Leader Sequences of Eukaryotic mRNA Can Be Simultaneously Bound to Initiating 80S Ribosome and 40S Ribosomal Subunit
E. A. Sogorin, N. E. Shirokikh, A. M. Ibragimova, V. D. Vasiliev, S. Ch. Agalarov, and A. S. Spirin*
Institute of Protein Research, Russian Academy of Sciences, 142290 Pushchino, Moscow Region, Russia; E-mail: spirin@vega.protres.ru* To whom correspondence should be addressed.
Received December 27, 2011
Binding of mRNA leader sequences to ribosomes was studied in conditions of a cell-free translation system based on wheat germ extract. Leader sequence of TMV mRNA (the so-called omega-RNA sequence) was able to bind simultaneously 80S ribosome and 40S ribosomal subunit. It was found that nucleotide substitutions in omega-RNA resulting in destabilization of RNA structure have no effect on the complex formation with both 80S ribosome and 40S ribosomal subunit. Leader sequence of globin mRNA is also able to form a similar joint complex. It is supposed that the ability of mRNA leader sequences to bind simultaneously 80S ribosome and 40S subunit is independent of leader nature and may reflect previously unknown eukaryotic mechanisms of translation initiation.
KEY WORDS: omega-RNA, initiation of protein biosynthesis, ribosomal complexes, electron microscopy of ribosomesDOI: 10.1134/S0006297912040049
The rate of mRNA translation by ribosomes and the protein synthesis efficiency directly depend on initiation rate. In eukaryotes 5′-untranslated regions (5′-UTR), called also leader sequences, may play the role of translation enhancers due to their ability to attract ribosomes and initiation factors and thus provide for effective translation initiation on mRNA. Leader sequences of most plant viral RNA are such enhancers. One of the best studied is 5′-unntranslated sequence of tobacco mosaic virus (TMV) RNA, the so-called omega-sequence [1-5]. This sequence consists of approximately 70 nucleotides (from 63 to 73, depending on the viral strain) and contains from 10 to 13 repeating CAA nucleotide motifs. These motifs, frequently as their tandems, comprise about half of total omega-sequence [1].
Earlier it was shown in our laboratory that isolated omega-RNA is a rather compact molecule with well-developed spatial structure [6]. At the same time, theoretical analysis of RNA nucleotide sequence is indicative of impossibility of formation of somewhat extended Watson–Crick type helical regions. To explain the apparent contradiction, a model was proposed for polyribonucleotide stacking with repeating (CAA)n-sequence in the form of triple helix, elements of which are C-A-C and A-C-A triads [7]. The absence of extended Watson–Crick-type helices was confirmed experimentally by chemical and enzymatic probing techniques [8], while results of investigation of target-altered forms of omega-RNA indirectly favored the existence of triple helix [9]. It was shown that nucleotide substitutions both in central (CAA)n-containing and in 3′-proximal omega-RNA regions result in significant structural destabilization and decompactization.
In this work we have decided to check how altered omega-RNA forms, devoid of high stability and compactness, will form complexes with 80S ribosomes. It appeared that the loss of RNA stability and compactness itself does not result in significant change of its ribosome-biding ability. However, most surprising is that in the course of our study we found an unusual, formerly unknown complex including both complete 80S ribosome and 40S ribosomal subunit. Formation of the complex seems to be independent of the nature of the leader. According to the generally accepted model of initiation in eukaryotes, 40S ribosomal subunit, originally binding to 5′-terminus of mRNA, begins scanning 5′-UTR until it stops at an initiating codon. Here it joins 60S ribosomal subunit, thus forming 80S ribosome, which is the complete initiating complex ready for translation. However, it should be noted that the scanning 40S subunit was never observed by physical methods.
METHODS OF INVESTIGATION
Isolation of omega-RNA and its altered forms was described earlier [9]. RNA was labeled at the 3′-end with fluorescein-5-thiosemicarbazide using a published technique [10]. RNA was bound to ribosomes in a cell-free translation system based on wheat germ extract [11]. The protocol of extract preparation is described in the same article [11]. RNA was incubated at 25°C for 30 min in 25 µl translation mixture, then 50 µl buffer (37.5 mM Hepes-KOH, pH 7.5, 150 mM KOAc, 15 mM MgOAc) was added, mixed, and the mixture applied onto a sucrose gradient. Sucrose solutions were prepared in buffer containing 18.5 mM Hepes-KOH, pH 7.6, 5 mM MgCl2, 100 mM KCl, 0.1 mM EDTA. The sample was centrifuged in a SW-41 rotor at 37,000 rpm and 4°C. Then the content of tubes was separated into 0.25 ml fractions. The optical density in the UV region was registered continuously using a flow-type cell. Fluorescence in fractions was measured on an RF-5103PC apparatus (Shimadzu, Japan). For electron microscopy, fractions were fixed in formaldehyde, negatively stained by 1% aqueous uranyl acetate according to Valentine et al. [12], and analyzed in a JEM-100C electron microscope (JEOL, Japan) at accelerating voltage 80 kV.
RESULTS AND DISCUSSION
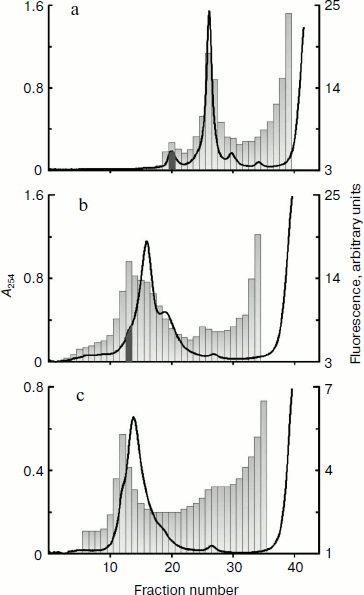
Omega-RNA can be simultaneously bound to both 80S ribosome and 40S ribosomal subunit. Fluorescence-labeled omega-RNA was added in a cell-free wheat germ translation system. After incubation with omega-RNA, the extract containing all components for protein synthesis was centrifuged in a sucrose concentration gradient. Figure 1a shows profiles of optical density and fluorescence distribution in 15-45% sucrose gradient. The character of optical density distribution at 254 nm shows in addition to the peak corresponding to 80S ribosomes, a “heavier” peak along with a “heavy” shoulder on the left slope of the 80S peak. This shoulder becomes clearly pronounced in a more flattened (10-30%) gradient. Data in Fig. 1b show that the main peak of fluorescence (omega-RNA) corresponds to this “heavy” shoulder of optical density.
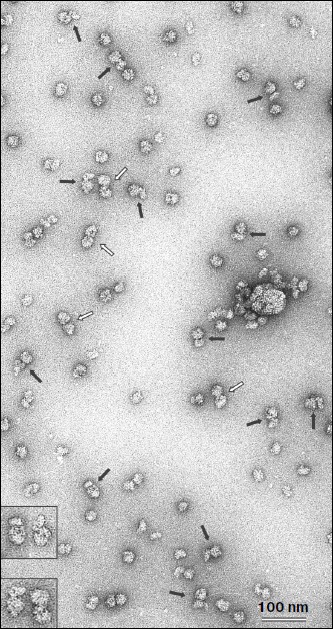
The fraction corresponding to the fluorescence peak and optical density shoulder (shown by dark gray in Fig. 1b) was analyzed by electron microscopy. Figure 2 shows an electron microphotograph of a preparation from this fraction. It is well seen that a significant fraction of the ribosomal components is represented in the form of 80S–40S complexes (black arrows). There are also 80S ribosomes and “free” 40S and 60S ribosomal subunits along with some examples of 80S–80S dimers (white arrows) in the field. This implies that omega-RNA is able to bind simultaneously the full 80S ribosome and the 40S subunit. Random mutual orientation of 80S ribosome and 40S subunit within the complex deserves special consideration. Microphotographs of two complexes at larger magnification are shown in the upper inset as examples of different mutual orientations of individual 40S subunit and 40S subunit within 80S ribosome. The revealed fact suggests the absence of specific interaction between two 40S subunits bound with each other within the complex only by means of an mRNA strand. Electron-microscopic analysis of a fraction from the “heavy” peak (dark gray in Fig. 1a) revealed mainly the presence of 80S–80S ribosome dimers (data not shown). The latter correlates with data of earlier works [13, 14], which showed that omega-RNA was able to form a complex with two 80S ribosomes. However, rather rough resolution of optical density profile and the absence of electron-microscopic data prevented authors of those works to identify the RNA complex with 80S ribosome and 40S subunit.Fig. 1. Binding of fluorescence-labeled RNA to ribosomes. Sucrose gradient centrifugation analysis. Sedimentation direction is from the right to the left. The continuous line shows optical density registered continuously at 254 nm. Columns show fluorescence measured in fractions. Fractions taken for electron-microscopic analysis are shown by dark gray. a, b) Omega-RNA; c) β-globin leader. Centrifuging conditions: a) 15-45% sucrose gradient, 2 h 50 min. b, c) 10-30% sucrose gradient, 4 h. Centrifugation rate in all cases was 37,000 rpm.
Nucleotide substitutions in omega-RNA have no effect on its ability to form complex simultaneously with 80S ribosome and 40S ribosomal subunit. Earlier we obtained and characterized target-altered forms of omega-RNA [9]. In one form three adenines were replaced by three cytosines in the CCA repeats following one after another at the central part of the sequence. In another form all adenines and uracils in the 3′-proximal AU-rich part were replaced by guanines and cytosines. Both forms were significantly less stable and not as compact as the original omega-RNA. At the same time, despite structural alterations they retained their ability for translation enhancement. Checking in the cell-free system of protein synthesis has shown that the altered RNA forms exhibit almost the same leader activity as the wild-type RNA (unpublished). One more much reduced omega-RNA form was studied in this work. Its sequence is completely devoid of the 5′-proximal and central parts, whereas only the 3′-adjacent AU-rich region is retained. It includes only 40 nucleotides (the sequence of AU-rich omega-RNA region is shown in bold): GGGAAAGCUUAUUACAAUUACUAUUUACAAUUACACCAUG. Despite the absence of a significant part of the omega-RNA sequence, this form also retained the translation enhancing ability in the cell-free synthesis system (unpublished). All three fluorescein-labeled omega-RNA forms were checked for their ribosome-binding ability using a similar technique described in the previous section. The optical density and fluorescence distribution profiles for two RNA forms with nucleotide substitutions in the central and 3′-adjacent regions and retained total length (87 nucleotides) were the same as in the original omega-RNA (Fig. 1, a and b), whereas the short form containing only AU-rich 3′-adjacent sequence had no peak corresponding to 80S ribosome dimer, but there was a “heavy” shoulder containing 80S–40S complex (not shown). Thus, the ability of omega-RNA to bind either two 80S ribosomes or 80S ribosome and individual 40S subunit does not directly depend on the extent of the RNA compactness. Besides, the joint binding of both 80S ribosome and 40S subunit is independent of leader length within the limits from 87 to 40 nucleotides.Fig. 2. Electron microphotograph of omega-RNA complexes with ribosomes. Preparation from the fraction marked by dark gray in Fig. 1b. Black arrows point to omega-RNA complexes with 80S ribosome and 40S subunit, white arrows show complexes with two 80S ribosomes. The insets are microphotographs of corresponding complexes at higher magnification, clearly showing different orientations of individual 40S subunits relative to 40S subunits within 80S ribosome in 80S–40S RNA-ribosome complexes (above) and morphological distinction of 80S–40S and 80S–80S RNA-ribosome complexes (below). Negative staining by uranyl acetate.
Leader sequence of globin mRNA is also able to form complex simultaneously with 80S ribosome and 40S ribosomal subunit. A question arises whether only omega-RNAs (and related viral leaders) are able to form a similar complex or it is a common property of mRNA leaders in eukaryotic translation systems. To answer this question, we carried out an analogous experiment where a leader of completely different nature – 71 nucleotides long 5′-untranslated region of rabbit β-globin mRNA – was used as RNA [15]. The data are shown in Fig. 1c, where one can see that the fluorescence peak (i.e. that of leader RNA) coincides with the peak of “heavy” shoulder of optical density. This implies that the globin mRNA leader is also capable of forming joint complex with both 80S ribosome and individual 40S subunit. No peak corresponding to 80S ribosome dimer was found in this case (data not shown). Summarizing all observations, we conclude that the ability of leader mRNA sequences to form the joint complex both with 80S ribosome and individual 40S subunit in the cell-free translation system from wheat germ is, most probably, independent of the nature of the leader.
However, so far it is not clear whether the existence of such a complex is an indication of a still unknown mechanism of translation initiation in eukaryotes or its discovery is the result of some unspecific interaction between ribosomal particles and RNA under our conditions. One can suppose that the complete 80S ribosome, localized on leader mRNA sequence, has already passed the stage of translation initiation, whereas the revealed 40S ribosomal subunit is in the course of scanning. Further experiments will be aimed at the answer to this question.
This work was supported by the Russian Foundation for Basic Research (grants 09-04-01729a and 09-04-00537a), the program of the Russian Academy of Sciences “Molecular and Cell Biology”, and the program for Support of Leading Scientific Schools of Russian Federation (grant NSh-8488.2010.4).
REFERENCES
1.Coulet, P., Lomonosoff, G. P., Butler, P. J. G.,
Akam, M. E., Gait, M. J., and Karn, J. (1982) Proc. Natl. Acad. Sci.
USA, 79, 5818-5822.
2.Gallie, D. R., Sleat, D. E., Watts, J. W., Turner,
P. C., and Wilson, T. M. (1987) Nucleic Acids Res., 15,
3257-3273.
3.Sleat, D. E., Gallie, D. R., Jefferson, R. A.,
Bevan, M. W., Turner, P. C., and Wilson, T. M. (1987) Gene,
60, 217-225.
4.Sleat, D. E., Hull, R., Turner, P. C., and Wilson,
T. M. (1988) Eur. J. Biochem., 175, 75-86.
5.Takamatsu, N., Watanabe, Y., Iwasaki, T., Shiba,
T., Meshi, T., and Okada, Y. (1991) J. Virol., 5,
1619-1622.
6.Kovtun, A. A., Shirokikh, N. E., Gudkov, A. T., and
Spirin, A. S. (2007) Biochem. Biophys. Res. Commun., 358,
368-372.
7.Efimov, A. V., and Spirin, A. S. (2009) Biochem.
Biophys. Res. Commun., 388, 127-130.
8.Shirokikh, N. E., Agalarov, S. Ch., and Spirin, A.
S. (2010) Biochemistry (Moscow), 75, 405-411.
9.Agalarov, S. C., Sogorin, E. A., Shirokikh, N. E.,
and Spirin, A. S. (2011) Biochem. Biophys. Res. Commun.,
404, 250-253.
10.Seki, M., Carninci, P., Nishiyama, Y.,
Hayashizaki, Y., and Shinozaki, K. (1998) Plant J., 15,
797-720.
11.Shirokov, V. A., Kommer, A., Kolb, V. A., and
Spirin, A. S. (2007) Methods Mol. Biol., 375,
19-55.
12.Valentine, R. S., Shapiro, B. M., and Stadman, E.
R. (1968) Biochemistry, 7, 2143-2152.
13.Konarska, M., Filipowicz, W., Domdey, H., and
Gross, H. J. (1981) Eur. J. Biochem., 114, 221-227.
14.Tyc, K., Konarska, M., Gross, H. J., and
Filipowicz, W. (1984) Eur. J. Biochem., 140, 503-511.
15.Baralle, F. E. (1977) Cell,
10, 549-558.