Biochemical Characterization of Iron–Sulfur Cluster Assembly in the Scaffold IscU of Escherichia coli
Genfu Wu* and Lingfei Li
College of Life Science, Zhejiang University, Hangzhou 310058, China; E-mail: wugenfu@zju.edu.cn* To whom correspondence should be addressed.
Received October 6, 2011; Revision received October 28, 2011
Iron–sulfur cluster is one of the most common prosthetic groups, and it functions in numerous biological processes. However, little is currently known about the mechanisms of iron–sulfur cluster biosynthesis. In this study, we cloned and purified iron–sulfur cluster assembly proteins from Escherichia coli and assembled the cluster in vitro. The results showed that the assembly of iron–sulfur cluster is completed in about 20 min. Although iron or sulfur binds with IscU equivalently, 2-fold amount of iron or cysteine compared with that of IscU is better for the cluster formation, while high concentrations of IscS (IscS/IscU > 1 : 10) do not facilitate the cluster formation. Environmental pH plays an important role in iron–sulfur cluster assembly; the cluster was well assembled at pH 7.6-8.0, but was inhibited at pH less than 7.4. On supply of a catalytic amount of IscS (1/50 of IscU) and excess of other substrates, with increasing each of IscU, iron, or cysteine concentration, the iron–sulfur cluster assembly process developed from first order reaction, mixed order reaction to zero order reaction, and up to 64% of apo-IscU was converted to the [2Fe–2S] cluster-bound IscU under the optimal laboratory conditions.
KEY WORDS: iron–sulfur cluster, IscS, IscU, IscA-Fe, iron, cysteineDOI: 10.1134/S0006297912020034
Abbreviations: DTT, dithiothreitol; FPLC, fast protein liquid chromatography; Isc, iron–sulfur cluster.
Iron–sulfur cluster is one of the most versatile prosthetic groups
in nature, and it functions in numerous biological processes such as
substrate binding and activation, regulation of enzyme activity and
gene expression, sensing of reactive species, radical generation,
disulfide cleavage, sulfur donation, and storage of iron or electrons
[1, 2]. It has been reported
that an iron–sulfur cluster is first assembled on a scaffold and
then transferred to a target protein [3, 4]. The mechanism of iron–sulfur cluster
assembly is still not completely understood despite many recent
discoveries on the assembly components. In prokaryotes, three distinct
operons for iron–sulfur cluster biosynthesis, termed Nif, Isc,
and Suf, have been identified [1]. The Nif system
exists in nitrogen-fixing bacteria and is thought to play a major role
in the maturation of nitrogenase [5]. The Isc and
Suf systems exist in most prokaryotes, where Isc has a housekeeping
function expressed in normal physiological conditions [4], and Suf is expressed in harsh environmental
conditions, such as oxidative stress or iron starvation [6].
The Isc system consists of at least seven proteins (IscR, IscS, IscU, IscA, HscB, HscA, and ferredoxin), which are encoded by a highly conserved gene cluster named iscRSUA-hscBA-fdx [7]. Among these proteins, IscS and IscU are two essential elements since an iron–sulfur cluster can be assembled with just these two proteins in the presence of Fe2+ and cysteine in vitro [8]. IscS is a pyridoxal phosphate-dependent cysteine desulfurase that catalyzes desulfurization of L-cysteine and provides sulfur for iron–sulfur cluster formation [9]. IscU appears to act as a scaffold that serves as a template in the initial iron–sulfur cluster assembly, and it subsequently transfers the assembled cluster to a target protein [10]. IscA acts either as an alternative scaffold [11, 12] or as an iron carrier to recruit and deliver “free” iron for cluster assembly [13]. HscB and HscA, a pair of heat shock cognate proteins, appear to have chaperone functions involved in modulating the iron–sulfur protein maturation [14]. Ferredoxin may play a role in electron transfer during cluster biosynthesis [15].
Iron–sulfur cluster biosynthesis is a cysteine desulfurase-mediated process [16]. It has been reported that an absorbance peak at 456 nm occurs when an iron–sulfur cluster is assembled in IscU, and the peak amplitude reflects the amount of assembled clusters [8, 17]. This study reports the effect of assembly substrates and environmental pH on iron–sulfur cluster biosynthesis using recombinant E. coli IscU as a scaffold protein.
MATERIALS AND METHODS
Cloning and cultivation of recombinant E. coli strains. The coding regions of E. coli IscA, IscU, and IscS were amplified from wild-type E. coli genomic DNA using polymerase chain reaction [18, 19]. After digestion with NcoI and HindIII, the PCR products were ligated to expression vector pET28b+ to yield recombinant plasmids. The plasmids were then introduced into competent cells of E. coli strain BL21, and the transformants were testified with plasmid sequencing.
The recombinant strains were cultivated in Luria–Bertani (LB) media (with 50 μg/ml kanamycin) overnight, and 5 ml of the culture was inoculated into 500 ml fresh LB media. After incubation in a shaker (250 rpm) at 37°C for about 4 h (to A600 = 1.0), 0.2 mM isopropyl-β-D-thiogalactopyranoside (IPTG) was added, and the incubation was continued for 2 h to induce the expression of the recombinant proteins.
Protein purification. The E. coli cells containing overproduced recombined proteins were harvested and resuspended into 30 ml of pre-chilled buffer A (500 mM NaCl, 20 mM Tris-HCl, pH 8.0). Cells were disrupted using an ultrasonic processor and centrifuged at 17,500g for 60 min to remove cell debris. The supernatants were treated with 10 μg/ml DNase to degrade the DNA before filtering through a 0.45 μm cellulose acetate membrane, and the filtrates were applied to a Superflow nickel-agarose column (2 ml) attached to an FPLC system. The column was washed with three column volumes of buffer A and three column volumes of buffer B (15 mM imidazole, 500 mM NaCl, 20 mM Tris, pH 8.0). The proteins were then eluted with buffer C (0.5 M imidazole, 500 mM NaCl, 20 mM Tris, pH 8.0). The eluted proteins were applied to a Hi-Trap desalting column (5 ml) equilibrated with buffer A to remove imidazole. All purification processes were performed in a 4°C refrigerated chamber. The prepared IscA was then diluted to 100 μM and mixed with an equivalent amount of ferrous ammonium sulfate in the presence of 1 mM dithiothreitol (DTT), and the prepared IscS was mixed with an equivalent amount of pyridoxal phosphate. After incubating at room temperature for 10 min, the iron-binding IscA (IscA-Fe) or pyridoxal phosphate-binding IscS was re-purified using a Hi-Trap desalting column. The purity of protein was greater than 95% as judged by electrophoretic analysis on a 15% polyacrylamide gel containing SDS followed by staining with Coomassie blue. The concentrations of IscA and IscA-Fe were determined using extinction coefficients at 260 nm of 2.4 and 6.0 mM–1·cm–1, respectively. The concentrations of IscU and IscS were determined using extinction coefficients at 280 nm of 11.2 and 39.7 mM–1·cm–1, respectively. All protein concentrations in the text refer to the monomeric species.
Iron–sulfur cluster assembly in IscU. Purified IscU was incubated with IscS, iron-loaded IscA, or ferrous ammonium sulfate in reaction buffer (500 mM NaCl, 20 mM Tris-HCl, pH 8.0) in the presence of 2 mM DTT. The mixture was purged with pure argon gas and preincubated at 37°C for 5 min before L-cysteine was added to initiate the reaction. Iron–sulfur cluster assembly kinetics were monitored in a Beckman DU-640 UV-Vis absorbance spectrometer equipped with a Peltier temperature controller. The formation of IscU[Fe–S] was estimated by the amplitude of the peak at 456 nm or the sharpness of the peak at 456 nm using the formula [A456 – (A430 + A482)/2]. All the assembly tests were performed at least in triplicate.
Purification of IscU[Fe–S]. Assembled IscU[Fe–S] was purified using a Hi-Trap desalting column or a Mono-Q column. Buffer A was used for the Hi-Trap desalting column, and buffer D (20 mM Tris-HCl, pH 8.0) and buffer E (1 M NaCl, 20 mM Tris, pH 8.0) were used for the Mono-Q column (with a linear gradient of 0-0.5 M NaCl in 10 column volumes at a flow rate of 1 ml/min). All buffers were degassed before use. Eluted fractions were collected immediately and stored on ice. The analyses were done as soon as possible.
Measurement of iron and sulfide. The iron-containing protein solutions were heated at 85°C for 15 min in the presence of 2 mM L-cysteine, and released iron was measured using the iron indicator ferrozine (3-(2-pyridyl)-5,6-diphenyl-1,2,4-triazine-4,4′-disulfonic acid) [20]. The amount of iron in solution was calculated based on a standard curve of ferrous ammonium sulfate. The sulfide bound on protein was measured following the method described by Siegel [21]. In brief, 160 μl of protein solution was mixed with 20 μl of 0.02 M N,N-dimethyl-p-phenylenediamine sulfate solution in 7.2 N HCl and 20 μl of 0.03 M FeCl3 solution in 1.2 N hydrochloric acid. After incubating at room temperature for 30 min, the reaction mixture was centrifuged at 14,000 rpm for 10 min, and the 667 nm absorbance of the supernatant was determined. Sulfide was calculated according to a standard curve generated with Na2S.
RESULTS AND DISCUSSION
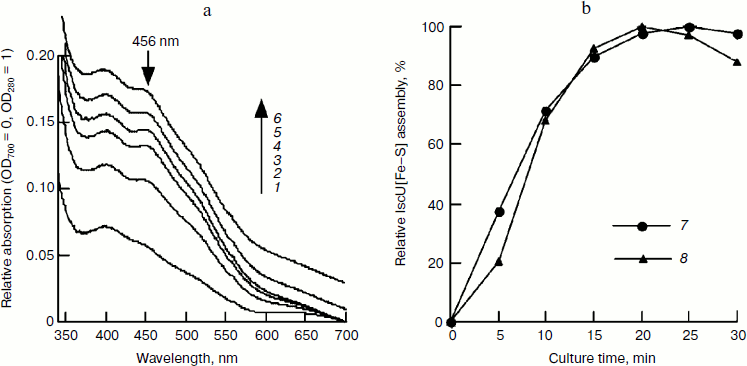
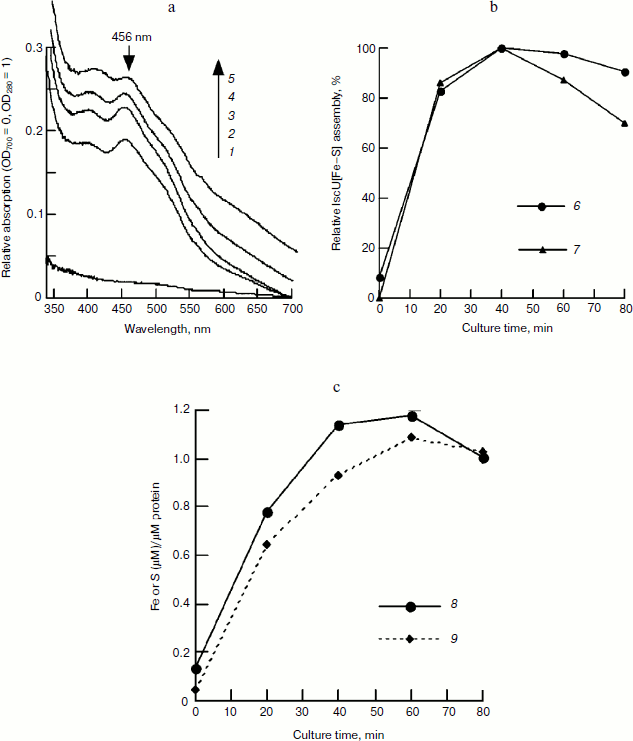
Time course of iron–sulfur cluster assembly in IscU. To determine the time course of cluster assembly, IscU was mixed with an equivalent amount of Fe2+ and a catalytic amount of IscS (1/50 of IscU); an excess of L-cysteine was added to initiate cluster assembly. UV-Vis spectra determination showed that iron–sulfur cluster assembly was completed in 20 min based on 456 nm absorbance. The formed IscU[Fe–S] had an A456/A280 of 0.15 (Fig. 1a), much lower than that (0.31) in Azotobacter vinelandii [22], suggesting the cluster may not be saturated on the IscU scaffold. Continuing the incubation did not increase this absorbance value and instead led to bleaching the cluster (Fig. 1). Based on an extinction coefficient at 456 nm of 5.8 mM–1·cm–1 for the IscU[2Fe–2S] cluster [22], it was estimated that about 52% of apo-IscU was converted to the [2Fe–2S] cluster-bound form. To test whether more iron–sulfur clusters can be assembled at higher iron concentrations, a subsequent experiment was done with IscU and a two-fold excess of ferrous ammonium sulfate and the assembled IscU[Fe–S] was re-purified using a Hi-Trap desalting column to exclude the interference of other substrates. As shown in Fig. 2, the A456/A280 reached 0.23 in the presence of a two-fold excess of ferrous iron and the assembly process continued for about 40 min, although more than 80% of IscU[Fe–S] was formed in 20 min based on the amplitude or the sharpness of 456 nm absorbance (Fig. 2b). Iron and sulfide determination also showed that about 70% of the iron–sulfur clusters were assembled in 20 min (Fig. 2c), agreeing well with the results obtained in the spectral determination (Fig. 1).
Fig. 1. Spectral studies of the time course of IscU[Fe–S] assembly. IscU (50 μM) was incubated with 1 μM IscS, 50 μM Fe2+, and 2 mM DTT at 37°C for 5 min, and 1 mM cysteine was added to initiate IscU[Fe–S] cluster assembly. UV-Vis spectra (a) were recorded every 5 min, and relative cluster assembly (b) was calculated based on the amplitude or the sharpness of the 456-nm absorbance peak. Labels 1-6 represent the incubation times of 5, 10, 15, 20, 25, and 30 min, respectively; curve 7 represents the amplitude of 456-nm absorbance, and curve 8 represents the sharpness of the 456-nm peak. The tests were performed at least in triplicate and similar results were obtained.
Figure 2a also shows that in the control sample without incubation (0 min), the eluted fraction has an absorbance of 0.2441 at 280 nm, indicating that there is about 21.8 μM IscU in this fraction based on an extinct coefficient of 11.2 mM–1·cm–1 [13]. At 40 min, when the iron–sulfur cluster was assembled completely, the absorbance at 456 nm dropped to 0.0783, indicating that there is 13.5 μM of assembled IscU[Fe–S] in this fraction. Assuming no loss of protein during chromatography, about 62% of apo-IscU was converted to IscU[Fe–S], demonstrating that iron concentration significantly influences the iron sulfur cluster assembly.Fig. 2. FPLC studies on the time course of IscU[Fe–S] assembly under the conditions used for Fig. 1. An IscU aliquot was re-purified every 20 min. a) UV-Vis spectra of re-purified IscU; 1-5 represent the culture times of 0, 20, 40, 60, and 80 min, respectively. b) Relative IscU[Fe–S] assembly evaluated from the amplitude of 456-nm absorbance (6) or the sharpness of the 456-nm peak (7). c) Iron (8) and sulfide (9) bound on IscU.
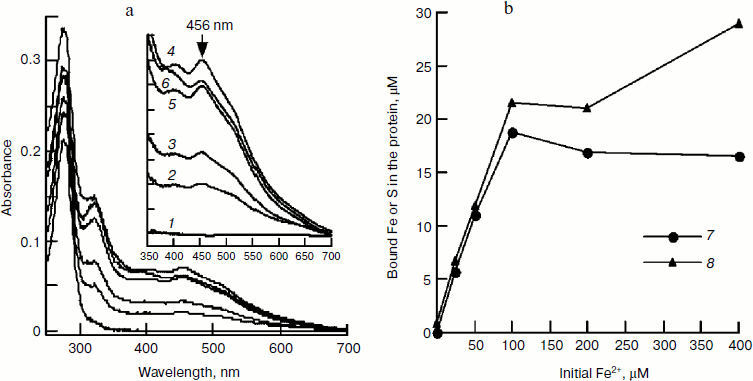
Effect of iron concentration on iron–sulfur cluster assembly. In Fig. 2c, it can be seen that iron and IscU bind equivalently; consistent with the finding that a [2Fe–2S] cluster was assembled in an E. coli IscU dimer [13]. However, in Figs. 1 and 2a, it can be seen that a 2-fold excess of Fe2+ over IscU is better for cluster formation. To determine the optimal iron concentration, clusters were assembled under different concentrations of Fe2+ followed by re-purifying the IscU[Fe–S] using a Hi-Trap desalting column. The results show that in the presence of 25, 50, 100, 200, and 400 µM Fe2+, the A456/A280 value of re-purified IscU[Fe–S] was 0.08, 0.13, 0.22, 0.21, and 0.18, respectively (Fig. 3a), suggesting 100 μM Fe2+ is optimal for iron–sulfur cluster assembly in 50 µM IscU. Based on UV absorbance at 280 nm, 18.7 µM IscU was eluted in the main fraction, and iron and sulfide concentrations in this fraction were 21.6 and 18.8 μM, respectively (Fig. 3b), confirming that a [2Fe–2S] cluster has assembled in an IscU dimer. Figure 3a also shows that with increasing amounts of iron, the absorbance at 456 nm increases. A456 reached 0.0692 upon addition of 100 μM Fe2+, indicating that there is 12 μM IscU[Fe–S] in this fraction. Thus, about 64% of apo-IscU was converted to IscU[Fe–S] under these conditions, consistent with the data presented in Fig. 2.
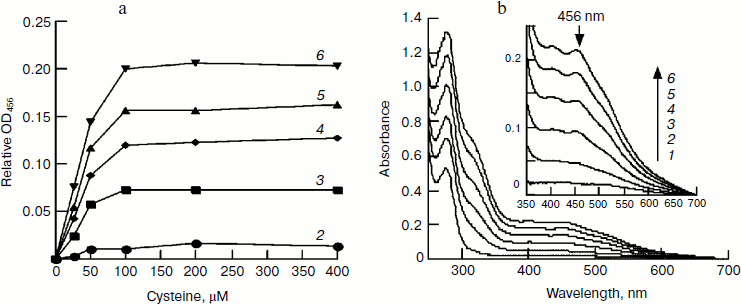
Effect of cysteine on iron–sulfur cluster assembly. Cysteine is another important component for iron–sulfur cluster assembly. To determine its optimal concentration, we assembled IscU[Fe–S] with different amounts of cysteine. Since the free iron level in living cells is very low and the iron for cluster assembly is to be recruited by IscA [23], IscA-Fe was used instead of Fe2+ in this experiment. Spectral determination showed that 100 μM cysteine is sufficient for the cluster assembly in 50 μM IscU (Fig. 4a). Further increase in the cysteine level did not accelerate the reaction; on the contrary, cluster assembly was inhibited when cysteine reached 4 mM. Cysteine is the only substrate for desulfurase IscS. From the half-maximum velocity of cluster assembly, the Michaelis constant for cysteine desulfurase (Km,cys) was calculated to be about 40 μM (Fig. 4a). Figure 4b shows that 100 μM IscA-Fe was not sufficient for the full assembly of iron–sulfur cluster in 50 μM IscU; this result corresponds well with the results in Figs. 1 and 2 demonstrating that 100 μM Fe2+ was necessary for full assembly of the cluster in 50 μM IscU, since only 50 μM iron was bound on 100 μM IscA [24].Fig. 3. Effect of Fe2+ on IscU[Fe–S] assembly under the conditions used for Fig. 1. IscU[Fe–S] was re-purified after 20 min reaction, its spectrum (a) was recorded, and iron and sulfide concentrations (b) were determined. Curves 1-6 represent the initial iron concentrations of 0, 25, 50, 100, 200, and 400 μM, respectively. Curve 7 represents the sulfur content and curve 8 represents the iron content in the re-purified IscU[Fe–S].
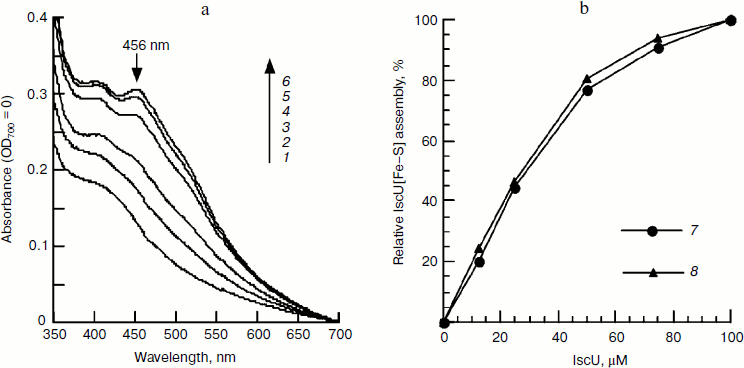
Effect of IscU on iron–sulfur cluster assembly. To further elucidate the optimal ratio of IscU to iron, the iron–sulfur cluster was assembled with a fixed concentration of IscA-Fe and increasing amounts of IscU. In the presence of 100 μM IscA-Fe, 2 mM DTT, and 1 μM IscS, the iron–sulfur cluster assembly exhibited first order reaction kinetics when IscU was less than 25 µM, while it exhibited a mixed order reaction when IscU was between 50 and 100 µM (Fig. 5). Since half of the clusters were assembled in the presence of 25 μM IscU (Fig. 5b), it is concluded that 100 μM IscA-Fe only meets the iron demand for cluster assembly in 50 μM IscU and a [2Fe–2S] cluster, not a [4Fe–4S] cluster, is assembled in an E. coli IscU dimer. Different from apo-IscU, which has no absorbance at 456 nm (Fig. 4b), IscA-Fe absorbs at 456 nm (Fig. 5a), suggesting that it is incorrect to calculate the amount of IscU[Fe–S] from the extinction coefficient at 456 nm when IscA-Fe is as an iron supplier.Fig. 4. Effect of IscA-Fe and cysteine on IscU[Fe–S] assembly under the conditions used for Fig. 1. a) Relative IscU[Fe–S] assembly in first 10 min (A456 = 0 for the control cuvette without addition of cysteine); b) UV-Vis spectra after a 10-min incubation in the presence of 100 μM cysteine. Curves 1-6 refer to the initial IscA-Fe concentration of 0, 12.5, 25, 50, 75, and 100 μM, respectively.
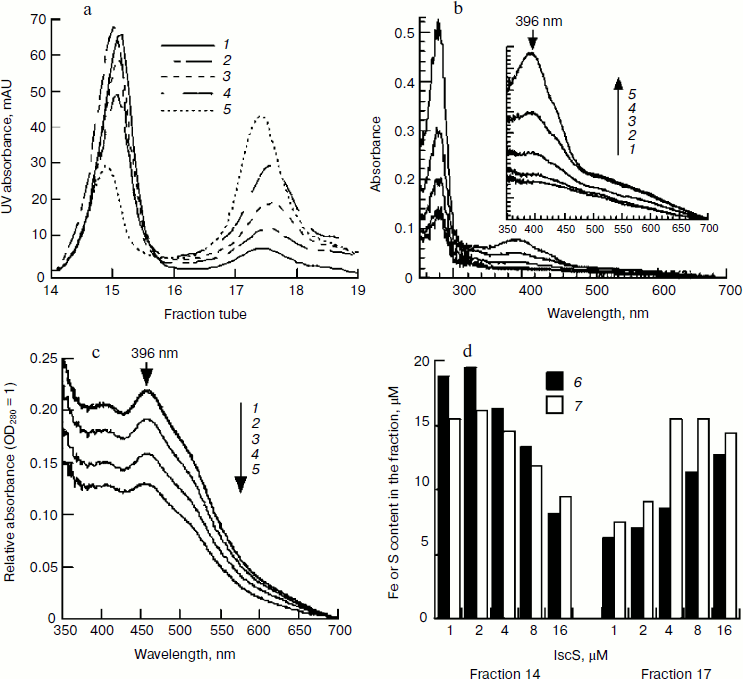
Effect of IscS on IscU[Fe–S] assembly. IscS is a cysteine desulfurase that transfers sulfide from cysteine to a scaffold during cluster assembly. It is commonly thought that the higher the enzyme concentration, the faster the reaction. However, the spectral determination showed that the characteristic 456-nm peak of IscU[Fe–S] disappeared when IscU/IscS reached 10 : 1, suggesting that a high concentration of IscS may not be beneficial for cluster assembly. To test this proposal and to exclude the absorbance interference of IscS, the assembled IscU[Fe–S] was re-purified using a Mono-Q column. As shown in Fig. 6a, there were two peaks in the FPLC profile. SDS-PAGE analysis confirmed that fractions 14 and 15 were IscU, while fraction 17 was an IscU–IscS complex (data not shown). Spectral determination of fraction 14 showed that the assembled IscU[Fe–S] concentrations were 12.12, 12.31, 9.03, 6.67, and 4.97 μM, respectively, in the presence of 1, 2, 4, 8, and 16 μM IscS; correspondingly, the A456/A280 decreased from 0.22 to 0.13 with increasing IscS concentrations (Fig. 6c). These results demonstrate that a high concentration of IscS inhibited the cluster assembly. Iron and sulfide determination confirmed this conclusion; 0.92, 1.00, 0.86, 0.76, 0.59 μM iron and 0.78, 0.82, 0.83, 0.78, 0.68 μM sulfide, respectively, were bound to 1 μM IscU when 1, 2, 4, 8, and 16 μM IscS was added in the reaction mixture (Fig. 6d). IscS and IscU entered the complex in equivalent amounts based on the changes in UV absorbance (Figs. 6b and 6c), corresponding well with the former report that IscU and IscS formed a heterodimeric complex [25]. The IscS–IscU complex is able to bind iron and sulfide (Fig. 6d), but no 456-nm peak was found in its spectral profile (Fig. 6b), suggesting that the complex may be an intermediate product. Prolonging the reaction time could not free IscU[Fe–S] from the complex, implying that additional apo-IscU may be necessary for the release of [Fe–S] cluster.Fig. 5. Effect of apo-IscU on IscU[Fe–S] assembly. Apo-IscU (0-100 μM) was mixed with 1 μM IscS, 100 μM IscA-Fe, and 2 mM DTT. After pre-warming at 37°C for 5 min, 1 mM cysteine was added to initiate iron–sulfur cluster assembly. a) UV-Vis spectra after a 20-min incubation. Curves 1-6 refer to the initial apo-IscU concentration of 0, 12.5, 25, 50, 75, and 100 μM, respectively. b) Relative IscU[Fe–S] assembly. Curve 7 represents the sharpness of the 456-nm peak and curve 8 represents the amplitude of the 456-nm absorbance.
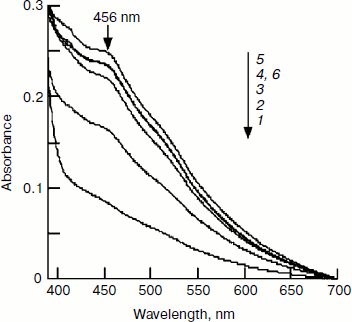
Effect of pH on IscU[Fe–S] assembly. In determining the optimal substrate concentration, 4 mM cysteine was found to inhibit the iron–sulfur cluster assembly. One possible explanation for this inhibition is the change in pH, since L-cysteine was added as hydrochloride as cysteine is instable in neutral solution. The experiment confirmed that the pH of the reaction mixture dropped to 7.4 when 4 mM L-cysteine hydrochloride was added. To further investigate the effect of pH on IscU[Fe–S] assembly, the pH of the reaction buffer was adjusted to 7.0, 7.2, 7.4, 7.6, 7.8, and 8.0. Spectral determination showed that the cluster was well assembled at pH 7.6-8.0, but the assembly was inhibited at pH 7.4 (Fig. 7). Such an inhibition may have an important physiological implication: a cell is able to control its sugar metabolism by negative regulation of enzyme activities because at least two enzymes in the citric acid cycle, aconitase and fumarase, hold iron–sulfur clusters as their prosthetic groups [26, 27].Fig. 6. Effect of IscS on IscU[Fe–S] assembly. IscU (50 μM) was incubated with 50 μM Fe2+, 2 mM DTT, and 1-16 μM IscS at 37°C for 5 min, then 1 mM cysteine was added; 20 min later, IscU and IscS were re-purified. a) Profile of Mono-Q FPLC; b) spectra of fraction 17; c) spectra of fraction 14; curves 1-5 refer to the reactions with 1, 2, 4, 8, and 16 μM IscS, respectively; d) the iron (6) and sulfide (7) contents in the fractions.
Proteins containing iron–sulfur clusters have important redox, catalytic, or regulatory functions, but the mechanism by which their clusters are formed is still not clear. In this study, E. coli IscU, IscS, and IscA were purified, and the iron–sulfur cluster was assembled in vitro. Since our former tests and other reports showed that His-tag did not influence the cluster assembly [17], all the experiments were performed using His-tagged proteins. This study verified that the iron–sulfur cluster assembly in E. coli IscU is an IscS-mediated biosynthetic process. IscU, iron, and cysteine are three essential assembly components, with increasing amounts of each stimulating the cluster assembly (Figs. 3-5). Unlike most enzymatic reactions, high concentrations of IscS inhibit the cluster formation. The FPLC test and electrophoresis analysis revealed that IscS and IscU form a heterodimeric complex (Fig. 6). Such a complex is able to bind iron and sulfide, but has no characteristic IscU[Fe–S] peak and only a low absorbance at 456 nm, suggesting the complex may be an intermediate product. In addition, extending the reaction time did not release IscU[Fe–S] from the complex. Therefore, for efficient assembly of iron–sulfur clusters in the scaffold, IscS should be maintained at a low concentration, such as 1-2% of IscU.Fig. 7. Effect of pH on IscU[Fe–S] assembly under the conditions used for Fig. 1. The spectrum obtained after a 20-min incubation is shown. Curves 1-6 refer to buffer pH of 7.0, 7.2, 7.4, 7.6, 7.8, and 8.0, respectively.
This project was supported by the Scientific Research Foundation for Returned Overseas Chinese Scholars of the State Education Ministry under grant No. J20100019.
REFERENCES
1.Ayala-Castro, C., Saini, A., and Outten, F. W.
(2008) Microbiol. Mol. Biol. Rev., 72, 110-125.
2.Filimonenkov, A. A., Zvyagilskaya, R. A.,
Tikhonova, T. V., and Popov, V. O. (2010) Biochemistry
(Moscow), 75, 744-751.
3.Zheng, L., Cash, V. L., Flint, D. H., and Dean, D.
R. (1998) J. Biol. Chem., 273, 13264-13272.
4.Barras, F., Loiseau, L., and Py, B. (2005) Adv.
Microb. Physiol., 50, 41-101.
5.Dos Santos, P. C., Johnson, D. C., Ragle, B. E.,
Unciuleac, M. C., and Dean, D. R. (2007) J. Bacteriol.,
189, 2854-2862.
6.Fontecave, M., Choudens, S. O., Py, B., and Barras,
F. (2005) J. Biol. Inorg. Chem., 10, 713-721.
7.Tokumoto, U., and Takahashi, Y. (2001) J.
Biochem. (Tokyo), 130, 63-71.
8.Qi, W., and Cowan, J. A. (2011) Coord. Chem.
Rev., 255, 688-699.
9.Li, K. Y., Tong, W. H., Hughes, R. M., and Rouault,
T. A. (2006) J. Biol. Chem., 281, 12344-12351.
10.Adinolfi, S., Rizzo, F., Masino, L.,
Nair, M., Martin, S. R., Pastore, A., and Temussi, P. A.
(2004) Eur. J. Biochem., 271, 2093-2100.
11.Morimoto, K., Yamashita, E., Kondou, Y., Lee, S.
J., Arisaka, F., Tsukihara, T., and Nakai, M. (2006) J.
Mol. Biol., 360, 117-132.
12.Wollenberg, M., Berndt, C., Bill, E., Schwenn, J.
D., and Seidler, A. (2003) Eur. J. Biochem., 270,
1662-1671.
13.Vinella, D., Brochier-Armanet, C., Loiseau, L.,
Talla, E., and Barras, F. (2009) PLoS Genet., 5,
e1000497.
14.Chandramouli, K., and Johnson, M. K.
(2006) Biochemistry, 45, 11087-11095.
15.Kakuta, Y., Horio, T., Takahashi, Y., and
Fukuyama, K. (2001) Biochemistry, 40, 11007-11012.
16.Smith, A. D., Agar, J. N., Johnson, K. A.,
Frazzon, J., Amster, I. J., Dean, D. R., and Johnson, M. K.
(2001) J. Amer. Chem. Soc., 123, 11103-11104.
17.Layer, G., Ollagnier-de Choudens, S., Sanakis,
Y., and Fontecave, M. (2006) J. Biol. Chem., 281,
16256-16263.
18.Wu, G. F., Li, P., and Wu, X. C. (2008)
Biochem. Biophys. Res. Commun., 374, 399-404.
19.Zeng, J., Zhang, K., Liu, J., and Qiu, G. Z.
(2008) J. Microbiol. Biotechnol., 18, 1672-1677.
20.Cowart, R. E., Singleton, F. L., and Hind, J. S.
(1993) Anal. Biochem., 211, 151-155.
21.Siegel, L. M. (1965) Anal. Biochem.,
11, 126-132.
22.Agar, J. N., Krebs, C., Frazzon, J., Huynh, B.
H., Dean, D. R., and Johnson, M. K. (2000) Biochemistry,
39, 7856-7862.
23.Jang, S., and Imlay, J. A. (2010) Mol.
Microbiol., 78, 1448-1467.
24.Bilder, P. W., Ding, H., and Newcomer, M. E.
(2004) Biochemistry, 43, 133-139.
25.Kato, S., Mihara, H., Kurihara, T., Takahashi,
Y., Tokumoto, U., Yoshimura, T., and Esaki, N. (2002) Proc. Natl.
Acad. Sci. USA, 99, 5948-5952.
26.Beinert, H., Holm, R. H., and Munck, E. (1997)
Science, 277, 653-659.
27.Matasova, L. V., and Popova, T. N. (2008)
Biochemistry (Moscow), 73, 957-964.