosphorus removal process,
Environ. Microbiol., 3, 110-122.
96.Ashford, A. E., Vesk, P. A., Orlovich, D. A.,
Markovina, A. L., and Allaway, W. G. (1999) Dispersed polyphosphate in
fungal vacuoles in Eucalyptus pilularis/Pisolithus tinctorius
ectomycorrhizae, Fungal Genet. Biol., 28, 21-33.
97.Yu, T., Nassuth, A., and Peterson, R. L. (2001)
Characterization of the interaction between the dark septate fungus
Phialocephala fortinii and Asparagus officinalis roots,
Can. J. Microbiol., 47, 741-753.
98.Ohtomo, R., and Saito, M. (2005) Polyphosphate
dynamics in mycorrhizal roots during colonization of an arbuscular
mycorrhizal fungus, New Phytologist, 167,
571-578.
99.Tani, C., Ohtomo, R., Osaki, M., Kuga, Y., and
Ezawa, T. (2009) ATP-dependent but proton gradient-independent
polyphosphate-synthesizing activity in extraradical hyphae of an
arbuscular mycorrhizal fungus, Appl. Environ. Microbiol.,
75, 7044-7050.
100.Plassard, C., and Dell, B. (2010) Phosphorus
nutrition of mycorrhizal trees, Tree Physiol., 30,
1129-1139.
101.Docampo, R., and Moreno, S. N. (2001) The
acidocalcisomes, Mol. Biochem. Parasitol., 114,
151-159.
102.Ruiz, F. A., Lea, C. R., Oldfield, E., and
Docampo, R. (2004) Human platelet dense granules contain polyphosphate
and are similar to acidocalcisomes of bacteria and unicellular
eukaryotes, J. Biol. Chem., 279, 44250-44257.
103.Omelon, S., Georgiou, J., Henneman, Z. J.,
Wise, L. M., Sukhu, B., Hant, T., Wynnyckyj, S., Holmyard, D.,
Bielecki, R., and Grynpas, M. D. (2009) Control of vertebrate skeletal
mineralization by polyphosphates, PLoS ONE, 4, e5634.
104.Pavlov, E., Aschar-Sobbi, R., Campanella, M.,
Turner, R. J., Gomez-Garcia, M. R., and Abramov, A. Y. (2010) Inorganic
polyphosphate and energy metabolism in mammalian cells, J. Biol.
Chem., 285, 9420-9428.
105.Morimoto, D., Tomita, T., Kuroda, S., Higuchi,
C., Rato, S., Shiba, T., Nakagami, H., Morishita, R., and Yoshikawa, H.
(2010) Inorganic polyphosphate differentiates human mesenchymal stem
cells into osteoblastic cells, J. Bone Miner. Metab., 28,
418-423.
106.Usui, Y., Uematsu, T., Uchihashi, T.,
Takahashi, M., Takahashi, M., Ishizuka, M., Doto, R., Tanaka, H.,
Komazaki, Y., Osawa, M., Yamada, K., Yamaoka, M., and Furusawa, K.
(2010) Inorganic polyphosphate induces osteoblastic differentiation,
J. Dent. Res., 89, 504-509.
107.Muller, W. E., Wang, X., Diehl-Seifert, B.,
Kropf, K., Schloßmacher, U., Lieberwirth, I., Glasser, G., Wiens,
M., and Schroder, H. C. (2011) Inorganic polymeric
phosphate/polyphosphate as an inducer of alkaline phosphatase and a
modulator of intracellular Ca(2+) level in osteoblasts (SaOS-2 cells)
in vitro, Acta Biomater., 7, 2661-2671.
108.Smith, S. A., Mutch, N. J., Baskar, D.,
Rohloff, P., Docampo, R., and Morrissey, J. H. (2006) Polyphosphate
modulates blood coagulation and fibrinolysis, Proc. Natl. Acad. Sci.
USA, 103, 903-908.
109.Smith, S. A., and Morrissey, J. H. (2008)
Polyphosphate as a general procoagulant agent, J. Thromb.
Haemost., 6, 1750-1756.
110.Smith, S. A., Choi, S. H., Davis-Harrison, R.,
Huyck, J., Boettcher, J., Reinstra, C. M., and Morrissey, J. H. (2010)
Polyphosphate exerts differential effects on blood clotting, depending
on polymer size, Blood, 116, 4353-4359.
111.Van der Meijden, P. E., and Heemskerk, J. W.
(2010) Polyphosphates: a link between platelet activation, intrinsic
coagulation and inflammation? Expert Rev. Hematol., 3,
269-272.
112.Mackman, N., and Gruber, A. (2010) Platelet
polyphosphate: an endogenous activator of coagulation factor XII, J.
Thromb. Haemost., 8, 865-867.