Gene Expression for VEGF-A, VEGF-C, and Their Receptors in Murine Lymphocytes and Macrophages
O. I. Stepanova, A. V. Krylov, V. I. Lioudyno, and E. P. Kisseleva*
Institute of Experimental Medicine, Russian Academy of Medical Sciences, ul. Akademika Pavlova 12, 197376 St. Petersburg, Russia; fax: (812) 234-9489; E-mail: ekissele@yandex.ru; iem@iem.spb.ru* To whom correspondence should be addressed.
Received May 10, 2007; Revision received June 21, 2007
Vascular endothelial growth factors VEGF-A and VEGF-C are the main angiogenic factors that control growth of new blood and lymphatic vessels in the organism, and they also possess several immunoregulatory activities. Expression of VEGF-A and VEGF-C mRNA as well as mRNA for VEGF receptors in lymphocytes and macrophages of naive mice was investigated. Using reverse transcription and subsequent polymerase chain reaction, we found that peritoneal macrophages, thymocytes, and lymph node cells constitutively expressed VEGF-A and VEGF-C mRNA. In addition, macrophages were positive for VEGFR-1, VEGFR-2, VEGFR-3, NRP-1, and NRP-2 mRNA, whereas thymocytes and lymph node cells expressed mRNA of the same receptors except VEGFR-1. These data expand our knowledge concerning gene distribution of VEGF receptors in the organism, in particular, among the cells of the immune system. This suggests that, along with their major angiogenic properties, VEGF family members additionally might also perform important mediatory functions within the immune system.
KEY WORDS: VEGF-A, VEGF-C, receptors of VEGF, angiogenesis, NRP-1, NRP-2DOI: 10.1134/S0006297907110041
The growth of new vessels in adults is a complex and coordinated process, and various molecules are involved in its regulation. Vascular endothelial growth factor (VEGF) is considered to be the main angiogenic factor. The VEGF family includes five major factors: VEGF-A, VEGF-B, VEGF-C, VEGF-D, and placental growth factor. VEGF-A is mostly important for the growth of blood vessels, while VEGF-C - for lymphatic vessels. The biological effect of the VEGF family members on endothelial cells is transduced via specific tyrosine kinase receptors: VEGF-A binds to VEGFR-1 (Flt-1) and VEGFR-2 (KDR/Flk-1), whereas VEGF-C interacts with VEGFR-2 and VEGFR-3. There are also two non-tyrosine kinase receptors that enhance activating signal but do not transduce it inside the cell. These two receptors are neuropilin 1 and 2 (NRP-1, NRP-2), where NRP-1 is a co-receptor to VEGFR-2, and NPR-2 is a co-receptor to VEGFR-3 [1].
Apart from its major angiogenic effect, VEGF also exhibits several immunoregulatory activities [2, 3] and affects hematopoiesis, differentiation, and maturation of dendritic cells [4], as well as of T and B lymphocytes [5]. Because of this, it seems important to study both the presence of VEGF receptors on the cells of immune system and the ability of lymphocytes and macrophages to synthesize VEGF molecules and be involved in their autocrine regulation. Here we studied mRNA expression of all defined VEGF receptors as well as VEGF-A and VEGF-C mRNA in lymphocytes and macrophages from naive mice.
MATERIALS AND METHODS
Male C3HA mice weighing 18-20 g were purchased from “Rappolovo” Animal Farm, Russian Academy of Medical Sciences. Peritoneal exudate cells were obtained by peritoneal lavage. To obtain enriched macrophage population, cells were cultured for 24 h at 37°C in RPMI-1640 media supplemented with 10% fetal calf serum (Biolot, Russia), followed by removal of non-adherent lymphocyte fraction. Inguinal lymph nodes and thymus were removed and homogenized, followed by filtering through nylon cell strainer to separate stroma from lymphocytes. Later isolated thymocytes, lymph node lymphocytes, and stroma of thymus were studied along with pieces of whole thymus and lung margin. In addition, the cell line of transplantable hepatoma 22A was studied; it was provided by O. N. Pogodina from the Cell Line Collection, Institute of Cytology, Russian Academy of Sciences. Hepatoma cells were cultured in DMEM media supplemented with 10% fetal calf serum until obtaining a confluent monolayer.
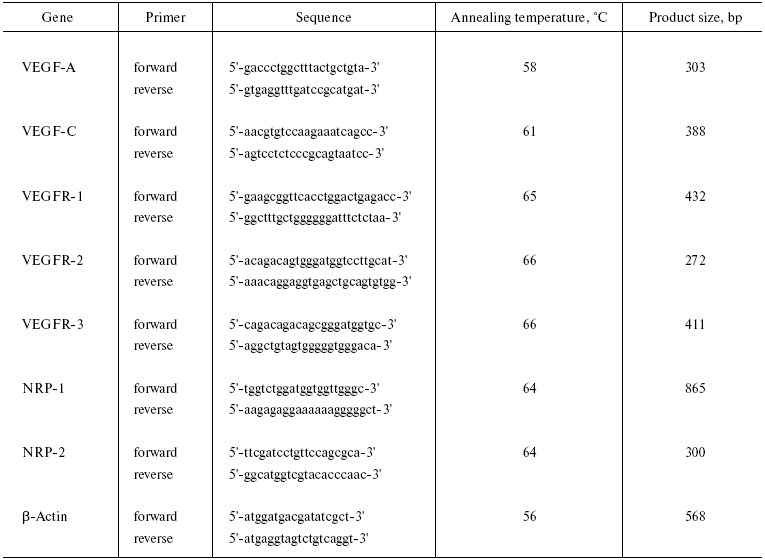
The mRNA expression was analyzed by reverse transcription followed by polymerase chain reaction (RT-PCR) with specific primers. For this, 5·106 cells or 50 mg of lung or thymus tissue were taken per sample. Total mRNA was isolated by a single-step method using TRI-reagent (Sigma, USA) according to the manufacturer's instructions. To perform reverse transcription reaction, 2 µg of RNA was supplemented with 200 U of reverse transcriptase M-MLV (Promega, USA), a mixture of dNTP (1.25 mM each), and specific reverse primers (25 pM each) (Table 1), in final volume 50 µl. Specific primers were designed using Primer-Master-1 software to obtain a product size that would be different for reactions on cDNA and nuclear DNA. beta-Actin was used as an internal RT standard. To perform PCR, 2 µl of cDNA was supplemented with 5 U of Taq-polymerase (Medigene, Russia), forward and reverse primers (10 pM each), dNTP mixture (0.8 mM each), and 2-6 mM MgCl2 (depending on the primers used), in final volume 25 µl. Initial denaturation was carried out for 5 min at 94°C. Each PCR cycle included denaturation at 94°C for 30 sec, hybridization at 56-66°C for 30 sec; the temperature was empirically selected for each pair of primers (Table 1), and extension at 72°C for 30 sec for all targets tested. beta-Actin amplification was repeated by 25 cycles, and 35 cycles for all other factors in a thermal amplifier (Techne, GB). PCR products were visualized by electrophoresis through 1% agarose gels stained with ethidium bromide. Gels were photographed by using a digital camera.
Table 1. Specific primers for RT-PCR
RESULTS AND DISCUSSION
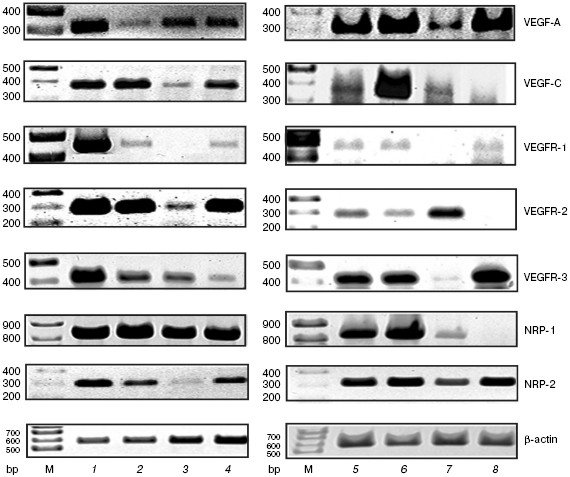
During investigation of VEGF-A and VEGF-C mRNA expression, we found that they were present in all cell types and tissues studied (resident peritoneal macrophages, peripheral lymph node lymphocytes and thymocytes, as well as thymus and lung of naive mice) (figure). The synthesis of VEGF-A and VEGF-C by macrophages is known [6, 7], whereas in the case of lymphocytes this question remained uncertain. Earlier it was described that VEGF-A mRNA may be found in mouse splenocytes [8], antigen-stimulated T cells from lymph nodes in rats [3], and activated T cells from peripheral blood in humans [9]. Moreover, VEGF-A and VEGF-C mRNAs were detected in whole thymus [3, 10]. However, we were able for the first time to demonstrate VEGF-A and VEGF-C mRNA synthesis by isolated thymocytes as well as VEGF-C mRNA expression in lymphocytes.
While studying mRNA expression of VEGF receptors, we used highly vascularized lung tissue as a positive control and hepatoma cells as an example of non-endothelial cells. In lungs, we found expression of all tested receptors, namely, VEGFR-1, VEGFR-2, VEGFR-3, NRP-1, and NRP-2 (figure). In addition, the mRNA expression of these receptors was detected in the whole thymus and its stroma, which might be explained by the presence of vessels in this tissue. A similar result was obtained during investigation of peritoneal exudate cells and macrophages.mRNA expression for VEGF-A, VEGF-C, and their receptors. M, molecular mass marker. 1) Lung; 2) whole thymus; 3) isolated thymocytes; 4) thymus stroma; 5) peritoneal exudate cells; 6) peritoneal macrophages cultures for 24 h; 7) isolated lymph node lymphocytes; 8) hepatoma 22A cells
Our data are consistent with those from the literature on expression of VEGFR-1, VEGFR-2, and VEGFR-3 mRNA in murine and human peritoneal macrophages as well as in macrophage-like cell lines [11-13]. We have obtained for the first time data on NRP-1 and NRP-2 mRNA expression in macrophages.
The figure shows that within the sensitivity of the method used, mRNAs of all studied receptors except VEGFR-1 were expressed in lymph node cells and thymocytes. Earlier it was shown [3] that VEGFR-2 mRNA was present and VEGFR-1 was absent in lymph node T cells in rats, which agrees with our data. On the other hand, the expression of both receptors was described in murine splenic T lymphocytes [8]. Perhaps this reflects the population differences in T cell content between peripheral lymph nodes and spleen. It is relevant to mention that the expression of VEGFR-2, VEGFR-3, NRP-1, and NRP-2 mRNA in thymocytes and VEGFR-3, NRP-1, and NRP-2 mRNA in lymph node cells was found for the first time.
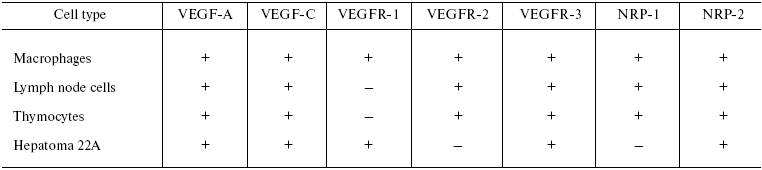
Thus, we have analyzed the pattern of constitutive mRNA expression for VEGF receptors in immune cells of naive mice (Table 2). Our data substantially expand our knowledge on distribution of VEGF receptor mRNA in different cell types of the organism, in particular, in the cells of the immune system. However, the role of the VEGF family in functioning of these cells is poorly understood.
Table 2. mRNA expression for VEGF-A, VEGF-C,
and their receptors in different cell types
VEGF-A and VEGF-C are known to be chemoattractants for monocytes/macrophages. VEGF-A recruits blood monocytes to the focus of inflammation and vessel growth via interaction with VEGFR-1 [2], whereas in tissue macrophages both receptors, VEGFR-1 and VEGFR-2, mediate chemotactic activity [11, 13]. VEGF-C affects macrophage migration via VEGFR-3 [14]. Few studies addressed the impact of VEGF-A on lymphocytes and thymocytes [3, 5], and none of them investigated the effect of VEGF-C on these cells.
However, there are numerous publications describing a critical role of vascular growth factors in regulation of migratory activity of different cell types. For example, VEGF receptor expression on tumor cells promotes their survival and enhances their migratory potential and capacity to metastasize [15]. On the other hand, the expression of VEGF receptors may lead to opposite effects. In particular, it was found that the enhanced expression of NRP-1 in human pancreas adenocarcinoma cells inhibited their migration and proliferation, whereas down-regulation of this co-receptor, on the contrary, stimulated tumor growth [16]. There are also data showing that the presence of VEGF receptors in breast cancer cells is an important clinical feature of tumor growth and has prognostic value [17, 18].
Here we investigated gene expression of VEGF-A, VEGF-N, and their receptors in murine tumor cell line hepatoma 22A. We found that the mRNA of these factors as well as VEGFR-1, VEGFR-3, and NRP-2 was detected, whereas VEGFR-2 and NRP-1 mRNAs were absent (figure). By comparing the pattern of mRNA expression of studied VEGF receptors in hepatoma cells versus lymphocytes we found remarkable distinctions.
Thus, we showed for the first time that thymocytes and lymph node cells of naive mice constitutively expressed mRNA of angiogenic factors VEGF-A and VEGF-C together with their main receptors. This suggests that VEGF family factors, along with their major angiogenic activities, might carry out important mediator functions within the immune system.
This work was supported by the Russian Foundation for Basic Research (grant No. 06-04-48250).
REFERENCES
1.Ferrara, N., Gerber, H.-P., and LeCouter, J. (2003)
Nature Medicine, 6, 669-676.
2.Barleon, B., Sozzani, S., Zhou, D., Weich, H. A.,
Mantovani, A., and Marme, D. (1996) Blood, 87,
3336-3343.
3.Mor, F., Quintana, F. J., and Cohen, I. R. (2004)
J. Immunol., 172, 4618-4623.
4.Gabrilovich, D., Ishida, T., Oyama, T., Ran, S.,
Kravtsov, V., Nadaf, S., and Carbone, D. P. (1998) Blood,
92, 4150-4166.
5.Huang, Y., Chen, X., Dikov, M. M., Novitskiy, S.
V., Mosse, A., Yang, L., and Carbone, D. P. (2007) Blood,
110, 624-631.
6.Crowther, M., Brown, N. J., Bishop, E. T., and
Lewis, C. E. (2001) J. Leukoc. Biol., 70, 478-490.
7.Maruyama, K., Masaaki, I., Cursiefen, C., Jackson,
D. G., Keino, H., Torrita, M., van Rooijen, N., Takeriaka, H., D'Amore,
P. A., Stein-Streilein, J., Losordo, D. W., and Streilein, J. W. (2005)
J. Clin. Invest., 115, 2363-2372.
8.Owen, L., Iragavarapu-Charyulu, V., Gunja-Smith,
Z., Herbert, L. M., Grosso, J. F., and Lopez, D. M. (2003) J.
Immunol., 171, 4340-4351.
9.Ijima, K., Yoshikawa, N., and Nakamura, H. (1996)
J. Immun. Meth., 196, 199-209.
10.Partanen, T. A., Arola, J., Saaristo, A.,
Jussila, L., Ora, A., Miettinen, M., Stacker, S. A., Achen, M. G., and
Alitalo, K. (2000) FASEB J., 14, 2087-2096.
11.Hiratsuka, S., Minowa, O., Kuno, J., Noda, T.,
and Shibuta, M. (1998) Proc. Natl. Acad. Sci. USA, 95,
9349-9354.
12.McLaren, J., Prentice, A., Charnock-Jones, D. S.,
Millican, S. A., Muller, K. H., Sharkey, A. M., and Smith, S. K. (1996)
J. Clin. Invest., 98, 482-489.
13.Yang, Z. F., Poon, R. T., Luo, Y., Cheung, C. K.,
Ho, D. W., Lo, C. M., and Fan, S. T. (2004) J. Immunol.,
173, 2507-2515.
14.Skobe, M., Hamberg, L. M., Hawighorst, T.,
Schirner, M., Wolf, G. L., Alitalo, K., and Detmar, M. (2001) Am. J.
Pathol., 159, 893-903.
15.Wey, J. S., Fan, F., Gray, M. J., Bauer, T. W.,
McCarty, M. F., Somcio, R., Lui, W., Evans, D. B., Wu, Y., Hicklin, D.
J., and Ellis, L. M. (2005) Cancer, 104, 427-438.
16.Gray, M. J., Wey, J. S., Belcheva, A., McCarty,
M. F., Trevino, J. G., Evans, D. B., Ellis, L. M., and Gallick, G. E.
(2005) Cancer Res., 65, 3664-3670.
17.Zhukova, L. G., Zhukov, N. V., and Lichinitser,
M. R. (2003) Byul. Eksp. Biol. Med., 135, 562-565.
18.Shcherbakov, A. M., Gershtein, E. S., Anurova, O.
A., and Kushlinskii, N. E. (2005) Vopr. Onkol., 51,
317-321.