Hydrophobic Interactions Are the Prevalent Force in Bromelain:Fab´ Complex
P. Gupta1,2, M. Saleemuddin1,3, and R. H. Khan3*
1Department of Biochemistry, Faculty of Life Sciences, Aligarh Muslim University, Aligarh 202002, Uttar Pradesh, India; fax: +91-571-272-1776; E-mail: pawan_g75@hotmail.com2Current address: Department of Pharmacology, Basic Science Biomedical Engineering, University of Minnesota, 6-120 Jackson hall, 321 Church Street SE, Minneapolis, MN55455, USA
3Interdisciplinary Biotechnology Unit, Faculty of Life Sciences, Aligarh Muslim University, Aligarh 202002, Uttar Pradesh, India; fax: +91-571-272-1776; E-mail: rizwanhkhan1@yahoo.com; msaleemuddin@rediffmail.com
* To whom correspondence should be addressed.
Received November 3, 2004; Revision received March 30, 2005
Antibromelain polyclonal antibodies against stem bromelain were raised in male albino rabbits and the Fab´ monomers isolated from the IgG of the immune sera as reported in our earlier communication (Gupta, P., Khan, R. H., and Saleemuddin, M. (2003) Biochim. Biophys. Acta, 1646, 131-135). Further, as evident from that communication bromelain:Fab´ complex has 1 : 1 stoichiometry. The stability of bromelain:Fab´ complex (1 : 1 stoichiometry) was investigated by far and near-UV CD and fluorescence measurements. Addition of up to 1.8 M NaCl caused no significant changes in fluorescence signals and near-UV CD peak pattern. However, the spectral studies together with gel filtration studies suggest dissociation of the complex beyond 5% (v/v) methanol. These results show that hydrophobic interactions play a pronounced role in the binding of Fab´ to bromelain while electrostatic interactions may be less crucial.
KEY WORDS: bromelain, Fab´, immunocomplexes, hydrophobic interaction, electrostatic interaction, methanol, NaClDOI: 10.1134/S0006297906130050
Abbreviations: CD) circular dichroism; CDR) complementarity determining region; Fab) antigen binding fragment of immunoglobulin; IgG) immunoglobulin G; UV) ultra violet.
The ability of protein to form specific, stable complexes with other
proteins is fundamental to many biological processes. Understanding the
molecular basis of protein-protein recognition requires knowledge of
individual interactions at the interface between the proteins [1-3]. One may define two broad
categories of protein-protein interfaces: (i) those which resemble
cross section through folded proteins in which hydrophobic residues are
in the interior and hydrophilic ones at the periphery and in which
productive binding is mediated largely by the former and (ii) ones in
which polar and non polar residues are interspersed throughout the
interface and in which both residue types make comparable contribution
to complex stabilization [4].
An antibody molecule combines with its antigen through the complementarity determining region (CDR). Since the repertoire of antigen to which it combines is large, it is understandable that the amino acid residues comprising the CDR have greater functional versatility. These residues have both polar and non-polar groups in them and generally the nonpolar end is buried and the polar end protrudes out. These polar atoms have the potential of forming hydrogen bonds and additional stability of the complex is provided by hydrophobic interactions. The interaction of the antibody with the antigen is highly specific and involves noncovalent interactions such as hydrophobic interactions and hydrogen bonding [5, 6].
With the availability of data on crystal structure of several antibody-antigen complexes [2, 7-11], it is possible to quantify these interactions. Some reports suggest preponderance of hydrogen bonds while others show hydrophobic [5, 12] suggesting universal principle of interactions in the immune complexes does not exist and each complex should be characterized de novo. The nature of the interactions between antigen and active center of the antibody plays an important role both in prognosis of immunochemical processes in vivo and in the development of different biotechnological model systems. Though there are a series of direct structural studies like site-specific mutations [13], docking thermodynamics [14, 15], X-ray crystallography [8-11, 16], and titration calorimetry devoted to the properties of these complexes, such work for each antigen-antibody pair is extremely labor-consuming and requires complicated equipment. In this connection development of methodically simple, universal approaches for screening of the interacting antigen-antibody pairs and rapid conclusion about probable nature of their interaction is required. It is often difficult to accurately quantify the contribution of the above-mentioned noncovalent interactions; however the preponderance of one can be established over another more easily. Furthermore, studies of antigen-antibody complex is often limiting inasmuch as study of precipitin reaction has to be taken into account [17, 18], which is a circuitous way of understanding the phenomenon. Here we present a stem bromelain:polyclonal Fab´ monomer (1 : 1) complex and have tried to study the nature of the interactions at the contact surface by studying behavior of the complex and individual components as a function of methanol and NaCl concentrations, with fluorescence and CD spectroscopy as tools. The system being soluble provides better understanding as the precipitin reaction is no longer required to be taken into account for understanding protein-protein interaction at the contact interface. Moreover, the use of polyclonal Fab´ monomer provides a collective composite nature of these interactions in this immune complex. The studies undertaken suggests preponderance of hydrophobic interactions over electrostatic. This complex has shown impressive thermal stability and stability in alkaline pH range of stem bromelain as reported in our earlier communication [1] and can serve as a model with potential in vivo and industrial application especially in homogenous reaction phases and development of subunit vaccines owing to preponderance of hydrophobic interaction and better diffusion potential of Fab´ than of intact IgG.
MATERIALS AND METHODS
Materials. Stem bromelain (3.4.22.32), Sephacryl S-300HR, and Sepharose 4B were purchased from Sigma (USA). Freund's complete and incomplete adjuvants were purchased from Difco Lab (USA). All the other chemicals used were of analytical grade.
Immunization. Bromelain preparation (300 µg/0.5 ml) in 20 mM sodium phosphate buffer, pH 7.0, was mixed with Freund's complete adjuvant in the ratio of 1 : 1 and administered subcutaneously to rabbits [19]. The animals were rested for 15 days and booster doses of antigen prepared by mixing 150 µg/0.5 ml solution of antigen with equal volumes of Freund's incomplete adjuvant, were given subcutaneously weekly for four weeks. Blood was withdrawn through an ear vein and clot formation allowed to take place at room temperature for 6 h. Serum was collected by centrifugation, decomplemented at 56°C for 30 min, and stored at -20°C. Titer value was calculated with ELISA as a tool. There was minimum variability of results except titer value between different immunization cycles and between different immunized animals.
Preparation of the affinity column. Bromelain was coupled to CNBr activated Sepharose 4B by method of Porath [20]. The preparation obtained thus contained 5 mg bromelain per ml gel [1].
Purification of specific IgG and Fab´ monomers. The affinity purification of specific IgG was achieved by employing bromelain coupled to Sepharose 4B following the published procedure [1, 21]. Briefly, bromelain specific antiserum was allowed to bind to the bromelain-Sepharose 4B affinity column. The column was washed thoroughly in the presence of 0.1 M Tris-HCl, pH 8.9, and 3 M NaCl to remove any unbound or nonspecifically associated protein. The bound IgGs were eluted with 0.1 M glycine HCl buffer, pH 3.5, and the eluate immediately neutralized with Tris buffer. While complete elution of antibody was obtained at pH 2.0, there were some irreversible conformational changes in antibody and binding capacity of affinity column was remarkably decreased. This was not so at pH 3.5, and this pH was chosen instead. Specific Fab´ fragments were similarly isolated by applying the papain treated IgG digest to the affinity column followed by washings and subsequent elution.
Preparation of bromelain:Fab´ complex. Stem bromelain and antibromelain Fab´ fragment were mixed in 1 : 1 molar ratio and incubated for 6 h at 4°C before investigating their stability.
Fluorimetry. Fluorescence measurements were performed using a Hitachi model F2000 spectrofluorometer (Japan). Excitation wavelength of 280 nm was used and emission spectra were recorded in the range from 300-400 nm. Excitation and emission slit widths were 5 nm. All solutions of protein were in the range of 0.1-0.5 mg/ml.
Circular dichroism. All the circular dichroism (CD) measurements were carried out at 25°C on a Jasco spectropolarimeter model J-720 using a SEKONIC XY plotter (model SPL-430A), with thermostatically controlled cell holder attached to a NESLAB model RTE 110 water bath with an accuracy of ±0.10°C. Far UV-CD spectra were recorded at a protein concentration range of 0.1-0.5 mg/ml using 1 mm pathlength cell and the near UV-CD spectra were measured in the range of 0.5-1 mg/ml concentration with a 10 mm pathlength cell. The CD results are expressed in millidegrees. CD spectra were recorded in wavelength range for far UV (210-250 nm) and for near UV (250-300 nm) at 5 millidegree sensitivity. All protein solutions were prepared in 10 mM sodium phosphate buffer, pH 7.0.
RESULTS AND DISCUSSION
Stem bromelain was quite immunogenic and rabbits challenged with the enzyme responded by producing precipitating antibodies in their sera. A high titer (~200,000) in the immune sera was observed as reported in our earlier communication [1]. Further, as evident from the communication Fab´ monomer isolated from specific polyclonal IgG form complex with stem bromelain with 1 : 1 stoichiometry and no high molecular weight adducts were observed.
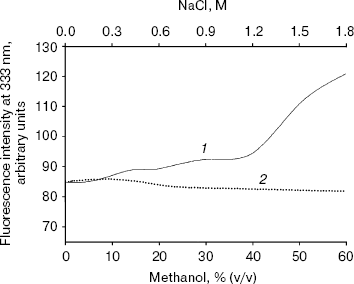
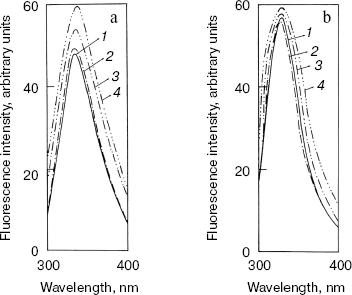
Effect of methanol and NaCl on fluorescence spectra of bromelain:Fab´ complex and the individual protein components. The fluorescence of the bromelain:Fab´ complex remained more or less unaltered in the presence of 0-1.8 M NaCl but was quite sensitive to high concentrations of methanol (Fig. 1). The gradual methanol concentration dependent increase in the fluorescence intensity of the complex was followed by a more rapid rise and at 60% (v/v) concentration of the solvent the fluorescence intensity increased over 40%. The observed solvent-induced increase in the fluorescence may be related to the exposure of chromophoric groups of bromelain or Fab´ to the solvent, resulting either from the dissociation of the complex or to the alteration/distortion of the structure of protein or both. However, while the fluorescence spectrum of bromelain (Fig. 2a) was highly solvent-sensitive, that of Fab´ (Fig. 2b) did not exhibit appreciable change in the fluorescence spectrum in the presence of up to 60% (v/v) methanol. The bromelain spectrum (Fig. 2a) was unaffected up to 20% (v/v) methanol but showed very significant increase (25%) in fluorescence at 60% (v/v) methanol concentration suggesting perturbation/alteration in bromelain conformation which is also evident by correspondence with near-UV CD studies as explained in the following section. Considering that increase in the fluorescence of bromelain:Fab´ complex between 20 and 60% (v/v) methanol is 40%, it is likely that exposure of chromophoric groups at protein-protein interface contributes towards the difference in increase in fluorescence intensity. Bromelain contains five tryptophan residues [22] and extensive sequence homology with papain suggests that three of these are buried in hydrophobic core whereas two may be located near the surface of the molecule [23, 24]. In high probability one or both of the surface tryptophans may be masked on formation of bromelain:Fab´ complex.
Fig. 1. Effect of methanol (1) and NaCl (2) on fluorescence intensity of bromelain:Fab´ complex at 333 nm. Bromelain and Fab´ were in equimolar amounts.
That complex formation with Fab´ also restricts alteration in bromelain structure resulting from high ionic strength was evident from the comparison between ionic strength induced alteration of free enzyme and the complex. While 1.8 M NaCl caused only a marginal increase in case of bromelain, there was no appreciable change in fluorescence intensity of Fab´ (data not shown), which is suggestive of some slight conformational perturbation in the former. Had the electrostatic interaction been crucial for complex formation, the complex would have dissociated into individual protein components at high ionic strength followed by an increase in fluorescence intensity inasmuch as contribution from bromelain or exposure of chromophoric groups (fluorescence spectrum of bromelain was quenched due to complex formation with Fab´, data not reported) or both. However, this does not happen to be the case, suggesting a preponderance of hydrophobic interaction. Analytical parameters for these additives were chosen in light of the effect of concentrations on the individual protein components.Fig. 2. Effect of methanol on fluorescence emission spectra of individual protein components: bromelain (a) and Fab´ (b) under native standard condition (pH 7.0) (1) and in presence of 20% (2), 40% (3), and 60% (v/v) methanol concentration (4).
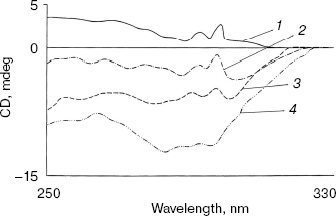
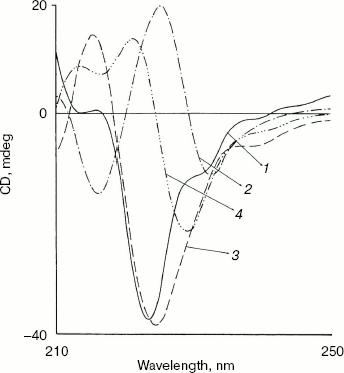
Effect of methanol and NaCl on near-UV CD spectra of bromelain:Fab´ complex and the individual protein components. As shown in Fig. 3 the near-UV CD spectrum of the complex has peaks at 298 nm and at 293 nm and seems to be characteristic of the complex (bromelain incubated with nonspecific Fab´ monomer fails to give a similar spectrum). There was little alteration in the peak pattern of the spectrum at 1.8 M NaCl suggesting remarkable stability of the complex. At 5% (v/v) methanol the spectrum was identical to native complex and at 20% (v/v) methanol spectral features showed a very little alteration in peak pattern (data not reported). However, in the presence of 40% (v/v) methanol both the peaks decreased markedly while at 60% (v/v) methanol inversion and loss of all spectral features of the complex was observed. While perturbation in the tertiary structure of bromelain (Fig. 4a) was marked even at 40% (v/v) methanol and increased further at high solvent concentration, the near-UV CD spectrum of the Fab´ (Fig. 4b) was stable up to 40% (v/v) methanol and only moderate disruption in tertiary structure was observed when the solvent concentration was raised to 60%. Disruption of the tertiary structure of either bromelain or Fab´ was marginal up to 1.8 M NaCl (data not shown). It is interesting to note that despite retention of characteristic feature of the complex at 1.8 M NaCl there was a shift in CD magnitude, which may presumably be due to different behavior of solvent towards individual protein components and complex; furthermore, a contribution could be from secondary structure alterations as detailed in next section. This study, while suggesting the sensitivity of the complex to methanol, cannot discriminate between the solvent effects on bromelain and dissociation of the complex.
Fig. 3. Effect of NaCl and methanol on near-UV CD spectra of bromelain:Fab´ complex: 1) native complex; 2-4) treatment with 1.8 M NaCl (2), 40% (v/v) methanol (3), and 60% (v/v) methanol (4), respectively. Bromelain and Fab´ were in equimolar amounts.
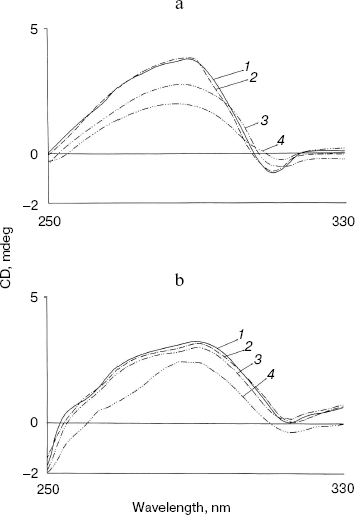
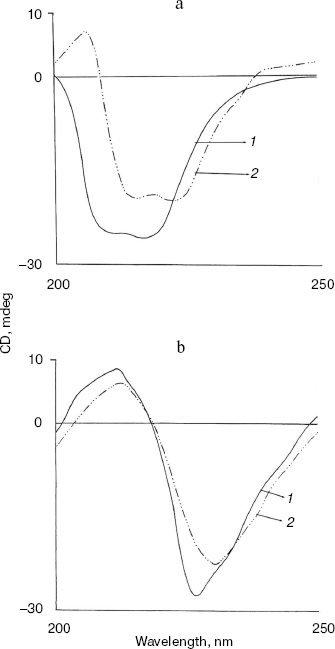
Effect of methanol and NaCl on far-UV CD spectra of bromelain:Fab´ complex and the individual protein components. The far-UV CD spectrum (Fig. 5) of the complex is characteristic with a sharp negative peak at 223 nm and a broad shoulder around 232 nm. There was no marked influence on the negative peak in the presence of 5% (v/v) methanol, but a clear 6 nm shift in the negative peak at 223 nm, a prominent decrease in ellipticity, and appearance of two positive peaks at 213 and 221 nm were observed in presence of 20% (v/v) methanol. This suggests major alterations/rearrangement in the secondary structure [25]. Considering that 20% (v/v) methanol has little effect on the tertiary structure of the proteins and little effect on the secondary structure of either bromelain or Fab´ (data not shown), rearrangement of the secondary structure in the complex is the likely explanation. Comparable effects on secondary structure are also apparent in the presence of 0.9 M NaCl. Only moderate alterations in secondary structure of the individual protein components bromelain (Fig. 6a) and Fab´ (Fig. 6b) were observed in the presence of 0.9 M NaCl. Interestingly, the bromelain:Fab´ complex appears to remain stable in the presence of up to 1.8 M NaCl as revealed by fluorescence and near-UV CD spectroscopy. It therefore seems likely that considerable interaction between bromelain and the Fab´ may remain intact even after significant alteration in the secondary structure of the two proteins. This may be due to induction of local interactions, which in good probability cause alterations in secondary structure without causing remarkable changes in the tertiary structure. These local interactions causing changes in the secondary structure were however reversible in bromelain:Fab´ complex and the individual protein components. The CD and fluorescence spectra of bromelain and Fab´ are comparable with those reported elsewhere [23, 26-29].Fig. 4. Effect of methanol on near-UV CD spectra of individual protein components: bromelain (a) and Fab´ (b) under native standard condition (pH 7.0) (1) and in presence of 20% (2), 40% (3), and 60% (v/v) methanol concentration (4).
Fig. 5. Effect of NaCl and methanol on far-UV CD spectra of bromelain:Fab´ complex: 1) native complex; 2-4) treatment with 0.9 M NaCl (2), 5% (v/v) methanol (3), and 20% (v/v) methanol (4), respectively. Bromelain and Fab´ were in equimolar amounts.
All these results taken together are suggestive of methanol sensitive hydrophobic interactions [29] rather than salt linkages playing a crucial role in the binding of bromelain to Fab´.Fig. 6. Effect of NaCl on far-UV CD spectra of individual protein components: bromelain (a) and Fab´ (b) under native standard condition (pH 7.0) (1) and in presence of 1.8 M NaCl (2) (spectra of individual protein components at 0.9 and 1.8 M NaCl are almost overlapping).
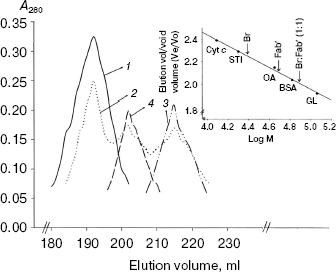
Size exclusion chromatography of bromelain:Fab´ complex against 1.8 M NaCl and 5 and 10% (v/v) methanol concentration. Spectral findings were reconfirmed and substantiated by a hydrodynamic method (size exclusion chromatography). Bromelain:Fab´ complex was subjected to size exclusion chromatography (Sephacryl S-300HR) under one salt concentration (1.8 M) and two methanol concentration (5 and 10%). While 1.8 M NaCl and 5% (v/v) methanol condition had no effect on elution position of the complex, at 10% (v/v) methanol bromelain:Fab´ complex tends to undergo dissociation (Fig. 7). The dissociation is not complete inasmuch as use of polyclonal Fab´ monomers having different affinity for protein epitope(s) on bromelain. Complete dissociation was observed at 40% (v/v) methanol concentration.
Conclusion and implications of the study. The spectral studies point towards significant dissociation of complex beyond 20% (v/v) methanol concentration along with some possible dissociation between 5-20% (v/v) methanol as well. The hydrodynamic study suggests stability of the complex at 5% methanol, however at higher concentration it tends to dissociate with complete dissociation taking place at 40% (v/v). Blending these studies implicates dissociation of complex beyond 5% (v/v) methanol, which is gradual till 20%, followed by significant dissociation which is almost complete at 40% (v/v) methanol concentration. This gradual dissociation can be explained in light of the fact that Fab´ monomers have been derived from polyclonal antiserum and therefore have different affinity towards epitope(s) on the bromelain surface. Spectral studies and hydrodynamic studies are complementary/supplementary and are suggestive of preponderance of hydrophobic interactions. Preponderance of hydrophobic interactions along with the fact that Fab´ monomer has better diffusion capability than that of intact IgG half IgG or Fab´´ dimer makes it a potential model to explore for in vivo and industrial application as detailed below.Fig. 7. Size exclusion chromatography (Sephacryl S-300HR) of bromelain:Fab´ complex under different NaCl and methanol concentrations: bromelain:Fab´ under native standard condition (1) and in presence of 1.8 M NaCl (1) or 5% (1) and 10% methanol (2). Elution profiles of individual protein components bromelain (3) and Fab´ (4) are shown. Inset shows elution positions of bromelain:Fab´ complex, Fab´. and bromelain (Br) against various marker proteins: beta-galactosidase (GL, 116 kD), bovine serum albumin (BSA, 66 kD), ovalbumin (OA, 45 kD), soybean trypsin inhibitor (STI, 20 kD), cytochrome c (Cyt c, 12.4 kD).
Earlier attempts were focused on stabilization of enzyme by formation of immunocomplexes [30-34] and binding on antibody support [32] rendering enzyme preparations resistant to solvent induced inactivation [35]. However these methodologies had limitations inasmuch as they provide insoluble preparations, which can well be overcome by using single chain Fv [36] or Fab´ monomer instead. This provides a soluble model and therefore can be used in homogenous reaction phases. As reported in our earlier communication [1] bromelain:Fab´ complex shows impressive thermal and pH stability profiles.
Preponderance of hydrophobic interaction makes it a suitable model to be used in enzyme therapy and preparation of subunit vaccines that require a specific, stable, and rapidly diffusible carrier in otherwise polar body fluids. Anti-edemic, anti-inflammatory, and anti-neoplastic properties of bromelain have been exploited [37-40], but intravenous and intraperitoneal administration of bromelain, despite their merits, have been discontinued and the enzyme is currently administered orally [41]. Complexing bromelain/other vital enzymes or antigenic peptides with Fab´ derived from humanized IgG [42, 43] will provide a further better model to solve several problems associated with enzyme therapy and subunit vaccines, respectively.
The disposition of this study was towards investigating in general collective, averaged, and composite nature of interactions between Fab´ monomer and antigen (antibromelain Fab´ and bromelain in particular). Polyclonal antiserum by default provides us the required system with variety of molecular interactions between the abovementioned protein moieties inasmuch as Fab´ monomers derived from polyclonal antiserum have different affinities towards epitope(s) on the bromelain surface. Use of polyclonal antibodies is explainable inasmuch as they are biotechnologically practical and reflect natural conditions prevalent in the body.
We further intend to study interaction of Fab´ monomer with the molten globule state of stem bromelain [24, 44]. Many proteins are said to exist and traverse body fluids in molten globule like states. The intended study may therefore provide valuable information.
Facilities provided by Aligarh Muslim University are gratefully acknowledged. P. G. was Senior Research Fellow of the Council of Scientific and Industrial Research, New Delhi, India. Financial assistance provided in form of lab facilities by DST (FIST) and UGC-DRS is also acknowledged.
REFERENCES
1.Gupta, P., Khan, R. H., and Saleemuddin, M. (2003)
Biochim. Biophys. Acta, 1646, 131-135.
2.Chen, Y., Wiesmann, C., Fuh, G., Li, B.,
Christinger, H. W., McKay, P., DeVos, A. M., and Lowman, H. B. (1999)
J. Mol. Biol., 293, 865-881.
3.Jiang, L., and Lai, L. (2002) J. Biol.
Chem., 277, 37732-37740.
4.Goldman, E. R., Acqua, W. D., Braden, B. C., and
Mariuzza, R. A. (1997) Biochemistry, 36, 49-56.
5.Davies, D. R., Padlan, E. A., and Sheriff, S.
(1990) Ann. Rev. Biochem., 59, 439-473.
6.Oss, C. J. V., and Absolom, D. R. (1984) in
Molecular Immunology (Atassi, M. Z., Oss, C. J. V., and Absolom,
D. R., eds.) Marcel Dekker Inc., New York, p. 337.
7.Chitarra, V., Aizari, P. M., Bentley, G. A., Bhat,
T. N., Eisele, J. L., Houdusse, A., Lescar, J., Souchon, H., and
Poljak, R. J. (1993) Proc. Natl. Acad. Sci. USA, 90,
7711-7715.
8.Li, Y., Li, H., Smith-Gill, S. J., and Mariuzza, R.
A. (2000) Biochemistry, 39, 6296-6309.
9.Dimasi, N., Moretta, L., Biassoni, R., and
Mariuzza, R. A. (2003) Acta Crystallogr. D. Biol. Crystallogr.,
59, 1856-1858.
10.Smith-Gill, S. J. (2002) J. Mol.
Recognit., 15, 44-52.
11.Li, Y., Li, H., Yang, F., Smith-Gill, S. J., and
Mariuzza, R. A. (2003) Nat. Struct. Biol., 10,
482-488.
12.Bhat, T. N., Bentley, G. A., Boulot, G., Greene,
M. I., Tello, D., Acqua, W. D., Souchon, H., Schwarz, F. P., Mariuzza,
R. A., and Poljak, R. J. (1994) Proc. Natl. Acad. Sci. USA,
91, 1089-1093.
13.Li, Y., Lipschultz, C. A., Mohan, S., and
Smith-Gill, S. J. (2001) Biochemistry, 40, 2011-2022.
14.Sundberg, E. J., and Mariuzza, R. A. (2002)
Adv. Protein Chem., 61, 119-160.
15.Lipschultz, C. A., Yee, A., Mohan, S., Li, Y.,
and Smith-Gill, S. J. (2002) J. Mol. Recognit., 15,
44-52.
16.Mohan, S., Sinha, N., and Smith-Gill, S. J.
(2000) Biophys. J., 85, 3221-3236.
17.Ahmad, A. (1995) Ph. D. Thesis, Aligarh
Muslim University, Aligarh, India.
18.Ansari, A. A., and Salahuddin, A. (1973)
Immunology, 25, 377-383.
19.Freunds, J. (1947) Annu. Rev. Microbiol.,
1, 291-308.
20.Porath, J., Axen, R., and Ernback, S. (1967)
Nature, 215, 1491-1492.
21.Stults, N. L., Asta, L. M., and Lee, Y. C. (1989)
Analyt. Biochem., 180, 114-119.
22.Ritonja, A., Rowan, A. D., Buttle, D. J.,
Rawlings, N. D., Turk, V., and Barett, A. J. (1989) FEBS Lett.,
247, 419-424.
23.Haq, S. K., Rasheedi, S., and Khan, R. H. (2002)
Eur. J. Biochem., 269, 47-52.
24.Gupta, P., Khan, R. H., and Saleemuddin, M.
(2003) Arch. Biochem. Biophys., 413, 199-206.
25.Khan, F., Khan, R. H., and Muzammil, S. (2000)
Biochim. Biophys. Acta, 1481, 229-236.
26.Reyna, A. A., Arana, A. H., and Espinosa, R. A.
(1994) Biochem. J., 300, 107-110.
27.Jaton, J. C., Huser, H., Blatt, Y., and Pecht, I.
(1975) Biochemistry, 14, 5308-5311.
28.Welfl, K., Misselwitz, R., Hausdorf, G.,
Höhne, W., and Welfl, H. (1999) Biochim. Biophys. Acta,
1431, 120-131.
29.Waseem, A., and Salahuddin, A. (1983) Ind. J.
Biochem. Biophys., 20, 253-258.
30.Ramjeesingh, M., Zywulko, M., Rothstein, A., and
Shami, E. Y. (1992) Biotechnology (NY), 10, 442-445.
31.Cherednikova, T. V., Muronetz, V. I., and
Nagradova, N. K. (1981) Mol. Immunol., 18, 1055-1064.
32.Cherednikova, T. V., Muronets, V. I., and
Nagradova, N. K. (1982) Biokhimiya, 47, 361-373.
33.Newsted, W. J., Ramjeesingh, M., Zywulko, M.,
Rothstein, S. J., and Shami, E. Y. (1995) Enzyme Microb.
Technol., 17, 757-764.
34.Younus, H., Köditz, J., Saleemuddin, M., and
Hofmann, R. U. (2002) Biotechnol. Lett., 24,
1821-1826.
35.Jan, U., Husain, Q., and Saleemuddin, M. (2001)
Biotechnol. Appl. Biochem., 34, 13-17.
36.Guo, L., Yan, X., Qian, S., and Meng, G. (2000)
Biotechnol. Appl. Biochem., 31, 21-27.
37.Taussig, S. J., Yokoyama, M. M., Chinen, A.,
Onari, K., Yamakido, M., and Nishimoto, Y. (1975) Hiroshima
Med., 24, 185-193.
38.Kumakura, S., Yamashita, M., and Tsurufuji, S.
(1988) Eur. J. Pharmacol., 150, 295-301.
39.Taussig, S. J., Szekerczes, J., and Batkin, S.
(1985) Planta Med., 6, 538-539.
40.Taussig, S. J., and Batkin, S. (1988) J.
Ethnopharmacol., 22, 191-203.
41.Winter, H. L. (1990) Planta Med.,
56, 249-253.
42.Clark, M. (2000) Immunol. Today,
21, 397-402.
43.Clark, M. A. (2002) Meth. Mol. Biol.,
178, 39-58.
44.Gupta, P., Khan, R. H., and Saleemuddin, M.
(2003) Int. J. Biol. Macromol., 33, 167-174.