Characterization of a New RNase HII and Its Essential Amino Acid Residues in the Archaeon Sulfolobus tokodaii Reveals a Regulatory C-Terminus
Ke Zhan and Zheng-Guo He*
National Key Laboratory of Agricultural Microbiology, Center for Proteomics Research, College of Life Science and Technology, Huazhong Agricultural University, Wuhan 430070, China; fax: +86-27-8728-0670; E-mail: he.zhengguo@hotmail.com; hezhengguo@mail.hzau.edu.cn* To whom correspondence should be addressed.
Received March 30, 2010; Revision received May 14, 2010
The archaea possess RNase H proteins that share features of both prokaryotic and eukaryotic forms. Although the Sulfolobus RNase HI has been reported to have unique structural and biochemical properties, its RNase HII has not yet been investigated and its biochemical properties remain unknown. In the present study, we have characterized the ST0519 RNase HII from S. tokodaii as a new form. The enzyme utilized hybrid RNA/DNA as a substrate and had an optimal temperature between 37 to 50°C. The activity of wild-type protein was stimulated by Mn2+, whereas this cation significantly inhibited the activity of C-terminal truncated mutant proteins. A series of mutation assays revealed a regulatory C-terminal tail in the S. tokodaii RNase HII. One mutant, ST0519 (residues 1-195), retained only partial activity, while ST0519 (residues 1-196) completely lost its activity. Based on the presumed structure, the C-terminus might form a short α-helix in which two residues, I195 and L196, are essential for the cleavage activity. Our data suggest that the C-terminal α-helix is likely involved in the Mn2+-dependent substrate cleavage activity through stabilization of a flexible loop structure. Our findings offer important clues for further understanding the structure and function of both archaeal and eukaryotic RNase HII.
KEY WORDS: Sulfolobus tokodaii, RNase HII, nuclease, archaeaDOI: 10.1134/S0006297910070163
RNase H proteins are found widely in all three domains of life: Bacteria, Archaea, and Eucarya. These enzymes usually degrade only the RNA strand of RNA/DNA hybrids and thus play important roles during DNA replication, repair, and transcription [1-3]. Based on the sequence similarity to their Escherichia coli homolog, RNase H proteins have been classified into two types: RNase HI and RNase HII. Several conserved sites important for activity have been characterized in E. coli RNase HI [4-6]. Escherichia coli RNase HII shows a strong preference for degradation of RNA/DNA [7, 8]. Divalent metal ions play important roles in the activities of all forms of RNase H [9-11].
The archaeal RNase H proteins show features of both prokaryotic and eukaryotic forms. For example, the RNase HI gene from Halobacterium, Vng0255c, shares an amino acid sequence identity of 33% with E. coli RNase HI and has conserved metal ion binding and catalyzing residues relative to those of the E. coli enzyme [12]. On the other hand, Vng0255c also contains a eukaryote-like N-terminal extension in the RNase HI [13]. Based on its crystal structure, archaeal RNase HII has been proposed to represent a homolog of the major human RNase H [14, 15].
Sulfolobus species are very important organisms for modeling key events in eukaryotic DNA replication and repair [16-18]. The S. tokodaii 7 RNase HI, encoded by ST0753, degrades both RNA/DNA and double RNA substrates and shares some conserved active site residues with E. coli RNase HI [19, 20]. Thus, S. tokodaii 7 RNase H enzymes possess unique structural and biochemical properties. However, the RNase HII from Sulfolobus has not yet been characterized and its biochemical properties remain unknown.
In the present study, we characterized the nuclease activity of a new S. tokodaii RNase HII, encoded by ST0519, against different substrates and examined regulatory effects of temperature and metal ions. A series of mutations revealed a regulatory C-terminal tail. Our data suggest that C-terminal α-helix can determine the Mn2+-dependent substrate cleavage activity.
MATERIALS AND METHODS
Strains, enzymes, plasmids, and reagents. Escherichia coli BL21 (F–ompT hsdSB(rB–mB–) gal–dcm (DE3)) cells and pET28a containing the T7 RNA polymerase promoter were purchased from Novagen (USA) and used to express archaeal proteins. Restriction enzymes, T4 ligase, DNA polymerase, dNTPs, and all antibiotics were obtained from TaKaRa Biotech (China). PCR primers were synthesized by Invitrogen (USA) (see table). Ni-NTA (Ni2+-nitrilotriacetate) and GST agarose were obtained from Qiagen (USA).
Primers used in this study
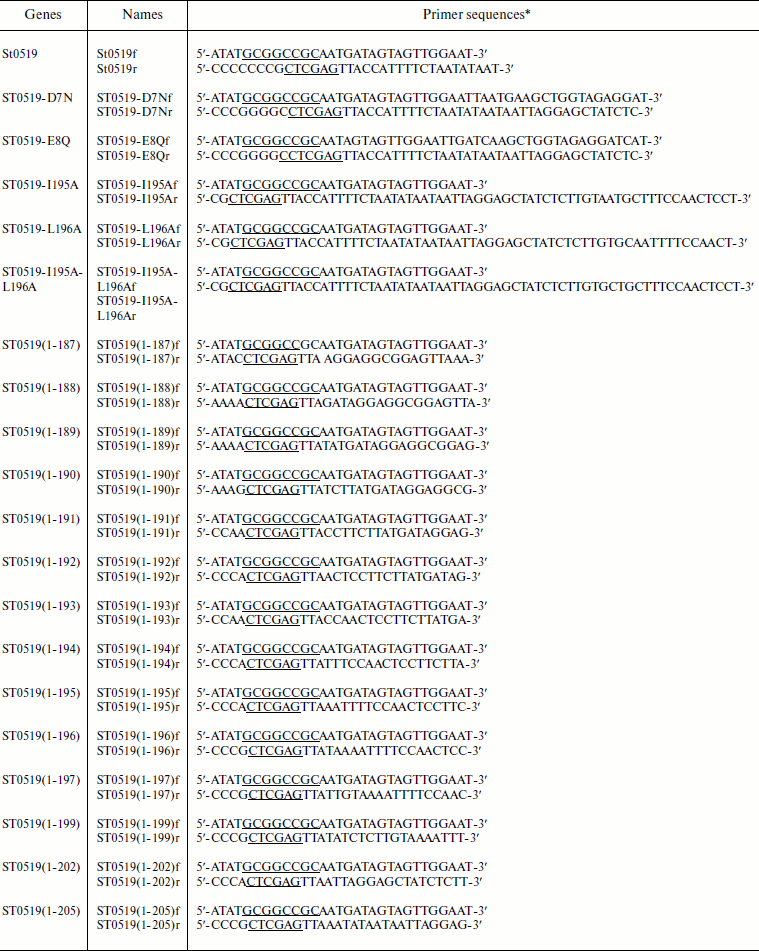
*Sites for restriction enzymes are underlined.
Site-directed gene mutagenesis, cloning, and purification of archaeal proteins. Prokaryotic recombinant vectors were constructed that expressed the genes for archaeal proteins ST0519 and ST0439. The genes encoding two mutant proteins, ST0519-D7N and ST0519-E8Q, were constructed by PCR. The specific forward primer for mutation was designed to change the codon of Asp7 (GAT) to Asn (AAT). A similar strategy was used to construct mutant ST0519-D7N, and these primer sequences are described in the table. The ST0519-I195A mutant gene was produced by PCR using a reverse primer of 5′-CGCTCGAGTTACCATTTTCTAATATAATAATTAGGAGCTATCTCTTGTAATGCTTTCCAACTCCT-3′ with the same forward primer as for the wild-type STY0519 gene. A similar strategy was used to construct mutant ST0519-L196A and ST0519-I195A/L196A proteins using the primer sequences described in the table. The C-terminal truncated mutant genes were produced by PCR using their respective reverse primer (table) with the same forward primer as used with the wild-type STY0519 gene. All of these mutant gene fragments were further cloned into pET28a yielding recombinant plasmids for protein expression and purification. Escherichia coli BL21 CodonPlus (DE3)-RIL cells (Novagen) were used as the host strain to express archaeal proteins as previously described [21]. Protein concentrations were determined from absorbance at 260 nm, according to Gill and Hippel [22].
RNase H activity assay. Oligoribonucleotides were 5′-end-labeled with [γ-32P]ATP and T4 polynucleotide kinase (NEB, USA). The mixture was treated at 68°C for 10 min to inactivate the protein kinase. The labeled oligonucleotide was purified as described previously [21], then 1.2-fold amount of unlabeled reverse oligonucleotide was added and incubated at 95°C for 10 min to allow complete annealing. Double substrates were stored at –20°C before use. The substrate was hydrolyzed in 10 mM Tris-HCl (pH 8.5) containing 10 mM NaCl, 1 mM DTT, and 50 mg/ml BSA in the presence of different divalent metal ions or under different reaction temperatures as indicated. The samples were mixed with ice-cold 2× loading buffer (95% deionized formamide, 100 mM EDTA, 0.02% bromphenol blue). Products were analyzed on a 20% polyacrylamide gel containing 7 M urea in 0.5× TBE. The gel was dried and analyzed using a published procedure [21].
Homology structure modeling. The structure of ST0519 was modeled computationally using the automated comparative protein modeling web server SWISS-MODEL [23]. The mouse RNaseH2 protein [24] was used as a template (PDB ID: 3kio).
RESULTS
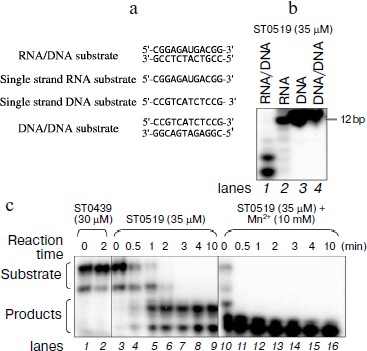
ST0519 specifically cleaves RNA/DNA hybrid substrate and the activity is stimulated by Mn2+. The function of Sulfolobus RNase HII has not yet been characterized. To identify the potential S. tokodaii 7 RNase HII, encoded by ST0519, several different substrates were designed, including single-stranded RNA, single-stranded DNA, double-stranded DNA, and hybrid RNA/DNA substrates (Fig. 1a). When 10 μM ST0519 protein was mixed with different substrates in the reaction mixtures, only 12-bp hybrid RNA/DNA substrates were hydrolyzed as indicated by the presence of shorter hydrolysis products on the gel (Fig. 1b, lane 1). This indicated a specificity of ST0519 towards cleavage of RNA/DNA substrate. In contrast, no cleavage occurred with an unrelated protein, ST0439, on the same RNA/DNA substrate (Fig. 1c, lanes 1 and 2). Cleavage of the RNA/DNA substrate by ST0519 increased with reaction time from 0.5 to 10 min (Fig. 1c, lanes 3-9). Addition of 50 mM Mn2+ to the reaction mixture clearly stimulated the cleavage activity of ST0519 (the substrates were completely degraded within 2 min) compared to the activity in the absence of Mn2+ (Fig. 1c, lanes 10-16). Therefore, ST0519 specifically cleaves RNA/DNA hybrid substrates and this activity can be stimulated by Mn2+.
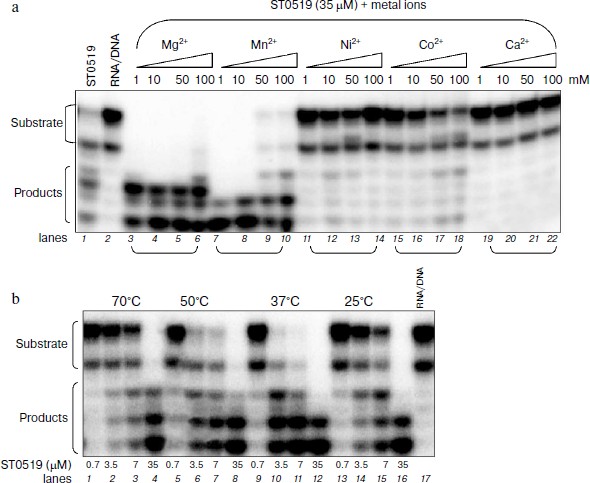
Cleavage activity of ST0519 depends on different ions and reaction temperatures. We characterized the effects of different divalent cations and reaction temperatures on the cleavage ability of ST0519 on the RNA/DNA substrate. As shown in Fig. 2a, when supplied from 1 to 100 mM, both Mg2+ and Mn2+ clearly stimulated the activity of ST0519 as increasingly more substrate was degraded (Fig. 2a, lanes 1-10). Addition of 1 to 100 mM Ni2+, Co2+, or Ca2+ appeared to inhibit the activity of ST0519 since substrate cleavage was reduced (Fig. 2a, lanes 11-22) and the activities were even lower than in the control reaction without any added metal ion (Fig. 2a, lanes 1 and 2).Fig. 1. Cleavage activity of S. tokodaii RNase HII with different oligomeric substrates. a) Four oligomeric substrates and their sequences. b) Cleavage activity of S. tokodaii ST0519 on the four different oligomeric substrates. Each substrate was incubated with ST0519 for 10 min. Products were separated on a 20% polyacrylamide gel containing 7 M urea. c) Time course of the cleavage of oligomeric substrate in the presence or absence of 10 mM Mn2+. ST0439 was used as a negative control for cleavage.
We also examined the effect of temperatures on the cleavage activity of ST0519. As shown in Fig. 2b, when an increasing amount of ST0519 protein (from 0.7 to 7 μM) was mixed with RNA/DNA substrate at a range of temperatures (from 25 to 70ºC), cleavage occurred at all temperature, but the optimum temperature was between 37 and 50°C (Fig. 2b, lanes 9-12).Fig. 2. Cleavage of oligomeric substrates in the presence of divalent metal ions. A 12 bp RNA/DNA hybrid was incubated with ST0519 for 10 min. a) Effects of different concentration (1-100 mM) of MnCl2, MgCl2, CoCl2, or NiCl2 on the cleavage activity. b) Effects of different temperatures (25-70°C) on the cleavage activity.
Therefore, our results show that Mg2+ and Mn2+ stimulated the cleavage activity of ST0519. The optimal temperature for the cleavage is between 37 and 50°C.
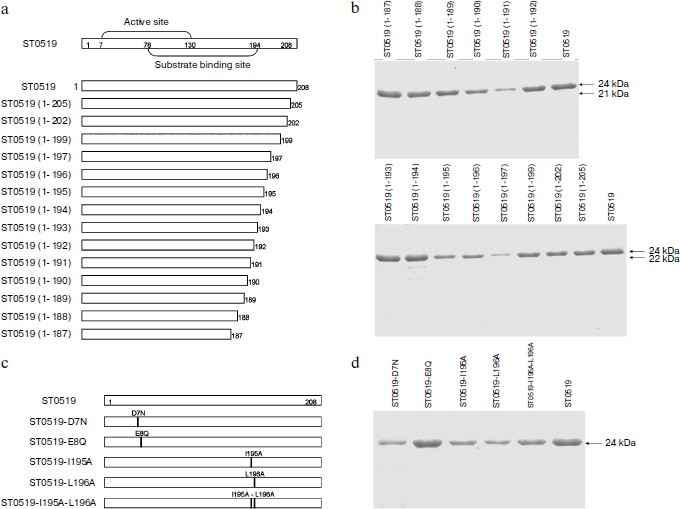
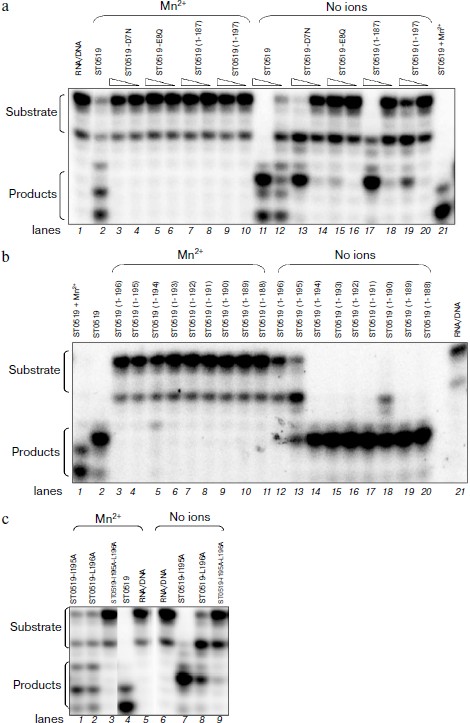
Essential amino acid residues for cleavage activity of ST0519. We produced several conserved single or double mutants and a series of C-terminal deleted mutants to characterize the amino acid residues essential for the activity of ST0519 (Figs. 3a and 3c). These mutant proteins were further purified (Figs. 3b and 3d). As shown in the activity assay (Fig. 4a), ST0519-D7N retained cleavage activity towards the RNA/DNA substrate (lanes 13 and 14) similar to that seen for the wild-type protein although to a lesser extent (lanes 11 and 12) and only in the absence of Mn2+. In contrast, ST0519-E8Q lost all activity (lanes 15 and 16). Neither ST0519-D7N (5 or 10 μM) nor ST0519-E8Q retained cleavage activity in the presence of 10 mM Mn2+ (lanes 3-6). This suggested that the conserved E8 residue serves an important function in the archaeal RNase HII. The mutant ST0519 (residues 1-187) (10 μM) was capable of cleaving the RNA/DNA substrate (lanes 17 and 18), whereas ST0519 (residues 1-197), containing an additional 10 amino acids, lost most of the cleavage activity (lanes 19 and 20) in the absence of Mn2+. This suggests that the additional protein fragment (from ST0519-188 to ST0519-196) contains some unknown inhibitory residues. Cleavage activities of ST0519(1-187) and ST0519(1-197) were inhibited in the presence of Mn2+ (lanes 7-10).
Fig. 3. Schematic representation of S. tokodaii ST0519 and purifications of its various mutant proteins. a) Schematic diagrams of the full-length mutants and different C-terminal deletion mutants. b) Purified proteins of different C-terminal deletion mutants are indicated on the top of the panels. c) Schematic diagrams of several site-specific mutants, ST0519-D7N, ST0519-E8Q, ST0519-I195A, ST0519-L196A, and ST0519-I195A/L196A. d) Purified proteins of these site-specific mutants.
ST0519-195/196 residues negatively regulate its cleavage activity. C-Terminal deleted mutants were used to compare cleavage activities in the presence and absence of Mn2+. As shown in Fig. 4b, each of nine mutants from ST0519(1-194) to ST0519(1-188) (lanes 14-20) retained cleavage activity in the absence of Mn2+. However, the ST0519(1-195) mutant appeared to lose part of its activity (lane 13). ST0519(1-196) lost almost all of its cleavage activity (lane 12). All mutants were inhibited if Mn2+ was present in the reactions (lanes 3-11). Therefore, residues 195 and 196 might negatively regulate the activity of the protein in the absence of Mn2+.Fig. 4. Cleavage activities of ST0519-D7N, ST0519-E8Q, ST0519(1-187), and ST0519(1-197) (a), of various C-terminal truncated mutants (b), and of ST0519-I195A, ST0519-L196A, and ST0519-I195A/L196A (c) in the presence or absence of 10 mM Mn2+. The RNA/DNA hybrid was incubated with ST0519 for 10 min.
To further investigate the effects of these two residues, we examined the cleavage activities of their single/double mutants. As shown in Fig. 4c, when Mn2+ was absent in the reaction mixtures, ST0519-I195A retained a clear cleavage activity (lane 7) and ST0519-L196A retained only a partial activity (lane 8). The double mutant with I195 and L196 replaced, ST0519-I195A/L196A, lost almost all the cleavage activity (lane 9) as no significant shorter products were observed. These results indicate that two residues of ST0519, I195 and L196, negatively regulate the activity of the archaeal RNase HII. In addition, cleavage activity for the double mutant was also not observed even when Mn2+ was added to the reaction mixtures (lane 3). Therefore, I195 and L196 are essential for the activity of ST0519.
DISCUSSION
The S. tokodaii RNase H has demonstrated unique structural and biochemical characteristics [14, 15, 19, 20]. In the present study, we characterized a new S. tokodaii RNase HII, encoded by ST0519, and its essential amino acid residues. The RNase HII utilized hybrid RNA/DNA as its substrate. Mn2+ stimulated the activity of the wild-type protein but significantly inhibited that of its C-terminal truncated proteins. A series of further mutation assays revealed a regulatory C-terminal tail in the S. tokodaii RNase HII.
The thermoacidophilic archaeon S. tokodaii grows at high temperatures and low pH conditions. We found that RNase HII ST0519 has an optimal temperature between 37-50°C and appears to function even at a temperature as high as 70°C, which was consistent with its physiological environment. A previous study showed that S. tokodaii 7 RNase HI, encoded by ST0753, degraded both RNA/DNA and double RNA substrates [19]. In contrast, the RNase HI of Halobacterium sp. NRC-1 and E. coli only degraded RNA/DNA substrates [12]. In the present study, the S. tokodaii RNase HII, encoded by ST0519, was incapable of hydrolyzing single DNA, or RNA, or double stranded DNA, and only utilized hybrid RNA/DNA as a substrate. These characteristics clearly make it different from RNase HI and suggest that two forms of S. tokodaii RNase H serve different purposes in archaeal nucleic acid metabolism in the extreme growing conditions of this organism.
Of all RNase H enzymes, E. coli RNase HI is one of the best studied [4-6]. A double-site metal-binding catalytic mechanism has been proposed, in which binding of one Mn2+ ion at site 1 is required for enzyme activation and binding of a second Mn2+ ion at site 2 is inhibitory [25]. In E. coli RNase HI, four acidic residues—Asp10, Glu48, Asp70, and Asp134—constitute the DEDD catalytic motif [26]. However, the effects of the C-terminal domain of these RNase H enzymes on both the metal binding and function of the protein remain unknown. Metal ions modulate the activity of archaeal RNase H [19]. In the present study, Mn2+ clearly stimulated the activity of the S. tokodaii RNase HII. Two conserved residues within the DEDD motif, D7 and E8, were shown to be essential for the Mn2+-dependent activity of the archaeal RNase H (Fig. 4a). Unexpectedly, the activities of almost all of the C-terminal truncated mutant proteins were inhibited by Mn2+, suggesting that the C-terminal domain of the archaeal RNase HII was involved in the regulation of metal ion binding and significantly affected the function of the protein.
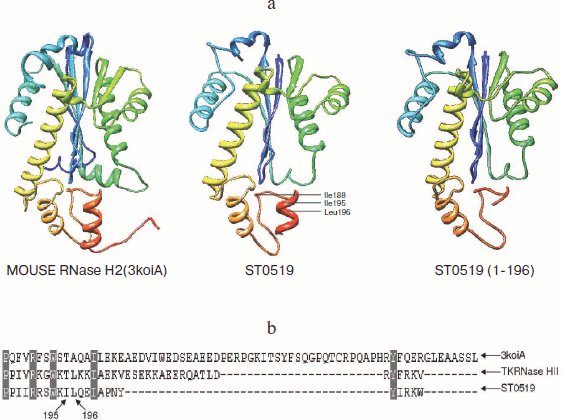
An important finding from the current study is that the C-terminal tail of S. tokodaii RNase HII has a regulatory function. L196 is an essential amino acid residue for RNA hydrolysis activity. The 3D structure of ST0519 is not yet known; however, a model structure (Fig. 5; see color insert) was obtained by an automated comparative protein modeling [23] using the mouse RNaseH2 protein as a template (PDB ID: 3kio) (Identities = 30.3%). From the model structure, the C-terminus forms a short α-helix, which includes the I195 and L196 residues (Fig. 5a). The α-helix has an important regulatory effect on the cleavage activity of RNase HII, as ST0519(1-195) only retained partial activity, and ST0519(1-196) completely lost its activity (Fig. 4b). This suggested that the entire C-terminal structure might be important for the function of the archaeal RNase HII. The two residues, I195 and L196, significantly contributed to the cleavage activity as ST0519-L196A mutant protein only partially retained its activity and the double mutant (ST0519-I195A/L196A) completely lost activity (Fig. 4c). The L196 residue is also conserved in another archaeal RNase HII, TkRNase HII, from T. kodakaraensis [15] (Fig. 5b). Since there is a long and flexible loop neighboring the C-terminus (Fig. 5a), the short α-helix most likely stabilizes the structure, which would be quite important for the function of the entire protein.
In conclusion, we characterized the function of RNA cleavage and identified the essential amino acid residues for the RNase HII from the archaeon S. tokodaii. The C-terminal structure is involved in the regulation of Mn2+-dependent substrate cleavage activity and its integrated function. These data offer important clues for further understanding the universal features underlying the structure and function of RNase HII from both archaea and eukaryotes.Fig. 5. Schematic diagram of the S. tokodaii ST0519 structure. a) The structure obtained using the homology modeling method. The structures for mouse RNase H2 and ST0519 or St0519(1-196) are indicated. b) The C-terminal sequence alignments of ST0519, TkRNase HII, and mouse RNase H2. TkRNase HII represents ST0753 from Thermococcus kodakaraensis. The conserved L196 is indicated.
This work was supported by the 973 Program (2006CB504402) and Doctoral Fund of Ministry of Education of China (200805040004).
REFERENCES
1.Qiu, J., Qian, Y., Frank, P., Wintersberger, U.,
and Shen, B. (1999) Mol. Cell. Biol., 198, 361-8371.
2.Arudchandran, A., Cerritelli, S., Narimatsu, S.,
Itaya, M., Shin, D. Y., Shimada, Y., and Crouch, R. J. (2000) Genes
Cells, 5, 789-802.
3.Chai, Q., Qiu, J., Chapados, B. R., and Shen, B.
(2001) Biochem. Biophys. Res. Commun., 286,
1073-1081.
4.Haruki, M., Noguchi, E., Nakai, C., Liu, Y.,
Oobatake, M., Itaya, M., and Kanaya, S. (1994) Eur. J. Biochem.,
220, 623-631.
5.Kanaya, S., and Crouch, R. J. (1983) J. Biol.
Chem., 25, 1276-1281.
6.Tadokoro, T., and Kanaya, S. (2009) FEBS J.,
76, 1482-1493.
7.Naoto, O., Mitsuru, H., Ayumu, M., Masaaki, M., and
Shigenori, K. (2000) J. Biochem., 127, 895-899.
8.Itaya, M. (1990) Proc. Natl. Acad. Sci. USA,
87, 8587-8591.
9.Kanaya, S., Oobatake, M., and Liu, Y. (1996) J.
Biol. Chem., 271, 32729-32736.
10.Tsunaka, Y., Haruki, M., Morikawa, M., Oobatake,
M., and Kanaya, S. (2003) Biochemistry, 42,
3366-3374.
11.Katayanagi, K., Miyagawa, M., Matsushima, M.,
Ishikawa, M., Kanaya, S., Ikehara, M., Matsuzaki, T., and Morikawa, K.
(1990) [javascript:AL_get(this,%20'jour',%20'Nature.');
Nature][javascript:AL_get(this,%20'jour',%20'Nature.'); ,] 6290,
306-309.
12.Ohtani, N., Yanagawa, H., Tomita, M., and Itaya,
M. (2004) Biochem. J., 381, 795-802.
13.Cerritelli, S. M., Fedoroff, O. Y., Reid, B. R.,
and Crouch, R. J. (1998) Nucleic Acids Res., 26,
1834-1840.
14.Lai, L., Yokota, H., Hung, L. W., Kim, R., and
Kim, S. H. (2000) Structure, 8, 897-904.
15.Muroya, A., Tsuchiya, D., Ishikawa, M., Haruki,
M., Morikawa, M., Kanaya, S., and Morikawa, K. (2001) Protein
Sci., 10, 707-714.
16.Tye, B. K. (2000) Proc. Natl. Acad.
Sci. USA, 97, 2399-2401.
17.Bernander, B. (2000) Trends Microbiol.,
8, 278-283.
18.Kelman, L. M., and Kelman, Z. (2003) Mol.
Microbiol., 48, 605-615.
19.Ohtani, N., Yanagawa, H., Tomita, M., and Itaya,
M. (2004) Nucleic Acids Res., 32, 5809-5819.
20.Kawarabayasi, Y., Hino, Y., Horikawa, H., Jin-no,
K., Takahashi, M., Sekine, M., Baba, S., Ankai, A., Kosugi, H.,
Hosoyama, A., Fukui, S., Nagai, Y., Nishijima, K., Otsuka, R.,
Nakazawa, H., Takamiya, M., Kato, Y., Yoshizawa, T., Tanaka, T., Kudoh,
Y., Yamazaki, J., Kushida, N., Oguchi, A., Aoki, K., Masuda, S.,
Yanagii, M., Nishimura, M., Yamagishi, A., Oshima, T., and Kikuchi, H.
(2001) DNA Res., 8, 123-140.
21.Zhang, L., Zhang, L., Liu, Y., Yang, S., Gao, C.,
Gong, H., Feng, Y., and He, Z. G. (2009) Proc. Natl. Acad. Sci.
USA, 106, 7792-7797.
22.Gill, S. C., and von Hippel, P. H. (1989)
Anal. Biochem., 182, 319-326.
23.Arnold, K., Bordoli, L., Kopp, J., and Schwede,
T. (2006) Bioinformatics, 22, 195-201.
24.Shaban, N. M., Harvey, S., Perrino, F. W., and
Hollis, T. (2010) J. Biol. Chem., 285, 3617-3624.
25.Keck, J. L., Goedken, E. R., and Marqusee, S.
(1998) J. Biol. Chem., 273, 4128-34133.
26.Kanaya, S. (1998) Ribonucleases H, INSERM,
Paris, pp. 1-38.