Structure and Functional Investigation of Ligand Binding by a Family 35 Carbohydrate Binding Module (CtCBM35) of β-Mannanase of Family 26 Glycoside Hydrolase from Clostridium thermocellum
A. Ghosh1#, A. K. Verma1#, S. Gautam2, M. N. Gupta2, and A. Goyal1*
1Department of Biotechnology, Indian Institute of Technology Guwahati, Guwahati, 781039, Assam, India; fax: 91 (361) 2690762; E-mail: arungoyl@iitg.ernet.in; amalarastar@gmail.com2Department of Chemistry, Indian Institute of Technology Delhi, New Delhi, 110016, Delhi, India
# These authors contributed equally.
* To whom correspondence should be addressed.
Received August 31, 2013; Revision received February 26, 2014
Functional attributes of recombinant CtCBM35 (family 35 carbohydrate binding module) of β-mannanase of family 26 Glycoside Hydrolase from Clostridium thermocellum were deduced by biochemical and in silico approaches. Ligand-binding analysis of expressed CtCBM35 analyzed by affinity-gel electrophoresis and fluorescence spectroscopy exhibited association constants Ka ~ 1.2·105 and 3.0·105 M–1 with locust bean galactomannan and mannotriose, respectively. However, CtCBM35 showed low ligand-binding affinity with insoluble ivory nut mannan with Ka of 5.0·10–5 M–1. Unfolding transition analysis by fluorescence spectroscopy explained the conformational changes of CtCBM35 in the presence of guanidine hydrochloride (5 M) and urea (6.25 M). This explained that CtCBM35 has good conformational stability and requires higher free energy of denaturation to invoke unfolding. The three-dimensional (3-D) model of CtCBM35 from C. thermocellum generated by Modeller9v8 displayed predominance of β-sheets arranged as β-jelly-roll fold. The secondary structure of CtCBM35 by PredictProtein showed the presence of two α-helices (3%), 12 β-sheets (45%), and 15 random coils (52%). Secondary structural element analysis of cloned, expressed, and purified recombinant CtCBM35 by circular dichroism also corroborated the in silico predicted secondary structure. Multiple sequence alignment of CtCBM35 showed conserved residues (Tyr123, Gly124, and Phe125), which are commonly observed in mannan specific CBMs. Docking analysis of CtCBM35 with manno-oligosaccharide displayed the involvement of Tyr26, Gln29, Asn43, Trp66, Tyr68, Leu69, Arg76, and Leu127 residues, making polar contact with the ligand molecules. Ligand docking analysis of CtCBM35 exhibiting higher binding affinity with mannotriose and galactomannan (Man-Gal-Man moiety) substantiated the affinity binding and fluorescence results, displaying similar values of Ka.
KEY WORDS: CtCBM35, homology modeling, docking, CD spectra, affinity electrophoresis, fluorescence spectra, unfolding transitionDOI: 10.1134/S0006297914070098
Hydrolytic enzymes and their enhanced polysaccharide specificity are often improved by a noncatalytic carbohydrate-binding module appended either at their N- or C-terminals. Polysaccharide recognition, binding, and enhanced catalysis by hydrolytic enzymes are facilitated by noncatalytic modular carbohydrate binding modules. Carbohydrate binding modules (CBMs) have been classified into 67 distinguished families based on sequence similarity (http://www.cazy.org/Carbohydrate-Binding-Module). Usually CBMs are annexed with hydrolytic enzymes such as cellulase, mannanase, and xylanase as scaffoldings and as peptides with noncatalytic functions [1]. Family 35 carbohydrate binding module is often associated with glycoside hydrolase family 26 (GH26) and GH5 mannanases [2-4], xylanases (GH30) [5], and GH39 [6], which significantly alter the polysaccharide specificity for galactomannan, glucomannan, mannan, and glucuronoxylan. The solved 3-D structures of family 35 CBMs have shown a dominance of β-sheet organized as jelly rolls [5]. CBM35 usually accommodates the polysaccharides utilizing a planer surface of aromatic side chains, which interact with the flat chains of manno-configured carbohydrates [5]. This form of conformation is known as type B module [5]. Polysaccharide binding significantly alters the conformation of CBM35, which creates a suitable binding space for polysaccharide accommodation [7]. Generally, it has been observed that bacterial CBMs have affinity towards the same or variable substrates other than their substrate-specific cognate catalytic modules [8]. Affinity gel electrophoresis and fluorescence spectroscopy are effective approaches that are popular for elucidating functional properties of a CBM [9].
It was reported earlier that CBM35 from Clostridium thermocellum possesses β-sandwich fold, forming a cavity that specifically accommodates the side chain galactose of galactomannan and Δ-4,5-anhydrogalaturonic acid [10, 11]. The present study describes the ligand binding properties of over-expressed recombinant CtCBM35 by affinity gel electrophoresis and fluorescence spectroscopy and the structural characterization of CtCBM35 by model prediction and its ligand binding by docking analysis. The gene encoding CtCBM35 present at the N-terminal of family GH26 glycoside hydrolase (β-mannanase) was cloned in pET28a(+) vector expressed in E. coli BL-21 cells [12]. The recombinant CtCBM35 was purified by immobilized metal ion affinity chromatography (IMAC). Structural stability and the unfolding transition of CtCBM35 were investigated in the presence of guanidine hydrochloride (GnHCl) and urea.
MATERIALS AND METHODS
Bacterial strains, plasmid, and fine chemicals. Escherichia coli DH5α cells were used for cloning of CtCBM35, and E. coli BL-21(DE3) cells were used as expression host. The plasmid used for cloning and expression was pET28a(+). All the above-mentioned items were procured from Novagen (USA). The chemicals required for polyacrylamide gel electrophoresis (PAGE), Bradford’s reagent, oat spelt xylan, birchwood xylan, carboxymethyl cellulose, and Avicel were purchased from Sigma-Aldrich (USA). Glycerol, methanol, concentrated hydrochloric acid, glacial acetic acid, urea, and guanidine hydrochloride were procured from Merck (India). Locust bean galactomannan, rye arabinoxylan and wheat arabinoxylan (insoluble), ivory nut mannan, glucuronoxylan, and mannotriose were purchased from Megazyme International (Ireland).
Production and purification of recombinant CtCBM35. In an earlier study we cloned, hyper-expressed, and biochemically characterized CtCBM35 from C. thermocellum ATCC 27405 [12]. For production of CtCBM35, 100 μl of the E. coli BL21(DE3) culture from glycerol stock was inoculated into 5 ml of LB medium containing 50 μg/ml kanamycin and incubated at 37°C for 16 h at 180 rpm. A 1% (v/v) inoculum of the culture was transferred to 200 ml of LB medium in a 500 ml flask containing 50 μg/ml kanamycin with incubation at 37°C, 180 rpm till the culture reached mid-exponential phase (A600nm = 0.6). Isopropyl-β-D-thiogalactopyranoside (IPTG) at 1 mM final concentration was added to this mid-exponential phase culture, and it was further incubated at 24ºC, 200 rpm, for 24 h for protein induction [12]. The cells were harvested by centrifugation at 9000g and 4ºC for 15 min, and the resulting pellet was resuspended in 50 mM sodium phosphate buffer, pH 7.0, containing 1 mM phenylmethanesulfonyl fluoride (PMSF). Then the cells were subjected to sonication (Sonics, Vibra cell) on an ice bath for 15 min (9 s on/9 s off pulse, 30% amplitude) and again centrifuged at 19,000g at 4ºC for 20 min. The cell free supernatant containing the soluble protein was purified by immobilized metal ion chromatography (IMAC). The recombinant CtCBM35 containing His6 tag was purified by 1 ml HiTrap chelating column (GE Healthcare, USA) following the protocol recommended by the manufacturer. The purity and molecular mass of the recombinant CtCBM35 was verified by SDS-PAGE [12].
Circular dichroism analysis of CtCBM35. The far-UV circular dichroism (CD) spectra of CtCBM35 were recorded on a JASCO J-815 spectropolarimeter (Jasco Corporation, Japan) equipped with a Peltier system for temperature control at 25°C using a cell with a pathlength of 0.1 cm. Typical spectral accumulation parameters were scanning rate of 50 nm/min with a 1 nm bandwidth over the wavelength range 195 to 250 nm with six scans averaged for each far-UV spectrum. The CD data are presented in terms of mean residue ellipticity (MRE, expressed as deg·cm2·dmol–1) as a function of wavelength, calculated according to the procedure described earlier [13] using a protein concentration of 10-15 μM in 10 mM Tris-HCl, pH 7.5. All CD spectra were corrected for buffer contributions, and secondary structures were calculated by using the web-based K2d neural network software package (kal-el.ugr.es/k2d/spectra.html) [14].
Affinity electrophoresis of CtCBM35 with soluble polysaccharides. Binding of CtCBM35 to soluble polysaccharides was assessed by affinity gel electrophoresis following the protocol of Takeo [15] on 7.5% native polyacrylamide gel in the absence and presence of varying concentration of mannotriose and locust bean galactomannan. Purified CtCBM35 (18 μg) was run on native gels containing soluble polysaccharides such as locust bean galactomannan, carboxymethyl cellulose, rye arabinoxylan, birchwood xylan, oat spelt xylan, and glucuronoxylan and oligosaccharide mannotriose. Native polyacrylamide gels (7.5%) were prepared containing varying polysaccharide concentrations ranging from 0.0 to 1.5% (w/v). A bovine serum albumin (BSA) sample (1.0 mg/ml) was run in native gel for assessment of nonspecific binding interaction.
Binding analysis of CtCBM35 with insoluble polysaccharides. Quantitative and qualitative assessment of CtCBM35 binding was carried out with insoluble polysaccharides such as ivory nut mannan, Avicel, and wheat arabinoxylan. For qualitative binding analysis, 30 µg of CtCBM35 in 50 mM sodium phosphate buffer, pH 7.0, was mixed with 1 mg of ivory nut mannan or Avicel or wheat arabinoxylan (insoluble) in a final reaction volume of 200 μl. The reaction mixture was incubated for 2 h at 4°C with gentle shaking. After that the insoluble ligand was precipitated by centrifugation at 13,000g at 4°C for 5 min. The supernatant, containing the unbound protein, was removed, and the pellet was washed three times with 200 μl of 50 mM sodium phosphate buffer, pH 7.0. The bound protein from the washed pellet was eluted by boiling the polysaccharides in 200 μl of 10% (w/v) SDS containing 10% (v/v) β-mercaptoethanol for 10 min. The pellet of bound protein and the supernatant of unbound protein were analyzed by SDS-PAGE (13%). A BSA (1 mg/ml) control was set in parallel to check for any nonspecific binding. For the quantitative analysis of ligand binding by adsorption isotherm, the protein concentration was varied as 1, 2, 5, 9, and 15 μM and mixed with 5 mg/ml of insoluble ivory nut mannan. Two hundred microliters of CtCBM35–ivory nut mannan mixtures were prepared in 50 mM sodium phosphate buffer, pH 7.0, and incubated at 4°C for 2 h. Different controls (native protein without any polysaccharide) were also kept for each set of reaction mixtures. The experiment was carried out in duplicate. The reaction mixture was centrifuged at 13,000g and at 4°C for 10 min. The supernatant containing the unbound fraction was removed, and its protein content was determined by Bradford’s method [16]. The adsorption parameters were calculated to determine the binding. If we consider [B] the bound protein concentration, [F] the unbound fraction of protein, [N] the number of binding site concentration, and Ka the association constant, then at equilibrium adsorptions were calculated as described earlier by Gilkes et al. [17].
Polysaccharide binding study of CtCBM35 by fluorescence spectroscopy. On binding to polysaccharide, carbohydrate-binding modules undergo conformational changes and behave differently than in its unbound native form [18]. To compare the results with affinity electrophoresis, 18 μg of CtCBM35 was incubated with polysaccharides viz., locust bean galactomannan and mannotriose at varying concentrations. Polysaccharide concentrations (0.01, 0.04, 0.06, 0.08, 0.15, and 0.2%, w/v) from 0.5% (w/v) stock solution in 100 μl reaction mixture were prepared in 50 mM Tris-HCl buffer, pH 7.0. The samples were incubated at 4°C for 2 h. The fluorescence measurements were carried out using a Fluoromax 3 fluorimeter (Horiba Scientific, USA). Emission and excitation slits were kept at 3.00 and 1.00 nm, respectively, with 0.5 s integration time. Three scans were taken per sample along with a control to reduce the noise created by buffer and polysaccharide. All the samples were excited at λmax = 295 nm with observance of emission spectra between λmax = 320-400 nm. The emission spectra of all the solutions were corrected against buffer and polysaccharide solution without CtCBM35 before setting the interaction study. The association constants Ka (M–1) of CtCBM35 complex with carob galactomannan and konjac glucomannan were derived using a modified Stern–Volmer equation as described by Belatik et al. [19].
Unfolding transition of CtCBM35 in guanidine hydrochloride and urea. The structural stability of recombinant CtCBM35 in guanidine hydrochloride and urea was studied under isothermal condition. Initially 30 μg of CtCBM35 was incubated with varying concentrations of guanidine hydrochloride (GnHCl) or urea at 25°C for 24 h. One milliliter of reaction mixture was prepared by the addition of 50 μl CtCBM35 (0.6 mg/ml) and varying concentrations of GnHCl or urea (1-8 M) were prepared in 50 mM Tris-HCl buffer, pH 7.0. The unfolding transition of CtCBM35 was monitored by the change in fluorescence intensity of tryptophan (Trp) emission spectra 320-400 nm by exciting the samples at 295 nm using the Fluoromax 3 fluorimeter. A sample without CtCBM35 was kept as control (buffer and denaturant). The free energy of denaturation ΔG was calculated by the method described by Ahmad et al. [20].
Molecular modeling of family 35 carbohydrate-binding module. The protein sequence of family 35 carbohydrate-binding module (CtCBM35) was retrieved from the NCBI protein sequence database with accession number ABN51273 (nucleotide accession number: CP000568) and UniProt ID A3DBE4. The molecular architecture deduced from amino acid sequence showed that CtCBM35 (134 a.a.) is located at the N-terminal, sandwiched between a 32-a.a. signal peptide and catalytic module CtGH26 (373 a.a.) at the C-terminus followed by a dockerin type 1 module. Modeller9v8 was used to build the 3-D model of CtCBM35. Modeller is a computer program used for comparative protein structure modeling by satisfaction of the spatial restraints (http://salilab.org/modeller/). In the first step, target query sequence is aligned with template sequences, and this alignment file was used as an input to generate models of CtCBM35. Loop refinement was done by using the loop model class in Modeller. The model with the lowest discrete optimized protein energy (DOPE) was chosen for further refinements.
Structure refinement and quality assessment. The structure of CtCBM35 with the lowest DOPE score obtained from the modeler was further improved by energy minimization at the YASARA Energy Minimization Server (www.YASARA.org/minimizationserver), in which molecular dynamics simulations of models were carried out in explicit solvent. It uses a new partly knowledge-based all atom force field derived from Amber [21]. The final structure after energy minimization was subjected for structure validation on structure analysis and verification server (SAVES) at NIH-MBI laboratory servers (http://nihserver.mbi.ucla.edu). Protein secondary structure prediction tool (PredictProtein) from the ExPASy SIB tool site was used to calculate the presence of α-helix, β-sheet, turns, coils, etc. at the Columbia University server (http://cubic.bioc.columbia.edu/predictprotein/).
Multiple sequence analysis. Generally the amino acid residues involved in ligand binding are conserved within the CBM35 family. To investigate these conserved amino acid residues, multiple sequence alignment was performed with representative members of CBM35 having different substrate specificities whose X-ray, crystal, or NMR structures were known. The protein sequence was retrieved from PDB (http://www.rcsb.org/pdb/) saved in FASTA format. Alignment was performed using the ClustalW program [22], and final alignment was generated by ESpript (http://espript.ibcp.fr) for better understanding of conserved residues and structure.
Docking study of modeled CtCBM35. Docking was studied with the help of AutoDock version 4.2.1. Ligands (mannotriose and galactomannan) for the docking study were obtained from PubChem (http://pubchem.ncbi.nlm.nih.gov) in 3D SDF format, which was converted and saved in Mol2 format using OpenBabel 2.2.3 [23]. Galactomannan has a mannan backbone which is decorated with highly substituted α-(1→6)-galactosyl moiety (Man-Gal-Man). Therefore, this trisaccharide moiety was considered as galactomannan ligand. The two other ligands, mannopentaose and mannohexose, were retrieved from PDB, crystal structure of Caldanaerobius polysaccharolyticus CBM16 bound to mannopentaose (PDB id: 3OEB), and CBM29 complex with mannohexose (PDB id: 1GWL), respectively. Ligand preparation was done by assigning Gasteiger partial charges, merging nonpolar hydrogens, and finally saved in extended PDBQT file format using AutoDock Tools (ADT) 1.5.4. CtCBM35 modeled protein was also saved as PDBQT file format after removing nonpolar hydrogen atoms and adding their charges with the carbon atoms. Address for ligands where to dock was fixed in a grid parameter file. The grid map dimensions were set in such a way that they fully cover the active site cavity and give enough space for the ligand molecule to rotate freely. For different conformation search of the ligand molecule, the Lamarckian Genetic Algorithm (GA) was implemented and the number of GA runs was set to 30. All other parameters were set to default values such as initial population size (150), maximum energy evaluation per run (2,500,000), and maximum number of generations (27,000). After 30 independent successful docking runs, the protein–ligand complex for each ligand having auto-generated lowest free energy of binding (ΔG) confirmation were saved [24]. Docking results were analyzed using PyMOL (www.PyMOL.org) (0.99) for possible polar and hydrophobic interactions, and the final figure was generated with the help of PyMOL and YASARAView (12.2.22). For better understanding of protein–ligand interaction in 2D, Ligplot was generated (http://www.ebi.ac.uk/pdbsum/).
RESULTS AND DISCUSSION
Cloning, overexpression and purification of recombinant CtCBM6A. The recombinant CtCBM35 was expressed as a soluble protein, and after purification it appeared as a homogenous single band of molecular size 15 kDa on SDS-PAGE [12]. That was close to its theoretical molecular mass of 14.74 kDa as calculated from the amino acid sequence.
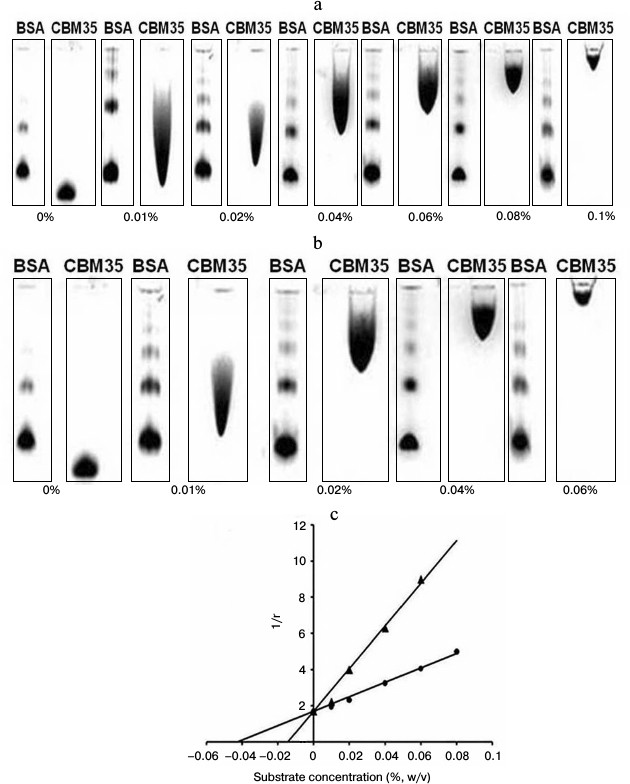
Affinity gel electrophoresis of family 35 carbohydrate binding module. The ligand binding of CtCBM35 was investigated by affinity gel electrophoresis, and the degree of association with locust bean galactomannan and mannotriose was determined as described by the method of Takeo [15]. The relative mobilities (r) of CtCBM35 in the presence of varying substrate concentrations were calculated from the polyacrylamide gels (Figs. 1a and 1b). The retardation of CtCBM35 was observed from 0.01% (w/v) with maximum retardation detected at 0.1% (w/v) concentration of locust bean galactomannan (Fig. 1a). In contrast, the retardation was achieved at much lower concentration (0.06%, w/v) of mannotriose (Fig. 1b) owing to simple and small molecular size (dp = 3). Mannotriose accesses the binding sites on CtCBM35 more easily than a galactose-substituted branched polysaccharide like locust bean galactomannan. This was similarly reported earlier by Mizutani et al. [25]. However, no significant binding of CtCBM35 was observed with carboxymethylcellulose, rye arabinoxylan, birchwood xylan, oat spelt xylan, and glucuronoxylan, showing that CtCBM35 is mannan-specific CBM (Table 1). The association constants (Ka) of CtCBM35 with different ligands were derived from linear regression plots between the inverse relative migration (1/r) versus substrate concentration (%, w/v) (Fig. 1c). CtCBM35 displayed higher association constant value with mannotriose (Ka = 3.2·105 M–1) than with locust bean galactomannan (Ka = 1.23·105 M–1) (Table 1). The free energy of binding (ΔG) with mannotriose was higher (7.0 kcal/mole) than with locust bean galactomannan (6.2 kcal/mole) (Table 1). While comparing these data with prediction outcome, it was observed that the association constant (Ka) and free energy of binding (ΔG) of CtCBM35 were similar in both cases (Table 1).
Fig. 1. Affinity gel electrophoresis using native polyacrylamide gel (7.5%) of CtCBM35 against (a) locust bean galactomannan and (b) mannotriose. c) Plot of 1/r versus polysaccharide concentration, where r is the relative migration distance of CtCBM35 in the presence of locust bean galactomannan (circles) and mannotriose (triangles) in the gel.
Table 1. Binding analysis of CtCBM35
with ligands by in silico prediction, affinity electrophoresis,
and fluorescence spectroscopy
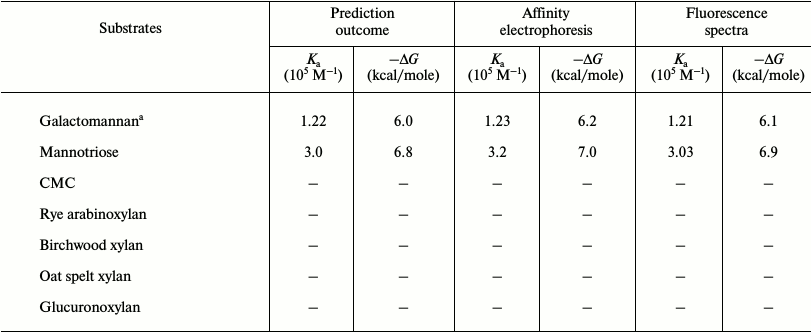
a Locust bean; –, no binding observed.
The recombinant CtCBM35 preferred binding manno-configured polysaccharides. CtCBM35 discriminated during carbohydrate selection, showing its affinity only with manno-configured ligands among the diverse polysaccharides tested. Family 35 CBMs often show affinity toward ligands other than manno-configured polysaccharides containing β-(1→4)-mannose chains, such as Δ-4,5-anhydrogalacturonic acid and glucuronic acid [26]. CtCBM35 showed higher affinity with mannotriose than locust bean galactomannan. The rationale behind this selective affinity is due to the galactose unit in locust bean galactomannan, which probably interferes with the CtCBM35 binding. Locust bean galactomannan is composed of a β-(1→4)-linked D-mannan backbone to which single D-galactosyl units are attached to C-6 of D-mannosyl residues, whereas mannotriose is a linear trisaccharide comprising β-(1→4)-linked D-mannosyl residues. However, no significant binding of CtCBM35 with carboxymethylcellulose, rye arabinoxylan, birchwood xylan, oat spelt xylan, and glucuronoxylan showing that it is a mannan specific binding CBM. Recently, we reported CtCBM35 from C. thermocellum displaying binding affinity towards both galacto- and gluco-mannan [12]. This finding gives a new insight into family 35 CBM when compared with other mannan specific CBM35 from Clostridium thermocellum and Cellvibrio japonicus, which were exo and endo acting to the D-mannan chain of galactomannan only [7, 10]. Therefore, it can be inferred that CtCBM35 was able to distinguish between mannotriose and highly galactosylated locust bean galactomannan by displaying better binding characteristics with mannotriose.
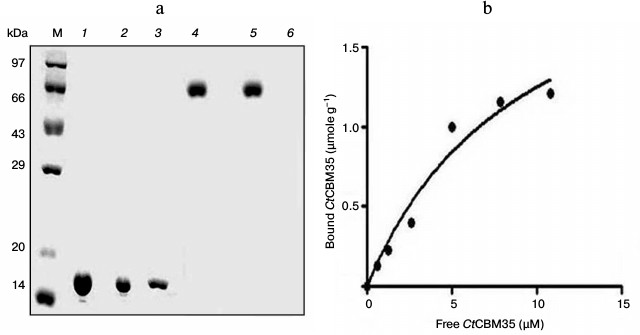
Binding analysis of CtCBM35 with insoluble polysaccharides. The quantitative and qualitative binding of CtCBM35 with insoluble polysaccharides was assessed by adsorption isotherm analysis. CtCBM35 displayed lower binding with insoluble mannan as analyzed by SDS-PAGE upon comparison of the bound protein fraction, unbound fraction, and the purified native protein (Fig. 2a). CtCBM35 displayed no binding with Avicel or wheat arabinoxylan (insoluble). The association constant Ka of CtCBM35 with insoluble mannan was observed to be 5.0·10–5 M–1 (Table 2). The relative equilibrium association constant Kr and the binding capacity (number of binding site concentration on mannan surface, No) were calculated from a nonlinear regression plot between bound CtCBM35 versus free CtCBM35 (Fig. 2b). The data were analyzed by GraphPad (Prism 2.0.1) software using nonlinear regression analysis based on one binding site equation as described by Gilkes et al. [17]. The estimated values of relative equilibrium constant Kr and concentration of binding sites [No] were 1.5 ± 0.2 liter/g and 0.45 ± 0.002 µmole/g, respectively. This suggested that CtCBM35 bound less significantly with insoluble ivory nut mannan. The low affinity against insoluble mannan has been reported for the inability of “trans” form (as a separate fold and not appended to a hydrolytic enzyme) of CBM to disrupt the interchain interactions of the polysaccharide [25]. This is in contrast to some CBMs that have the ability to disintegrate the surface of crystalline insoluble polysaccharides displaying higher binding to soluble fractions of insoluble polysaccharides [27, 28].
Fig. 2. Binding of CtCBM35 with insoluble ivory nut mannan. a) Qualitative binding analysis using 13% SDS-PAGE. Lanes: M, high range unstained molecular weight marker (14-97 kDa); 1) purified CtCBM35 as control; 2) unbound CtCBM35; 3) bound CtCBM35; 4) bovine serum albumin (BSA) as negative control; 5) unbound BSA; 6) bound BSA. b) Adsorption of CtCBM35 to insoluble ivory nut mannan. The plot shows the equilibrium adsorption isotherm (bound CtCBM35 [B] versus free CtCBM35 [F]). Adsorption assay was done at 4°C as described in “Materials and Methods”. Initial protein concentrations of CtCBM35 used were 1-15 μM.
Table 2. Binding parameters of
CtCBM35 with insoluble ligands derived from adsorption isotherm
analysis
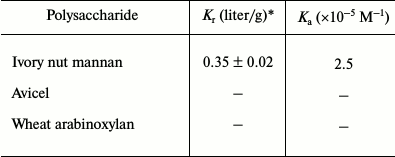
* Values are mean ± SD (n = 3); –, not
determined.
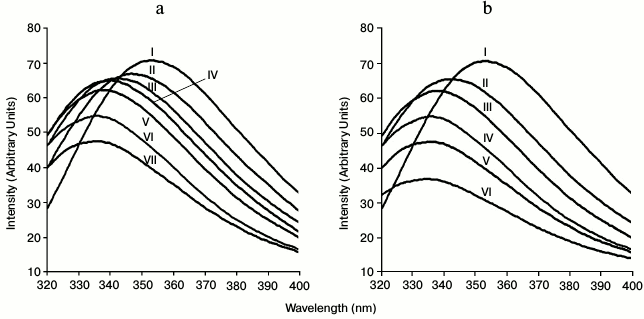
Polysaccharide binding study of CtCBM35 by fluorescence spectroscopy. In the presence of polysaccharide such as locust bean galactomannan and oligosaccharide (mannotriose) with their varying concentration from 0.01 to 0.08% (w/v), significant blue shifts were observed. Binding of locust bean galactomannan and mannotriose with CtCBM35 displayed 21-nm peak shifts towards shorter wavelength of tryptophan emission spectra from λmax 355 to 334 nm (Figs. 3a and 3b). Ligand binding changes the microenvironment of tryptophan due to conformational changes in protein [18, 29]. The higher binding affinity of CtCBM35 with mannotriose was due to simpler structure of the trisaccharide composed of β-(1→4) linked mannose moieties resulting in reduced fluorescence peak intensity, whereas the highly substituted α-(1→6)-galactose residues in locust bean galactomannan hindered the main chain of β-(1→4)-linked mannose residues from binding tryptophan of CtCBM35, thus causing comparatively higher fluorescence intensity (Figs. 3a and 3b). The decrease in the fluorescence intensity was also coupled with a peak shift by 21 nm for CtCBM35 binding mannotriose as well as locust bean galactomannan (Figs. 3a and 3b).
Fig. 3. Tryptophan fluorescence emission spectrum of CtCBM35 in the presence of (a) locust bean galactomannan (% w/v) (0.0 (I), 0.01 (II), 0.02 (III), 0.04 (IV), 0.06 (V), 0.08 (VI), 0.1 (VII)) and (b) mannotriose (0.0 (I), 0.01 (II), 0.02 (III), 0.04 (IV), 0.06 (V), 0.08 (VI)).
From the relative fluorescence intensities, the deduced affinity constant (Ka) of CtCBM35 with locust bean galactomannan was 1.21·105 M–1 and with mannotriose 3.03·105 M–1 (Table 1). The Ka values were similar to those determined by affinity electrophoresis. Therefore, the fluorescence studies of polysaccharide binding of CtCBM35 also confirmed the same binding affinity pattern as observed with the affinity electrophoresis experiment. The number of binding site concentration (n) were derived from Stern–Volmer equation, and it was found to be 0.79 ± 0.09 µmol/g with locust bean galactomannan, whereas with mannotriose the binding capacity was observed to be 0.90 ± 0.05 µmole/g. It means both the polysaccharides have a single binding site for CtCBM35. From the derived affinity constants, the Gibb’s free energies of binding were calculated using the equation:
ΔG = –RT·ln Ka,
where ΔG is Gibb’s free energy, R universal gas constant (J·K–1·mol–1), T temperature in Kelvin, Ka association constant (M–1). The free energy of binding of CtCBM35 with locust bean galactomannan was –6.1 kcal/mole and with mannotriose –6.9 kcal/mole (Table 1). The higher binding affinity (Ka) and free energy of binding indicated that mannotriose made an easier platform for CtCBM35 binding than locust bean galactomannan, although CtCBM35 has a similar number of binding site concentration for both of these ligands.
Polysaccharide specificity by members of the CtCBM35 family is probably due to the conserved hydrophobic aromatic residues that play a major role in polysaccharide binding [7]. Tryptophan, being one of such residues having an indole ring with intrinsic fluorescence property with higher quantum yield, displays fluorescence emission at 320 to 400 nm [18]. Polysaccharide binding alters the microenvironment of tryptophan due to conformational changes in the protein. Usually in CBMs, the aromatic residues responsible for polysaccharide binding are lying in the hydrophobic core. Due to polysaccharide binding and direct interaction with tryptophan, the fluorescence emission is gradually decreased. Thus, gradual fall in peak intensities were coupled with peak shifts (~21 nm) due to altered conformation of the native CtCBM35.
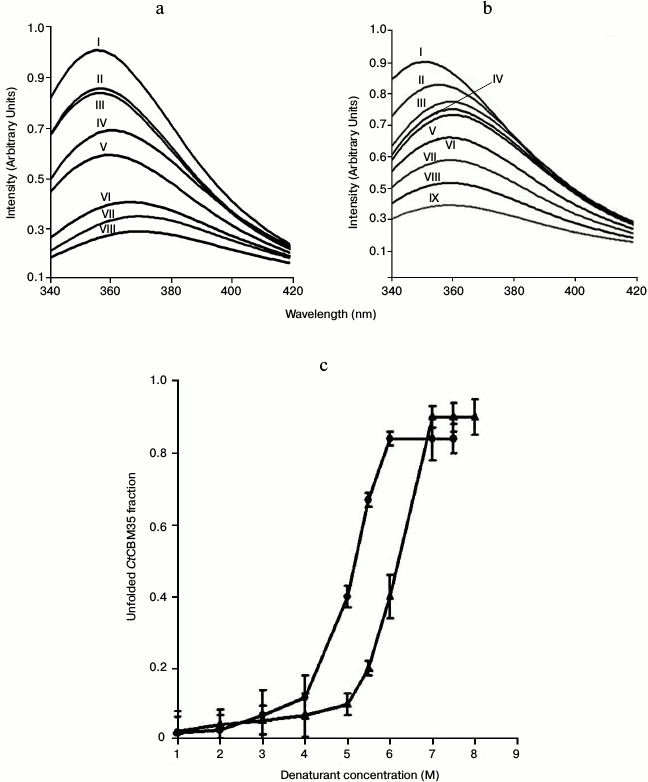
Unfolding transition of CtCBM35 in guanidine hydrochloride and urea. The unfolding transition of CtCBM35 was investigated in the presence of GnHCl and of urea. The addition of GnHCl (1-6 M) or urea (1-7 M) to CtCBM35 caused a decrease in the peak intensity as the denaturant concentration increased (Figs. 4a and 4b). The maximum fall in peak intensity of CtCBM35 was observed at 6 M GnHCl and 7 M urea (Figs. 4a and 4b). The lowering of peak intensities overlapped with a bathochromic shift (red shift, shift towards longer wavelength) of the fluorescence maxima from 355 nm (native form) to 370 nm (denatured form) in the presence of GnHCl and from 355 to 360 nm in the presence of urea. This was perhaps due to the Trp residue, which is otherwise buried in the native form, being exposed on the surface in the denatured CtCBM35. A similar unfolding mechanism of recombinant human interferon γ was described earlier [30]. The unfolding curves obtained at pH 7.0 for GnHCl and urea are shown in Fig. 4c. In both cases the curves displayed a sigmoid pattern, signifying that the unfolding of CtCBM35 in the presence of GnHCl or urea was a two-stage process, where low unfolded fractions of CtCBM35 were obtained till 4 and 5 M, respectively. The unfolding of CtCBM35 increased up to 5.5 M of GnHCl and 6.5 M of urea with a saturation phase observed till 8 M. The outcomes also indicated that the unfolding of CtCBM35 was more effective in the presence of GnHCl than of urea. As seen in Fig. 4c, the midpoint value of unfolding of CtCBM35 was 5 M for GnHCl and 6.25 M for urea. The free energies ΔG of CtCBM35 unfolding calculated from these curves by a linear extrapolation method at pH 7.0 were 2.19 kJ/mole for GnHCl and 1.97 kJ/mole for urea. The free energies clearly indicated that higher energy is required by both denaturants to destabilize the electrostatic interactions within CtCBM35. These results were similar to those with the unfolding phenomenon of human interferon γ as described by Christova et al. in presence of GnHCl and urea, which required lower energies to unfold [30].
Fig. 4. Tryptophan fluorescence emission spectrum of CtCBM35 in the presence of (a) guanidium hydrochloride (GnHCl) (control (I), 1 M (II), 2 M (III), 3 M (IV), 4 M (V), 5 M (VI), 5.5 M (VII), 6 M (VIII)), (b) urea (control (I), 1 M (II), 2 M (III), 3 M (IV), 4 M (V), 5 M (VI), 6 M (VII), 6.5 M (VIII), 7 M (IX)), (c) fraction of CtCBM35 unfolded as a function of (circles) GnHCl and (triangles) urea concentrations.
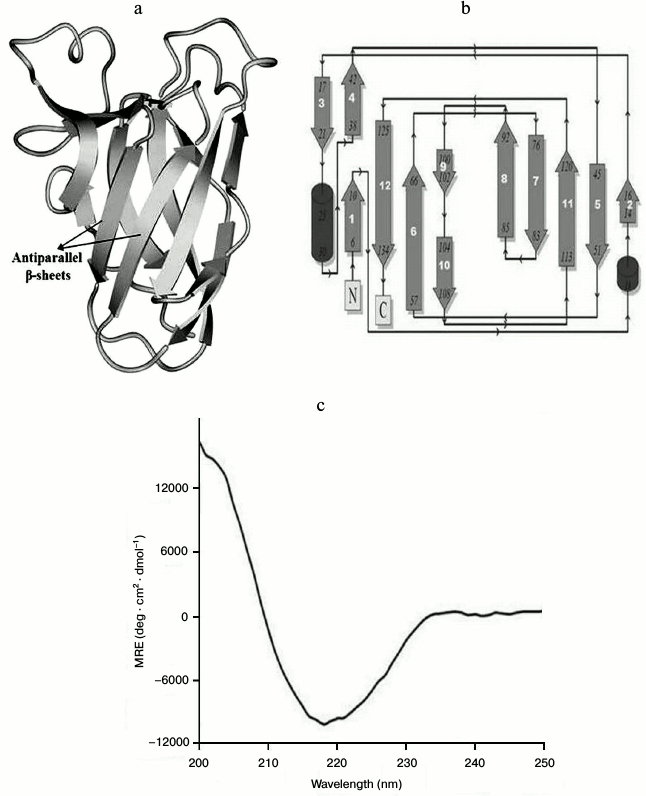
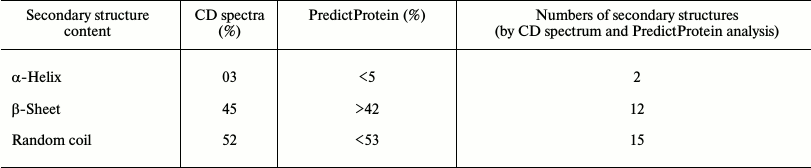
Structure characterization and quality assessment of modeled protein. A BLAST (Basic Local Alignment Search Tool) search for sequence similarity with default parameter in NCBI against PDB database displayed the number of hits of CtCBM35 belonging to the family 35 carbohydrate-binding module. Top hit X-ray crystallography structure of CBM35 from Amycolatopsis orientalis (PDB 2vzp) that covered full query and displayed 34% similarity (score of 39.7 and E-value 0.001) was acquired as a template to model the structure. The overall 3-D structure of CtCBM35 showed that it is rich in β-sheets, consisting of a pair of six-stranded antiparallel β-sheets having β-jelly-roll fold, which is quite common in many CBMs (Fig. 5a). The topology diagram displayed 12 β-strands (dark gray), only two small α-helices (black), and 15 random coils (arrow lines) forming a β-jelly roll topology (Fig. 5b). Secondary structure prediction by PredictProtein showed a pair of α-helices (<5%), but apart from this the dominance of a pair of six extended β-sheets (>42%) and 15 random coils (<53%) were also observed (Table 3) in the 3-D model of CtCBM35. The CD spectrum of CtCBM35 (Fig. 5c) was analyzed using the K2d neural network algorithm tool as described by Andrade et al. [14], which revealed that it contains 45% β-sheets (12 in number), 52% random coils (15 in number), and only 3% α-helices (2 in number) (Table 3). Therefore, the CD analysis substantiated that the prediction of secondary structure was acceptable. The presence of a high percentage of β-sheets and random coils are in agreement with the in silico prediction of the CtCBM35 3-D model, which also revealed β-jelly roll architecture. Structure similarity search of the modeled protein in the DaliLite server (www.ebi.ac.uk/Tools/dalilite/) showed similarity with the native structure of a family 35 CBM from C. thermocellum (2W1W chain B) and CBM35 from A. orientalis (2VZP chain B) with RMSD of 1.7 and 1.8 Å, respectively. Modeled CtCBM35 after energy minimization was validated by Ramachandran plot analysis. Out of a total of 134 residues, 93% of the residues lie in most favored regions, 7% of residues lie in additional allowed region, and there was no residue in generously allowed or disallowed regions. This indicated that amino acid residues in the modeled CtCBM35 occupied favorable phi (φ) and psi (ψ) backbone dihedral angles. Overall quality factor of CtCBM35 was 88% in the ERRAT plot analysis (data not shown). Verify 3D score was obtained as 95%, i.e. 95% of the residues had an averaged 3D-1D score >0.2. This determined the compatibility of an atomic model (3D) with its own amino acid sequence (1D). First it categorized each residue into structural class based on its location and environment (α, β, loop, polar, nonpolar, etc.), then it generates a score by comparing each residue with a collection of good structures as a reference [31].
Fig. 5. a) Overall 3-D representation of modeled carbohydrate-binding module family 35 from Clostridium thermocellum showing β-jelly-roll fold; b) topology diagram of CtCBM35 showing β-jelly roll topology; c) CD spectrum of CtCBM35 for secondary structure determination.
Table 3. Secondary structure analysis of
CtCBM35 by far-UV CD spectrum and the PredictProtein tool
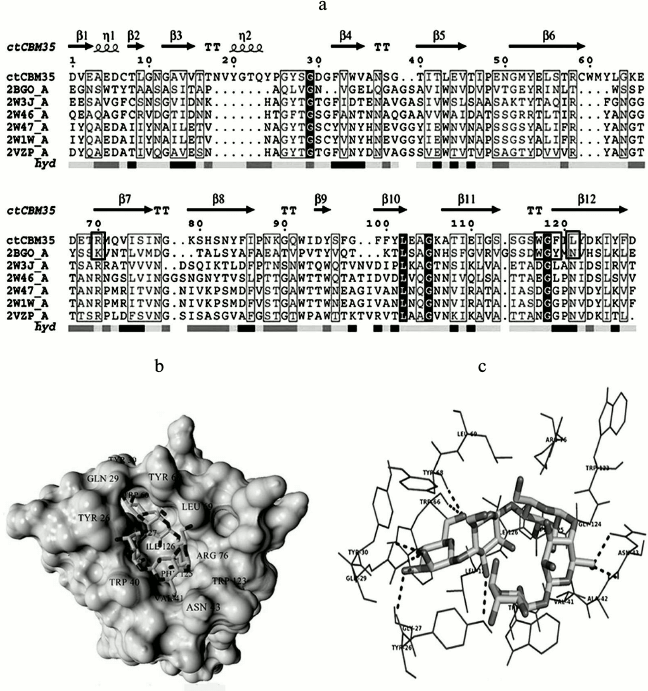
Substrate-binding site analysis. CBMs play a critical role in substrate targeting and substrate binding. It is very important to investigate the key residues playing significant roles in recognition and binding with the substrate and whether they are conserved within the family or are substrate-specific. Multiple sequence alignment (MSA) was performed by taking representative members of the CBM35 family having different substrate specificities. The MSA result showed many amino acid residues are conserved within the family 35 CBMs (Fig. 6a). We have identified that a Trp-Gly-Phe motif is conserved and is also present in CtCBM35 (shown in horizontal black box) (Fig. 6a). Moreover, a Trp-Gly-Tyr motif that is probably involved in the substrate specificity towards mannans was conserved within the mannan-specific CBMs [32]. It was demonstrated by NMR solution structure of mannan-specific CBM35 from C. japonicus that it bound to mannopentaose, involving amino acid residues Tyr60, Lys63, Trp109, Gly110, and Tyr111 showing maximum chemical shifts on ligand binding [33]. The involvement of aromatic amino acid residues Tyr22-Tyr53-Tyr129 in β-(1→4)-linked glucomannan recognition by CBM11 was reported earlier by Carvalho et al. [34]. We also found that the above-mentioned key amino acid residues were present in CtCBM35, except for Tyr60. The other amino acid residues like Arg76, Trp123, Gly124, and Phe125 were conserved within CtCBM35 (highlighted in black box in Fig. 6a).
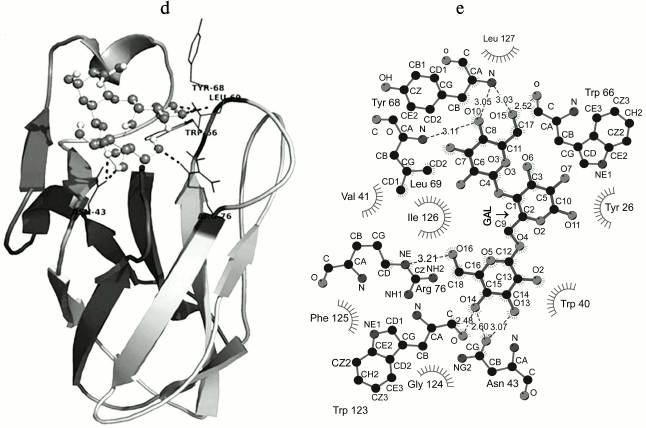
Fig. 6. a) Multiple sequence alignment of CBM35 family members having different substrate specificity with their respective PDB:IDs of Amycolatopsis orientalis (2VZP), Cellvibrio japonicus Abf62 (2W46), uncultured bacterium from environmental isolate (2W3J), Clostridium thermocellum (2W1W), and Cellvibrio japonicus (2BGO) were aligned with CtCBM35. The residues that are conserved and probably involved in substrate specificity and binding (Man-CBM35) are shown in rectangular boxes. The line below the sequence represents the relative hydrophobicity of the residues, dark gray for hydrophilic, black for hydrophobic, and light gray for intermediate. b) Model of binding site surface cavity with labeled residues forming a hydrophobic groove to accommodate mannopentaose. c) Best pose of docking of CtCBM35 with mannopentaose bound conformation shows polar interaction (dashed line) and residues probably involve in hydrophobic interaction present within 4 Å. d) Ribbon representation of CtCBM35 using PyMOL complex with galactomannan (ball-stick view) shows position of substrate binding site (shown in circle). Residues making polar contact with galactomannan are positioned mainly in the loop region (shown by dashed lines). e) Schematic 2-D depiction of galactomannan (GAL) binding site residues of CtCBM35. Dashed lines show hydrogen bonds, and connected atoms are shown with spokes radiating back. The amino acid residues displayed in the arc with spokes are involved in hydrophobic interactions.
Docking analysis of ligand binding interaction with CtCBM35. The molecular docking analysis of modeled CtCBM35 with various manno-oligosaccharides provided a better understanding about the key amino acid residues involved in making strong hydrophobic interactions with the ligand. There is much evidence that shows how hydrophobic stacking interactions and hydrogen bonds play an important role in ligand binding in the case of family 35 CBMs [35]. Our docking results showed that residues Tyr26, Gln29, Asn43, Trp66, Tyr68, Leu69, Arg76, and Leu127 participate in making polar interaction with mannopentaose, while residues Tyr30, Trp40, Val41, Ala42, Met67, Trp123, Phe125, and Ile126 make a hydrophobic pocket around the binding site cavity (Figs. 6b and 6c). Mannotriose showed higher binding affinity for CtCBM35, Ka = 3.0·105 M–1 as compared to galactomannan, Ka = 1.22·105 M–1 (Table 2). The free energy of binding (ΔG) of mannotriose was maximum (–6.8 kcal/mole), followed by galactomannan (–6.0 kcal/mole) (Table 1), while mannohexose displayed positive free binding energy on docking with CtCBM35 (data not shown). This is possibly due to the size of the binding site pocket, which is not large enough to accommodate bulkier mannohexose as compared to mannotriose. The putative protein–ligand binding site shown by CBMs having β-sandwich fold is mainly situated on either concave surface made by the antiparallel β-sheets or at the top cavity made by the loop that connects the two β-sheets [36]. Analysis of binding site topography of the CtCBM35 showed that it is made up by the amino acid residues present in loops connecting antiparallel β-sheets (Fig. 6d). The first loop started from Ser120 to Phe125 (between the 11-12 β-strand) making a base of the cavity, creating a flat hydrophobic platform involving two conserved aromatic amino acids Trp123 and Phe125. The second loop started from Cys65 to Arg76 (the 6-7 β-strand), and third loop from Val21 to Tyr33 (the 3-4 β-sheets) formed the side surface of the cavity and provided the solvent accessible surface for the ligands (Fig. 6d). Here, amino acid residues Trp66, Tyr68, Leu69, and Arg76 (from the loop second) and Tyr26 and Glu29 (from the loop third) were also present and made polar contact with the ligand molecule (Fig. 6e). Therefore, the functional and structural elucidation of mannan specific CtCBM35 from C. thermocellum may be useful to enhance activity by appending to a mannanase for higher degree of hydrolysis of complex manno-configured polysaccharides into simple sugars. Moreover, these findings might lead to understanding mannan-specific CBM35 from C. thermocellum, which may play a role in prebiotic oligosaccharide production in conjunction with mannanase from mannan rich polysaccharides.
The research work was supported in part by the Cutting-Edge Research Enhancement and Scientific Training (CREST) Fellowship from the Department of Biotechnology, Ministry of Science and Technology, to Arun Goyal.
REFERENCES
1.Cai, S., Zheng, X., and Dong, X. (2011) CBM3d, a
novel subfamily of family 3 carbohydrate-binding modules identified in
Cel48A exoglucanase of Cellulosilyticum ruminicola, J.
Bacteriol., 193, 5199-5206.
2.Taylor, E. J., Goyal, A., Guerreiro, C. I. P. D.,
Prates, J. A., Money, V. A., Ferry, N., Morland, C., Planas, A.,
Macdonald, J. A., Stick, R. V., Gilbert, H. J., Fontes, C. M., and
Davies, G. J. (2005) How family 26 glycoside hydrolases orchestrate
catalysis on different polysaccharides: structure and activity of a
Clostridium thermocellum lichenase, CtLic26A, J. Biol.
Chem., 280, 32761-32767.
3.Sunna, A. (2010) Modular organization and
functional analysis of dissected modular β-mannanase CsMan26 from
Caldicellulo siruptor Rt8B.4, Appl. Microbiol.
Biotechnol., 86, 189-200.
4.Bolam, D. N., Xie, H., Pell, G., Hogg, D.,
Galbraith, G., Henrissat, B., and Gilbert, H. J. (2004) X4 modules
represent a new family of carbohydrate-binding modules that display
novel properties, J. Biol. Chem., 279, 22953-22963.
5.Valenzuela, S. V., Diaz, P., and Pastor, F. I.
(2012) Modular glucuronoxylan-specific xylanase with a family CBM35
carbohydrate-binding module, Appl. Environ. Microbiol.,
78, 3923-3931.
6.Ahmed, S., Saraf, T., and Goyal, A. (2009) Homology
modeling of family 39 glycoside hydrolase from Clostridium
thermocellum, Curr. Trend. Biotechnol. Pharm., 3,
210-218.
7.Tunnicliffe, R. B., Bolam, D. N., Pell, G.,
Gilbert, H. J., and Williamson, M. P. (2005) Structure of a
mannan-specific family 35 carbohydrate-binding module: evidence for
significant conformational changes upon ligand binding, J. Mol.
Biol., 347, 287-296.
8.Johansson, R., Gunnarsson, L. C., Ohlin, M., and
Ohlson, S. (2006) Thermostable carbohydrate-binding modules in affinity
chromatography, J. Mol. Recognit., 19, 275-281.
9.Abbott, D. W., and Boraston, A. B. (2012)
Quantitative approaches to the analysis of carbohydrate-binding module
function, Methods Enzymol., 510, 211-231.
10.Correia, M. A., Abbott, D. W., Gloster, T. M.,
Fernandes, V. O., Prates, J. A., Montanier, C., Dumon, C., Williamson,
M. P., Tunnicliffe, R. B., Liu, Z., Flint, J. E., Davies, G. J.,
Henrissat, B., Coutinho, P. M., Fontes, C. M. G. A., and Gilbert, H. J.
(2010) Signature active site architectures illuminate the molecular
basis for ligand specificity in family 35 carbohydrate binding module,
Biochemistry, 49, 6193-6205.
11.Montanier, C., van Bueren, A. L., Dumon, C.,
Flint, J. E., Correia, M. A., Prates, J. A., Firbank, S. J., Lewis, R.
J., Grondin, G. G., Ghinet, M. G., Gloster, T. M., Herve, C., Knox, J.
P., Talbot, B. G., Turkenburg, J. P., Kerovuo, J., Brzezinski, R.,
Fontes, C M., Davies, G. J., Boraston, A. B., and Gilbert, H. J. (2009)
Evidence that family 35 carbohydrate binding modules display conserved
specificity but divergent function, Proc. Natl. Acad. Sci. USA,
106, 3065-3070.
12.Ghosh, A., Luis, A. S., Bras, J. L. A., Pathaw,
N., Chrungoo, N. K., Fontes, C. M. G. A., and Goyal, A. (2013)
Deciphering ligand specificity of a Clostridium thermocellum
family 35 carbohydrate binding module (CtCBM35) for gluco- and
galacto-substituted mannans and its calcium induced stability, Plos
One, 8, e80415.
13.Kelly, S. M., Jess, T. J., and Price, N. C.
(2005) How to study proteins by circular dichroism, Biochim.
Biophys. Acta, 1751, 119-139.
14.Andrade, M. A., Chacon, P., Merelo, J. J., and
Moran, F. (1993) Evaluation of secondary structure of proteins from UV
circular dichroism spectra using an unsupervised learning neural
network, Protein Eng., 6, 383-390.
15.Takeo, K. (1984) Affinity electrophoresis:
principles and applications, Electrophoresis, 5,
187-195.
16.Bradford, M. M. (1976) Rapid and sensitive method
for the quantitation of microgram quantities of protein utilizing the
principle of protein–dye binding, Anal. Biochem.,
72, 248-254.
17.Gilkes, N. R., Eric, J. S., Henrissat, B.,
Tekant, B., Miller, R. C., Jr., Warren, R. A., and Kilburn, D. G.
(1992) The adsorption of a bacterial cellulase and its two isolated
domains to crystalline cellulose, J. Biol. Chem., 267,
6743-6749.
18.Royer, C. A. (2006) Probing protein folding and
conformational transitions with fluorescence, Chem. Rev.,
106, 1769-1784.
19.Belatik, A., Hotchandani, S., Carpentier, R., and
Tajmir-Riahi, H. A. (2012) Locating the binding sites of Pb (II) ion
with human and bovine serum albumins, Plos One, 7,
1-9.
20.Ahmad, F., Yadav, S., and Taneja, S. (1992)
Determining stability of proteins from guanidinium chloride transition
curves, Biochem. J., 287, 481-485.
21.Krieger, E., Joo, K., Lee, J., Raman, S.,
Thompson, J., Tyka, M., Baker, D., and Karplus, K. (2009) Homology
modeling in YASARA, Proteins, 77, 114-122.
22.Thompson, J. D., Higgins, D. G., and Gibson, T.
J. (1994) ClustalW: improving the sensitivity of progressive multiple
sequence alignment through sequence weighting, position-specific gap
penalties and weight matrix choice, Nucleic Acids Res.,
22, 4673-4680.
23.O’Boyle, N. M., Banck, M., James, C. A.,
Morley, C., Vandermeersch, T., and Hutchison, G. R (2011) Open babel:
an open chemical toolbox, J. Chem. Inf. Model, 3,
1-14.
24.Morris, G. M., Goodsell, D. S., Halliday, R. S.,
Huey, R., Hart, W. E., Belew, R. K., and Carlson, A. J. (1998)
Automated docking using a Lamarckian genetic algorithm and an empirical
binding free energy function, J. Comput. Chem.,
19, 1639-1662.
25.Mizutani, K., Fernandes, V. O., Karita, S., Luis,
A. S., Sakka, M., Kimura, T., Jackson, A., Zhang, X., Fontes, C. M.,
Gilbert, H. J., and Sakka, K. (2012) Influence of a mannan binding
family 32 carbohydrate binding module on the activity of the appended
mannanase, Appl. Environ. Microbiol., 78, 4781-4787.
26.Montanier, C., van Bueren, A. L., Dumon, C.,
Flint, J. E., Correia, M. A., Prates, J. A., Firbank, S. J., Lewis, R.
J., Grondin, G. G., Ghinet, M. G., Gloster, T. M., Herve, C., Knox, J.
P., Talbot, B. G., Turkenburg, J. P., Kerovuo, J., Brzezinski, R.,
Fontes, C. M., Davies, G. J., Boraston, A. B., and Gilbert, H. J.
(2009) Evidence that family 35 carbohydrate binding modules display
conserved specificity but divergent function, Proc. Natl. Acad. Sci.
USA, 106, 3065-3070.
27.Din, N., Damude, H. G., Gilkes, N. R., Miller, R.
C., Jr., Warren, R. A., and Kilburn, D. G. (1994) C1-Cx revisited:
intramolecular synergism in a cellulose, Proc. Natl. Acad. Sci.
USA, 91, 11383-11387.
28.Din, N., Gilkes, N. R., Tekant, B., Miller, R.
C., Jr., Warren, R. A. J., and Kilburn, D. G. (1991) Non-hydrolytic
disruption of cellulose fibers by the binding domain of a bacterial
cellulase, Nat. Biotechnol., 9, 1096-1099.
29.Mohan, S., Thambi Dorai, D., Srimal, S.,
Bachhawat, B. K., and Das, M. K. (1983) Fluorescence studies on the
interaction of some ligands with carcinoscorpin, the sialic acid
specific lectin, from the horseshoe crab, Carcinoscorpius
rotundacauda, J. Biosci., 5, 155-162.
30.Christova, P., Todorova, K., Timtcheva, I.,
Nacheva, G., Karshikoff, A., and Nikolov, P. (2003) Fluorescence
studies on denaturation and stability of recombinant human
interferon-gamma, Z. Naturforsch. C, 58, 288-294.
31.Pace, N. C., Shirley B. A., and Thomson, J. A.
(1989) in Protein Structure: a Particular Approach (Creighton,
T. E., ed.) IRL Press, Oxford, pp. 311-329.
32.Pinheiro, B. A., Proctor, M. R.,
Martinez-Fleites, C., Prates, J. A. M., Money, V. A., Davies, G. J.,
Bayer, E. A., Fontesm, C. M., Fierobe, H. P., and Gilbert, H. J. (2008)
The Clostridium cellulolyticum dockerin displays a dual binding
mode for its cohesin partner, J. Biol. Chem., 283,
18422-18430.
33.Colovos, C., and Yeates, T. O. (1993)
Verification of protein structures: patterns of nonbonded atomic
interactions, Prot. Sci., 2, 1511-1519.
34.Carvalho, A. L., Goyal, A., Prates, J. A. M.,
Bolam, D. N., Gilbert, H. J., Pires, V. M. R., Ferreira, L. M. A.,
Planas, A., Romao, M. J., and Fontes, C. M. G. A. (2004) The family 11
carbohydrate-binding module of Clostridium thermocellum
Lic26ACel5E accommodates β-1,4- and β-1,3-1,4-mixed linked
glucans at a single binding site, J. Biol. Chem., 279,
34785-34793.
35.Bowie, J. U., Luthy, R., and Eisenberg, D. (1991)
A method to identify protein sequences that fold into a known
three-dimensional structure, Science, 5019, 164-170.
36.Fanutti, C., Ponyi, T., Black, G. W., Hazlewood,
G. P., and Gilbert, H. J. (1995) The conserved noncatalytic 40-residue
sequence in cellulases and hemicellulases from anaerobic fungi
functions as a protein docking domain, J. Biol. Chem.,
270, 29314-29322.