Molecular Analysis of Heavy Chain-Only Antibodies of Camelus bactrianus
S. V. Tillib1*, A. S. Vyatchanin1, and S. Muyldermans2,3
1Institute of Gene Biology, Russian Academy of Sciences, ul. Vavilova 34/5, 119334 Moscow, Russia; fax: +7 (499) 135-4105; E-mail: tillib@genebiology.ru2Laboratorium voor Cellulaire en Moleculaire Immunologie, Vrije Universiteit Brussel, Pleinlaan 2, B-1050 Brussel, Belgium; fax: +32 (0) 2-629-19-81; E-mail: svmuylde@vub.ac.be
3Structural Biology Research Center, Vlaams Interuniversitair Instituut voor Biotechnologie, Pleinlaan 2, B-1050 Brussel, Belgium
* To whom correspondence should be addressed.
Received August 30, 2014
In this work, IgG content and structures of antigen-binding domains and hinge regions of different IgG subtypes of Camelus bactrianus were analyzed in detail for the first time. Our data demonstrate that C. bactrianus contains a very large amount of heavy chain-only antibodies that can be used as a source of VHH domain-containing molecules. Despite some minor sequence differences identified in this study, C. bactrianus VHH domains possess principally the same unique features as those of C. dromedarius and the llama. These features are important for developing an efficient phage display-based antibody selection technology. We conclude that C. bactrianus is a very suitable animal to raise an immune response that serves as a source to identify antigen-specific VHHs selected after phage display.
KEY WORDS: antibody, IgG, variable domain, single-domain antibodyDOI: 10.1134/S000629791412013X
Abbreviations: CDR (complementarity determining regions), hypervariable region of the variable antibody fragment; CH and CL, constant (C) domains of immunoglobulin heavy (H) and light (L) chains, respectively; DTT, dithiothreitol; Fc (fragment crystallizable), antibody fragment comprising the constant domains CH2 and CH3 and responsible for effector functions; HCAb (heavy-chain antibody), unusual type of IgG comprising homodimer of shortened H-chains and devoid of L-chains; VH and VL, variable (V) domains of the heavy (H) and of the light (L) chains of the conventional antibody (immunoglobulin); VHH, variable domain of the H-chain of HCAb.
Classical antibodies (immunoglobulins G, IgG) throughout mammalian
species are composed of two identical heavy (H) chains and two
identical light (L) chains connected by disulfide bonds. The IgG
antibodies from species of Camelidae (and some chondrichthyan fish)
form a surprising exception to this paradigm as their serum contains
also a considerable fraction of unusual heavy-chain antibodies (HCAbs),
which lack the L-chain and are composed of a heavy-chain homodimer [1-3]. These HCAbs are expressed
after a V-D-J rearrangement and require dedicated constant
γ-genes. The H-chain within the HCAb is composed of three instead
of four globular domains. The two constant domains are highly
homologous to CH2-CH3, the Fc domains of classical antibodies [4, 5]. The domain corresponding to
the CH1 domain of classical antibodies is missing in HCAbs. Hence, the
antigen-binding fragment of a classical antibody, the Fab, is reduced
to a single variable domain of the H-chain of HCAbs (abbreviated as VHH
and also referred to as Nanobody®, single-domain antibody, or
nanoantibody). This variable domain, VHH, is adapted to become
functional in antigen binding in absence of a variable light-chain
domain (VL) [6-9]. It has been
repeatedly demonstrated that the VHH, cloned and expressed in bacteria,
is a strictly monomeric, single-domain antigen-binding entity [10]. An immune response is raised in these so-called
Heavy-Chain Antibodies (HCAb) following a classical immunization
protocol. These HCAb are easily purified from serum, and the
antigen-binding fragment interacts with parts of the target that
probably are less antigenic to conventional antibodies. Since the
antigen-binding site of the dromedary HCAb is comprised of one single
domain, VHH, a new very efficient and promising strategy was designed
to clone the VHH repertoire of an immunized animal (typically dromedary
Camelus dromedarius or llama) and to select the VHH with
specificity for particular target antigens. Some very useful
characteristic features of VHHs promise great potential for use in
immunobiotechnology and medicine [3, 11].
About 10 years ago, we decided to employ this technology to be our future platform for generating VHHs (nanobodies) against various antigens to address a wide spectrum of scientific questions and as a tool in multiple applications. Camelus bactrianus (Bactrian camel) was chosen as the most suitable (more resistant to cold winter) and most available Camelidae for our Moscow region of Russia. Bactrian camels, or two-humped camels, are Asian camels originating from the deserts and steppes of Central Asia, Kazakhstan, North China, and Mongolia. These camels have thick, warm, long, shaggy coats in winter and are equipped for extreme temperatures. They can withstand cold down to –40°C. In summer, they shed and tolerate heat up to 50°C. In contrast to Bactrians, dromedary camels have one hump. These camels live in North and East Africa. They have a shorter fiber coat even in winter. Dromedaries are not equipped to resist the extreme cold that Bactrians can withstand.
Whereas C. dromedarius (dromedary) or llamas have been used successfully many times to immunize and identify antigen-specific VHHs that were studied in great detail, C. bactrianus was a new and mainly unknown animal in this respect at the beginning of this work. The purpose of this paper was to fill this gap of knowledge by analyzing the IgG content and the HCAb mRNA nucleotide and deduced amino acid sequences specifically in C. bactrianus and to compare them with those of other camelids (primarily of C. dromedarius).
A new and large set of comparative data was generated. It is shown that both Old World camelids possess similar content of IgG, and the IgG sequences of all these camelids differ only slightly. Therefore, C. bactrianus is definitely a good animal for the generation and identification of single-domain antibodies (VHHs), which is also confirmed by the results of our other works on the generation of various nanoantibodies [12-16].
MATERIALS AND METHODS
Blood samples from two-humped camel and other blood sera. A one-year-old male camel (C. bactrianus) was bought from the Moscow Zoo and then kept at the Chernogolovka Biological Station of A. N. Severtsov Institute of Ecology and Evolution. Blood was taken from the jugular vein.
Other blood sera (from C. dromedarius, Lama glama, and Vicugna pacos) were obtained from Gentaur (Belgium) or were taken from C57Bl/6 mice or from a healthy donor (by qualified personnel in a medical facility).
IgG fractionation on protein A- and protein G-Sepharose. IgG fractionation from sera of C. bactrianus and other camelids was performed according to Hamers-Casterman et al. [1]. Serum was adsorbed on protein G-Sepharose and washed with 20 mM phosphate buffer, pH 7.0. IgG3 was eluted with 0.15 M NaCl, 0.58% acetic acid, pH 3.8. IgG1 was subsequently eluted with 0.1 M glycine-HCl, pH 2.7. The flow-through fraction was adsorbed on protein A-Sepharose. IgG2a and IgG2b/IgG2c were eluted with 0.15 M NaCl, 0.58% acetic acid, pH 3.5. Fractions were neutralized with 1 M Tris-HCl, pH 9.0.
Total IgGs were isolated on combined protein A/G-Sepharose with loading and washing in standard phosphate buffered saline (PBS), pH 7.2, followed by elution of the IgG in 0.1 M glycine-HCl, pH 2.7, and neutralization with 1 M Tris-HCl, pH 9.0.
Electrophoresis of proteins (according to Laemmli [17]) was performed either under “native” conditions (without the addition of dithiothreitol (DTT)) to maintain the integrity of the antibody or under reducing conditions (by adding 65 mM DTT to the sample), which leads to the reduction of the disulfide bonds.
RT-PCR, cloning, and sequencing of VH/VHH fragments. The lymphocyte-monocyte fraction of peripheral blood cells was obtained from the Bactrian camel before its immunization. For this, the blood (diluted 2-fold in PBS with 1 mM EDTA) was layered on a Histopaque-1077 (Sigma-Aldrich, USA) density cushion and centrifuged (800g for 20 min) in a table-top centrifuge, Eppendorf 5702R or 5415R (Eppendorf, Germany), at room temperature. RNA from the lymphocyte-monocyte fraction was isolated using TRIzol Reagent (Invitrogen, USA), and then poly(A)+ RNA (mRNA) was purified on an oligo(dT)-cellulose (Sigma-Aldrich) column. The mRNA was converted into single strand cDNA using Superscript II RNaseH– reverse transcriptase (Invitrogen) and an oligo(dT)18 primer. The PCR on the cDNA template was performed using primers annealing to highly conserved portions of the heavy chains of all dromedary and llama IgG mRNAs: forward primer CALL001 (5′-gtcctggctgctcttctacaagg-3′) corresponding to the leader peptide sequence, and reverse primer CALL002 (5′-ggtacgtgctgttgaactgttcc-3′) corresponding to a conserved region in the CH2 domain. The PCR program consisted of the following steps: 95°C – 2 min, (95°C for 30 s, 58°C for 300 s, 72°C for 90 s) × 5 cycles, (95°C for 30 s, 58°C for 60 s, 72°C – starting from 90 s and extending incubation time by 2 s for each following cycle) × 20 cycles, 72°C – 10 min.
The PCR products were fractionated by agarose gel electrophoresis, and the DNA from each zone of interest was purified using QIAquick PCR gel extraction kit (Qiagen). Amplified DNA fragments were cloned using a TOPO_TA cloning kit (Invitrogen) and then sequenced using M13 universal sequence primers (from the same kit) annealing at the vector sequence located near the insertion site of the PCR product.
RESULTS AND DISCUSSION
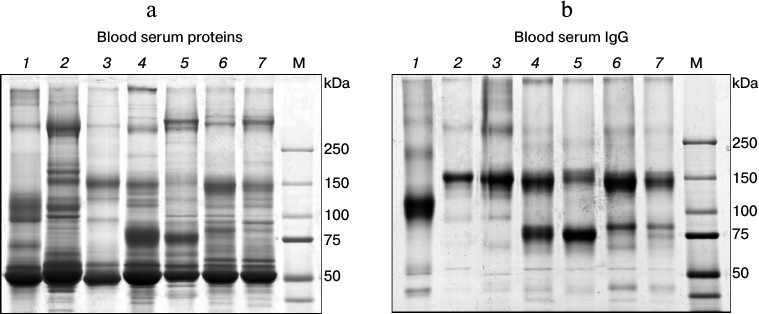
Heavy-chain antibodies (HCAbs) are the major IgG subclass in C. bactrianus serum. We assumed that HCAbs must be circulating in a significant amount in the C. bactrianus blood as in the case of other camelids [1]. To assess the quantitative distribution of different IgG subclasses in sera of C. bactrianus and other Camelidae species compared to sera of other mammals (human, mouse, rabbit), we analyzed the distribution of major blood proteins as well as purified serum IgGs on polyacrylamide gels under non-reducing conditions (Fig. 1). IgGs were isolated using mixed (1 : 1) protein A/G-Sepharose. From a comparison of Figs. 1a and 1b, it seems that the chromatography on protein A/G-Sepharose allows purifying proteins with electrophoretic mobilities expected for the case of classical IgGs (approximately 150 kDa) in almost all cases. We note that IgG from rabbit serum unexpectedly had a slightly increased electrophoretic mobility. Perhaps this anomalous mobility is due to rabbit IgGs having a special type of glycosylation, which includes an increased number of negatively charged groups of sialylated oligosaccharides [18]. More importantly, Fig. 1 further reveals that Camelidae sera contain significant amounts of smaller IgG molecules (migrating around 80 kDa) in addition to conventional IgGs. Relative quantities of IgG subclasses purified using the protein A/G-Sepharose chromatography might vary due to a more effective adsorption of classical antibodies to the bacterial proteins A and G than the noncanonical HCAb. Therefore, the relative amounts of these two IgG subclasses are better scored by comparing the band intensities for these respective IgG proteins in total serum (Fig. 1a). Although the relative intensity of bands around 150 and 80 kDa differ substantially in the different camelid species (Fig. 1a), it is clear that the Old World camelids (Camelus species) have relatively much more HCAbs than the New World camelids (Lama species). Figure 1 shows the results of a study of proteins from naive (preimmune) sera of individual animals. Relative quantities of IgG subclasses can vary significantly as a result of immunization. However, the relative proportion of noncanonical antibodies in camel sera remains significantly higher than that in sera of llamas and alpacas (this is also concluded from our other experiments when sera from several llamas and/or camels were tested). From these results, we assume that noncanonical antibodies, i.e. HCAbs, in camels may comprise initially a slightly broader spectrum of specificities than HCAbs in llama species, and that the use of dromedary or Bactrian camel instead of alpaca or llama for the immunization might be a more advantageous starting point to generate a diverse set of single-domain antibodies.
Fig. 1. Analysis of IgGs in sera of different mammals on SDS-polyacrylamide gel (according to Laemmli [17]) under non-reducing conditions. a) Separation of total proteins from sera; b) separation of proteins, mainly IgGs, isolated from these sera by affinity chromatography on a mixture (1 : 1) of protein A- and protein G-Sepharose. Numbers at the top mark lanes with fractionated protein samples from particular mammalian species: 1) rabbit; 2) mouse; 3) human; 4) Bactrian camel (C. bactrianus); 5) dromedary (C. dromedarius); 6) llama (Lama glama); 7) alpaca (Vicugna pacos). M, marker proteins with known molecular masses (in kDa).
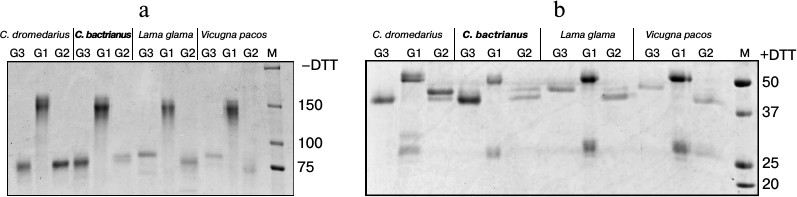
We performed a sequential affinity separation (as described in “Materials and Methods”) of serum IgGs from different Camelidae into three subclasses (Fig. 2) to confirm that the smaller IgGs in C. bactrianus, as in other Camelidae, really correspond to HCAb, and to analyze the relative amounts of the separated IgG subclasses. The first subclass, IgG1 (abbreviated G1), is represented by classical antibodies that effectively bind to the protein G as well as to the protein A adsorbent and can then be eluted from the column in a solution of pH 2.7. IgG1 antibodies migrate as a 150-kDa protein in SDS-polyacrylamide gel under non-reducing conditions (Fig. 2a). Under reducing conditions (with the addition of DTT to the sample), IgG1 antibodies split into four proteins (two of each H- and L-chains) due to rupture of the disulfide bonds, which gives protein bands of about 52-55 kDa (H-chain) and about 27 kDa (L-chain) (Fig. 2b).
Fig. 2. Electrophoretic analysis of IgG subfractions (G1, G2, and G3) isolated by sequential affinity fractionation of proteins from blood serum of various camelids (as described in “Materials and Methods”). The electrophoresis was carried out in SDS-polyacrylamide gel (a) under non-reducing conditions (–DTT) or (b) under reducing conditions (+DTT). M, marker proteins with known molecular masses (in kDa).
The second subclass, IgG2 (or G2), is represented by antibodies of noncanonical composition. IgG2 binds to the protein A but not to the protein G. These antibodies do not contain light chains, and they migrate as proteins with size of about 80 kDa in SDS-polyacrylamide gel under non-reducing conditions (Fig. 2a) and as proteins with size of about 40-47 kDa (shortened H-chains) under reducing conditions (Fig. 2b). In this antibody subclass, two H-chain variants of distinct size are clearly present in the case of camels and L. glama. This size variation is presumably due to differences in the length of the hinge regions in various representatives of this subclass: the longest in IgG2a, and shorter in the case of IgG2b or IgG2c.
Finally, the third subclass, IgG3 (G3), is represented by antibodies of noncanonical structure similar to that of IgG2. Unlike IgG2, IgG3 binds to both bacterial proteins A and G, although with lower affinity than IgG1. The elution of IgG3 from protein G occurs at pH 3.8 (IgG1 is not yet eluted under these conditions). As seen from Fig. 2b, IgG3 do not contain light chains and consist of a homodimer of two shortened H-chains, with size of about 40 kDa in camels or about 47 kDa in llamas.
Despite the principal similarity, notable variations are observed for the relative amounts and sizes of H-chains of noncanonical antibodies in different camelids. Whereas these differences are especially notable between camels and llamas, some minor differences are observed also when comparing the data for the two camel species. In the case of dromedary, major HCAb variants are G3 and G2a, and minor variants are G2b or G2c. In the case of Bactrian, the major variant is G3, whereas G2a is the minor variant (even less represented than G2b/G2c). Noncanonical HCAbs were detected in larger quantities than the classical IgG1 antibodies in the case of sera from both camel species.
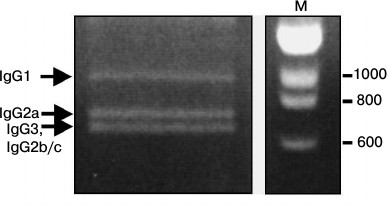
Sequence analysis of C. bactrianus VH/VHH-coding sequences. In a subsequent study, we aimed to analyze VH/VHH-coding sequences corresponding to various IgG subclasses of B-lymphocytes obtained from C. bactrianus peripheral blood before immunization (the naive repertoire). Initially, it was unclear whether primers CALL001 and CALL002 (adapted previously for specific amplification of VH/VHH-coding nucleotide sequences from B-lymphocytic cDNA of C. dromedarius or llama [19]) are suitable for the PCR amplification in the case of cDNA template made on mRNA from C. bactrianus B-lymphocytes. However, our data confirmed that the sequences of the primers are quite conserved and quite suitable for the amplification of the repertoire of VH/VHH-coding sequences of C. bactrianus, which is important for the further development of the technology. In Fig. 3, we show the result of a PCR amplification of cDNA (made from poly(A)+ RNA of Bactrian camel B-lymphocytes) using a forward primer for the well-conserved leader signal sequence of VH (of family 3) and VHH, and a reverse primer annealing to the beginning of a conserved region of the second constant domain CH2 (the CH1 constant domain is missing in HCAb).
Fig. 3. PCR products separated by electrophoresis in 1.5% agarose gel. PCR products corresponding to fragments of different IgG subclasses are marked by arrows. M, marker DNA fragments (in bp).
Three main amplicons can be identified after electrophoresis (Fig. 3): the longest products correspond to the size of the amplicon of the classical antibodies IgG1, and the shorter amplicons correspond to the sequences of noncanonical HCAbs lacking the CH1 region (IgG2a, IgG2b/IgG2c and IgG3) with variations in the hinge region length (confirmed by subsequent sequencing data).
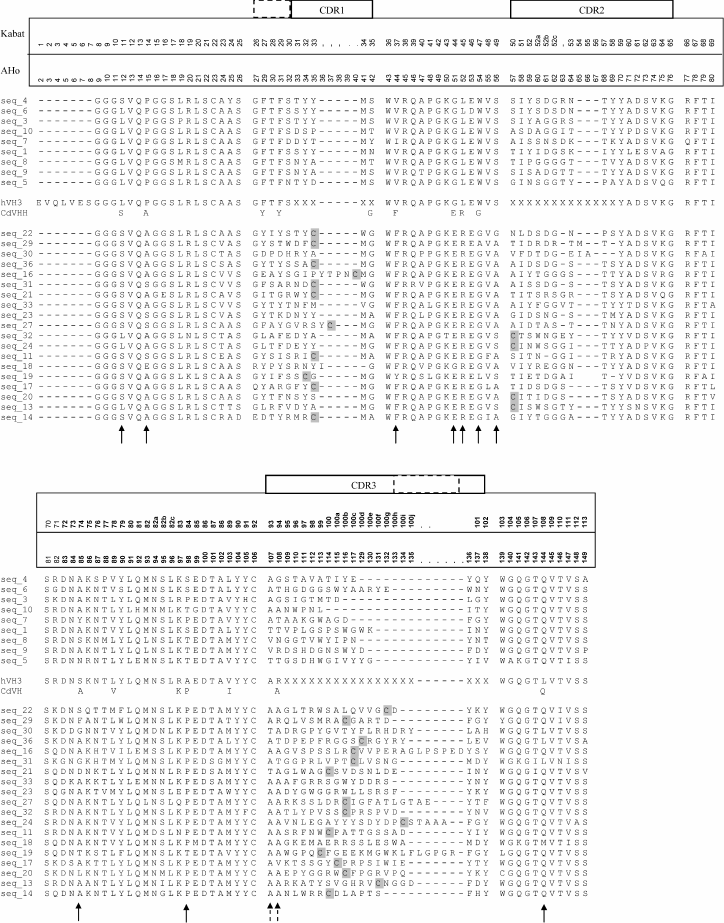
The amplified DNA fragments were separated, cloned individually, and about 10 cloned sequences from each of these three different size fractions were sequenced. Figure 4 shows different C. bactrianus VH/VHH amino acid sequences deduced from nucleic acid sequences of cloned PCR products. The general conclusion from these data is that C. bactrianus VH/VHH-sequences are very similar and in most parts are nearly identical to those of C. dromedarius. It is clear that practically all key amino acid residues determining the typical β-pleated sheet of an immunoglobulin fold [22] are very well conserved in both C. bactrianus VH and VHH sequences. The new set of sequences allows us to verify once more some important structural features of VHH antibodies. In the case of classical antibodies (IgG1) of C. bactrianus, heavy-chain variable domains (VH) have sequences that are almost identical to those of the human VH-3 family. From Fig. 4, one can infer possible differences only at three positions of the framework sequences (74, 84, and 108 according to Kabat numbering [20]) that are not essential for protein folding. Despite sharing a high degree of identity between these VH sequences, camel-specific VHH sequences are clearly distinguished. Several amino acids conserved in VHs are constitutively substituted in the VHHs in the case of C. bactrianus as well as in dromedary and llama. These residues (Leu11Ser, Val37Phe, Gly44Glu, Leu45Arg, Trp47Gly) discriminate the conventional VHs from the heavy-only specific VHHs (marked by arrows in Fig. 4). The substitution of Leu11Ser in most of Bactrian and dromedary camel VHHs is seen as an adaptation to accommodate the absence of the CH1 domain. In conventional VH domains, Leu11 makes a ball-and-socket joint with Phe149 and Pro150 of the CH1 [23]. These contacts are essential for the correct orientation of the VL–VH dimer relative to the CL–CH1. The absence of the CH1 domain in the HCAb makes this conserved hydrophobic contact residue unnecessary. The substitution of the hydrophobic side chain of Leu with the smaller hydrophilic Ser undoubtedly helps in the solubility behavior of the VHH [24]. While the Leu11Ser substitution is widespread among camel VHHs (in 16 out of 19 sequences, Fig. 4), it was noted earlier that a considerable fraction of llama VHHs (IgG2b subfraction) still maintained Leu at position 11 [7].
Fig. 4. Alignment of different C. bactrianus VH/VHH amino acid sequences (single letter code is used) deduced from nucleic acid sequences of cloned PCR products corresponding to IgG1- (seq_1 to seq_10, presented above the hVH3 sequence), IgG2a- (seq_11 to seq_20, presented at the lower part of the sequences set), or IgG3/IgG2c-subclasses (seq_21 to seq_36, placed between CdVHH and IgG2a sequences). Consensus VHH sequences from C. dromedarius HCAb (CdVHH [6]) and VH-sequence of the VH-3 family of human IgG (hVH3 [20]) are shown in the middle of Fig. 4 for comparison. In the case of CdVHH, only altered (compared to hVH3) amino acids are shown. In the figure, two different numbering schemes for immunoglobulin variable domains are used: “Kabat” – according to Kabat et al. [20], and “AHo” – according to Honegger and Pluckthun [21]. The Kabat numbering [20] is used throughout the text. Arrows indicate amino acid positions with substitutions in VHH domains relative to VH domains. Positions of additional VHH-specific cysteine residues are shown as shaded C. Extension of the CDR1 and CDR3 regions in the VHH domains is marked by dashed rectangles.
The residues at positions 37, 44, 45, and 47 constitute a key part of the large VH–VL interface [25]. The characteristic hydrophobic to hydrophilic amino acid substitutions at these positions in the VHH domain make it impossible to associate the VHH domain with a VL domain. These substitutions also explain the solubility of the HCAb and their recombinant VHH single domains [26].
Moreover, several additional VHH-specific amino acid positions are distinguished. For example, Pro14Ala/Ser and (Ala)/Thr/Ser84Pro amino acid VHH-specific substitutions are located in the turns between β-sheets at the antipode of the antigen-binding site. However, these substitutions (also found in dromedary and llama VHHs) were shown to have no significant effect on the structure in this region [27].
The Ser49Ala substitution was found in 14 of 19 C. bactrianus VHH sequences, whereas in all VHs only Ser was found in this position. This substitution was also observed for many dromedary and llama VHH domains. This residue buried between β-sheets and its influence on VHH structure or the CH2 loop is unknown.
Finally, 15 of 19 VHH sequences and only 2 of 9 VH sequences contain Met at position 89, and 15 of 19 VHH sequences and only 1 of 9 VH sequences contain Ala at position 94 (marked by dashed arrows in Fig. 4). These substitutions were less apparent but also observed in dromedary and llama VHHs. These substitutions seem to be indifferent for the immunoglobulin fold [22].
The alignment of the VH (seq_1 to seq_10) and VHH (seq_11 to seq_36) amino acid sequences (Fig. 4) revealed that the first and third hypervariable regions (CDR1 and CDR3) of the VHHs are more extended than that of VHs. This feature probably reflects mechanisms introduced to increase the diversity of the antigen-binding loops within one domain (VHH) in the absence of the VH–VL combinatorial diversity and thus partly compensate for the loss of antigen-interacting surface contributed by the hypervariable loops of the VL. Furthermore, VHH sequences typically (15 of 19) contain an additional Cys pair with the first Cys located in either the CDR1 or CDR2 loop and the second Cys always located in CDR3 loop. From crystal structures of VHH, we know that these Cys are forming a disulfide bond that possibly assists in shaping the loop structure and stabilizing the domain [28].
CDR1 of VHHs is extended by approximately four amino acid positions towards the N-terminal end in the case of C. bactrianus as well as all other studied camelids. The higher variability of the CDR1 loop in the camelid VHHs suggests a more direct participation of the loop in the interaction with the antigen.
CDR3 of a VHH is typically, on average, longer than that of VHs. In the case of C. bactrianus, this difference is very clear: 15-25 (an average of 18.6) amino acids in VHHs against 8-16 (an average of 12.3) amino acids in VHs. This difference is much higher than that reported for llama [7] and a little bit higher than that reported for dromedary [6]. It is known that a longer loop length increases the paratope repertoire. The CDR3 loop is the main contributor of antigen binding. It provides at least 60-80% of the contacts with antigen [28].
In addition to conserved cysteine residues at positions 22 and 92, which form the intra-domain disulfide bond that is characteristic for the immunoglobulin fold (both in VHs and VHHs), an additional pair of cysteines was identified in approximately 80% of C. bactrianus VHH sequences (this feature was not found in VHs). In each pair, one Cys is always located in the CDR3 region. Second Cys is typically (in 11 of 19 sequences) located in the CDR1 region (at positions 32-35) or less often (in 4 of 19 sequences) in the CDR2 region (at position 50). It was shown that the orientation of the Cys residues in the canonical structures for the CDR1 and CDR2 loops is compatible with the formation of a disulfide bond with the Cys of the CDR3 [29, 30], and it was directly shown that these Cys form a disulfide bond [28]. Thus, in the case of C. bactrianus VHHs, we have an intermediate situation between dromedary and llama VHHs. While in the case of dromedary VHH, typically only a CDR1–CDR3 inter-CDR disulfide bond is forming, in llama the inter-loop disulfide bond formation occurs equally between CDR1–CDR3 and CDR2–CDR3.
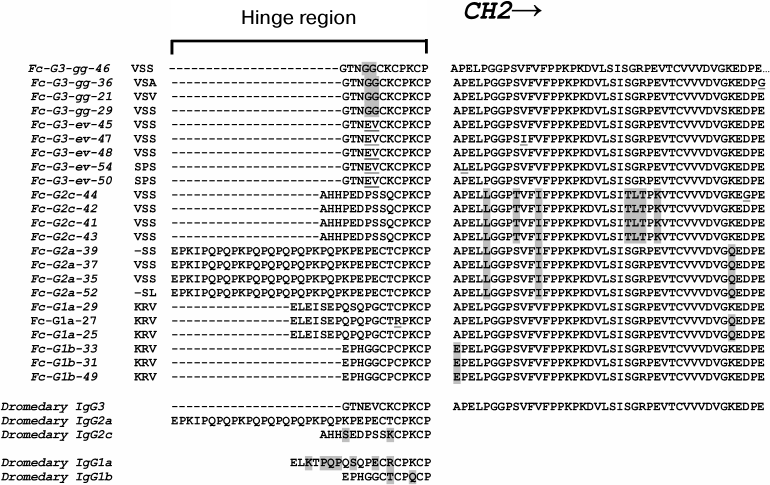
Besides the variable domains, special attention is also usually given to the hinge region sequences and to the sequences of corresponding constant regions for the classification of IgG subclasses. Figure 5 shows our generalized data of the identified sequences of hinge regions (and adjacent portions of the corresponding constant regions) of various C. bactrianus IgG subclasses resulting from sequence analysis of many cloned PCR products. We found two types of hinge regions for classical antibodies, IgG1 (G1a and G1b), and four types for noncanonical heavy-chain antibodies, HCAb (G2a, G2c, G3-ev, G3-gg) in the case of C. bactrianus. For comparison, the previously published data on similar sequences for C. dromedarius [31] are shown on the lower part of Fig. 5. The classification of IgG subclasses was taken from the same source [31]. Two identified hinge sequences for C. bactrianus classic antibodies (G1a and G1b) have the same length (12 and 19 amino acids) as the corresponding sequences from a dromedary IgG1. Although these hinge sequences are similar, they are not identical between these two camel species as some point mutations occur (Fig. 5). Two of four identified types of HCAb hinge sequences were identical to the hinges described previously: the 35-amino acid hinge sequence was identical to that of dromedary IgG2a antibody subtype, and the 12-amino acid hinge “GTNEVCKCPKCP” was identical to that of dromedary IgG3 antibody subtype. Interestingly, two other HCAb hinge types were found although similar but not identical to that of dromedary IgG (as in the case of IgG1 hinges). These two characteristic types of C. bactrianus HCAb hinge sequences are the 12-amino acid sequence “GTNGGCKCPKCP” of IgG3 antibody subtype and 15-amino acid sequence “AHHPEDPSSQCPKCP” marking IgG2c antibody subtype.
Fig. 5. Amino acid sequences of hinge regions and adjacent portions of the corresponding CH2 constant regions of various C. bactrianus IgG subclasses deduced from nucleic acid sequences of cloned PCR products. Two types of hinge regions for classic antibodies, IgG1 (G1a and G1b), and four types for noncanonical heavy-chain antibodies, HCAb (G2a, G2c, G3-ev, G3-gg), were found in the case of C. bactrianus. For comparison, the previous findings of similar sequences of dromedary [1, 31] are presented in the lower part of the figure.
Our preliminary analysis of partial constant region sequences adjacent to the hinge sequence (Fig. 5, from the right) also suggests their possible correspondence to particular subclasses of IgG. This is particularly evident in the case of IgG2c constant regions and to a lesser extent in the case of IgG2a constant regions. These sequences have a number of conserved amino acid substitutions in comparison with corresponding sequences of other subclasses. However, this issue requires more careful study over a larger number of animals.
In this work, we analyzed for the first time in detail the IgG content and composition of antigen-binding domains of different IgG subtypes of C. bactrianus. These sequences were compared with those of other camelids (primarily of dromedary). Amino acid sequences of VH/VHH-domains and hinge regions of different IgG subtypes of C. bactrianus were identified and compared with the corresponding sequences of C. dromedarius. Our data demonstrated that C. bactrianus contains a very high proportion of heavy chain-only antibodies (HCAb) in its blood, and these antibodies can be used as a valuable source to generate VHH domain-containing molecules. Camelus bactrianus antigen-binding VHH domains possess the same unique features as those of dromedary camel or llama. These features are important for an efficient phage display-based single-domain antibody selection technology. The Bactrian camel is more resistant to cold weather than the dromedary. We and other researchers repeatedly and successfully used a two-hump C. bactrianus camel for immunization and subsequent selection of specific VHH-domains to obtain new effective single-domain antibodies (Nanobodies®, nanoantibodies) [12-16, 32]. It is safe to conclude that C. bactrianus is a very suitable animal model for single-domain antibody generation technology.
We thank Dr. K. Conrath (Lab of S. M.) for introductory methodical advice and Dr. M. V. Rutovskaya for help in experimental work with the camel.
The work was partly supported by a DWTC Belgium grant (S. V. T.), by Program No. 24 of the Russian Academy of Sciences “Fundamentals of Technology of Nanostructures and Nanomaterials” (S. V. T.), and by the Russian Foundation for Basic Research (project No. HK-14-04-01189).
REFERENCES
1.Hamers-Casterman, C., Atarhouch, T., Muyldermans,
S., Robinson, G., Hamers, C., Bajyana Songa, E., Bendahman, N., and
Hamers, R. (1993) Naturally occurring antibodies devoid of light
chains, Nature, 363, 446-448.
2.Greenberg, A. S., Avila, D., Hughes, M., Hughes,
A., Mckinney, E. C., and Flajnik, M. F. (1995) A new antigen receptor
gene family that undergoes rearrangement and extensive somatic
diversification in sharks, Nature, 374, 168-173.
3.Muyldermans, S. (2013) Nanobodies: natural
single-domain antibodies, Annu. Rev. Biochem., 82,
775-797.
4.Nguyen, V. K., Hamers, R., Wyns, L., and
Muyldermans, S. (1999) Loss of splice consensus signal is responsible
for the removal of the entire CH1 domain of the functional camel IgG2A
heavy chain antibodies, Mol. Immunol., 36, 515-524.
5.Woolven, B. P., Frenken, L., van der Logt, P., and
Nicholls, P. J. (1999) The structure of the llama heavy chain constant
genes reveals a mechanism for heavy-chain antibody formation,
Immunogenetics, 50, 98-101.
6.Muyldermans, S., Atarhouch, T., Saldanha, J.,
Barbosa, J. A. R. G., and Hamers, R. (1994) Sequence and structure of
VH domain from naturally occurring camel heavy chain immunoglobulins
lacking light chains, Protein Eng., 7, 1129-1135.
7.Vu, K. B., Ghahroudi, M. A., Wyns, L., and
Muyldermans, S. (1997) Comparison of llama VH sequences from
conventional and heavy chain antibodies, Mol. Immunol.,
34, 1121-1131.
8.Harmsen, M. M., Ruuls, R. C., Nijman, I. J.,
Niewold, T. A., Frenken, L., and de Geus, B. (2001) Llama heavy
chain V-regions consist of at least four distinct subfamilies
revealing novel sequence features, Mol. Immunol., 37,
579-590.
9.Maass, D. R., Sepulveda, J., Pernthaner, A., and
Shoemaker, C. B. (2007) Alpaca (Lama pacos) as a convenient
source of recombinant camelid heavy chain antibodies (VHHs), J.
Immunol. Methods, 324, 13-25.
10.Muyldermans, S., and Lauwereys, M. (1999) Unique
single-domain antigen binding fragments derived from naturally
occurring camel heavy-chain antibodies, J. Mol. Recognit.,
12, 1-10.
11.Hassanzadeh-Ghassabeh, G., Devoogdt, N., De Pauw,
P., Vincke, C., and Muyldermans, S. (2013) Nanobodies and their
potential applications, Nanomedicine (Lond.), 8,
1013-1026.
12.Rothbauer, U., Zolghadr, K., Tillib, S., Nowak,
D., Schermelleh, L., Gahl, A., Backmann, N., Conrath, K., Muyldermans,
S., Cardoso, C. M., and Leonhardt, H. (2006) Targeting and tracing
antigens in live cells with fluorescent nanobodies, Nature
Methods, 3, 887-889.
13.Tillib, S. V. (2011) Camel nanoantibody is an
efficient tool for research, diagnostics and therapy, Mol. Biol.
(Moscow), 45, 77-85.
14.Tillib, S. V., Ivanova, T. I., Lyssuk, E. Y.,
Larin, S. S., Kibardin, A. V., Korobko, E. V., Vikhreva, P. N.,
Gnuchev, N. V., Georgiev, G. P., and Korobko, I. V. (2012)
Nanoantibodies for detection and blocking of bioactivity of human
vascular endothelial growth factor A165, Biochemistry
(Moscow), 77, 659-665.
15.Tillib, S., Ivanova, T. I., Vasilev, L. A.,
Rutovskaya, M. V., Saakyan, S. A., Gribova, I. Y., Tutykhina, I. L.,
Sedova, E. S., Lysenko, A. A., Shmarov, M. M., Logunov, D. Y.,
Naroditsky, B. S., and Gintsburg, A. L. (2013) Formatted single-domain
antibodies can protect mice against infection with influenza virus
(H5N2), Antiviral Res., 97, 245-254.
16.Tillib, S. V., Privezentseva, M. E., Ivanova, T.
I., Vasilev, L. F., Efimov, G. A., Gurskiy, Ya. G., Georgiev, G. P.,
Goldman, I. L., and Sadchikova, E. R. (2014) Single-domain
antibody-based ligands for immunoaffinity separation of recombinant
human lactoferrin from the goat lactoferrin of transgenic goat milk,
J. Chromatogr. B, 949-950, 48-57.
17.Laemmli, U. K. (1970) Cleavage of structural
proteins during the assembly of the head of bacteriophage T4,
Nature, 227, 680-685.
18.Raju, T. S., Briggs, J. B., Borge, S. M., and
Jones, A. J. S. (2000) Species-specific variation in glycosylation of
IgG: evidence for the species-specific sialylation and branch-specific
galactosylation and importance for engineering recombinant glycoprotein
therapeutics, Glycobiology, 10, 477-486.
19.Saerens, D., Kinne, J., Bosmans, E., Wernery, U.,
Muyldermans, S., and Conrath, K. (2004) Single domain antibodies
derived from dromedary lymph node and peripheral blood lymphocytes
sensing conformational variants of prostate-specific antigen, J.
Biol. Chem., 279, 51965-51972.
20.Kabat, E., Wu, T. T., Perry, H. M., Gottesman, K.
S., and Foeller, C. (1991) Sequence of Proteins of Immunological
Interest, US Public Health Services, NIH, Bethesda, MD, Publication
No. 91-3242.
21.Honegger, A., and Pluckthun, A. (2001) Yet
another numbering scheme for immunoglobulin variable domains: an
automatic modeling and analysis tool, J. Mol. Biol., 309,
657-670.
22.Chothia, C., Lesk, A. M., Tramontano, A., Levitt,
M., Smith-Gill, S. J., Air, G., Sheriff, S., Padlan, E. A., Davies, D.,
Tulip, W. R., Colman, P. M., Spinelli, S., Alzari, P. M., and Poljak,
J. (1989) Conformations of immunoglobulin variable regions,
Nature, 342, 877-883.
23.Lesk, A. M., and Chothia, C. (1988) Elbow motion
in immunoglobulins involves a molecular ball-and-socket joint,
Nature, 335, 188-190.
24.Nieba, L., Honnegger, A., Krebber, C., and
Pluckthun, A. (1997) Disruption of the hydrophobic patches at the
antibody variable/constant domain interface: improved in vivo
folding and physical characterization of an engineered scFv fragment,
Protein Eng., 10, 435-444.
25.Chothia, C., Novotny, J., Bruccoleri, R., and
Karplus, M. (1985) Domain association in immunoglobulin molecules. The
packing of variable domains, J. Mol. Biol., 186,
651-663.
26.Nguyen, V. K., Desmyter, A., and Muyldermans, S.
(2001) Functional heavy-chain antibodies in Camelidae, Adv.
Immunol., 79, 261-296.
27.Desmyter, A., Transue, T. R., Arbabi Ghahroudi,
M., Dao-Thi, M.-H., Poortmans, F., Hamers, R., Muyldermans, S., and
Wyns, L. (1996) Crystal structure of a camel single-domain VH antibody
fragment in complex with lysozyme, Nature Struct. Biol.,
3, 803-811.
28.De Genst, E., Silence, K., Decanniere, K.,
Conrath, K., Loris, R., Kinne, J., Muyldermans, S., and Wyns, L. (2006)
Molecular basis for the preferential cleft recognition by dromedary
heavy-chain antibodies, Proc. Natl. Acad. Sci. USA, 103,
4586-4591.
29.Chothia, C., Lesk, A. M., Gherardi, E.,
Tomlinson, I. M., Walter, G., Marks, J. D., Llewelyn, M. B., and
Winter, G. (1992) Structural repertoire of human VH segments, J.
Mol. Biol., 227, 799-817.
30.Davies, J., and Riechmann, L. (1996) Single
antibody domains as small recognition units: design and in vitro
antigen selection of camelids, human VH domains with improved protein
stability, Protein Eng., 9, 531-537.
31.De Genst, E., Saerens, D., Muyldermans, S., and
Conrath, K. (2006) Antibody repertoire development in camelids, Dev.
Compar. Immunol., 30, 187-198.
32.Rahbarizadeh, F., Rasaee, M. J., Forouzandeh
Moghadam, M., Allameh, A. A., and Sadroddiny, E. (2004) Production of
novel recombinant single-domain antibodies against tandem repeat region
of MUC1 mucin, Hybridoma and Hybridomics, 23,
151-159.