Academician Andrei Nikolaevich Belozersky

|
Enzymatic DNA Methylation Is an Epigenetic Control for Genetic Functions of the CellB. F. VanyushinBelozersky Institute of Physico-Chemical Biology, Lomonosov Moscow State University, 119992 Moscow, Russia; fax: (7-095) 939-3181; E-mail: vanyush@belozersky.msu.ru
|
Received December 24, 2004
In eukaryotic cells nuclear DNA is subjected to enzymatic methylation resulting in formation of 5-methylcytosine residues mainly in CG and CNG sequences. In plants and animals, this DNA methylation is species-, tissue-, and organelle-specific. It changes (diminishes) with age and is regulated by hormones. On the other hand, genome methylation can control hormonal signal. There are replicative and postreplicative DNA methylations. They are served by multiple DNA-methyltransferases with different site specificity. Replication is accompanied by appearance of hemimethylated sites in DNA; pronounced asymmetry of DNA chain methylation disappears at the end of the cell cycle; a model of regulation of replication by DNA methylation is suggested. DNA methylation controls all genetic processes in the cell (replication, transcription, DNA repair, recombination, gene transposition) and it is a mechanism of cell differentiation, gene discrimination, and silencing. Prohibition of DNA methylation stops development (embryogenesis), switches on apoptosis, and is usually lethal. Distortions in DNA methylations result in cancerous cell transformation, and the DNA methylation pattern is one of the safe cancer diagnostics at early stages of carcinogenesis. The malignant cell has a different DNA methylation pattern and a set of DNA-methyltransferase activities expressed as compared with normal cells. Inhibition of DNA methylation in plants is accompanied by induction of genes of seed storage proteins and flowering. In eukaryotes one and the same gene can be methylated both on cytosine and adenine residues; thus, there are, at least, two different and probably interdependent systems of DNA methylation in the cell. First higher eukaryotic adenine DNA-methyltransferase was isolated from plants; this enzyme methylates DNA with formation of N6-methyladenine residues in the sequence TGATCA → TGm6ATCA. Plants have AdoMet-dependent endonucleases sensitive to DNA methylation status; therefore, like microorganisms, plants seem to have a restriction-modification (R-S) system. Revelation of an essential role of DNA methylation in the regulation of genetic processes has laid a foundation for and materialized epigenetics and epigenomics.
KEY WORDS: apoptosis, DNA methylation, mitochondria, epigenetics, cancer, aging, regulation of replication, transcription, adenine-DNA-methyltransferase, methyl-DNA-binding proteins, plants
In fact, without methylation there would be no life at all.
Craig Cooney
“From small beginnings come great things” (proverb). More than 50 years ago it was already known that along with four classical bases (adenine, guanine, cytosine, and thymine) so-called “minor” methylated bases can occur in DNA. 5-Methylcytosine (m5C) was found first as a minor base in various DNAs [1, 2], and N6-methyladenine (m6A) was detected then in bacterial DNA [3, 4]. The origin of these bases in DNA was unknown for a long time. Only in 1963 the specific enzymes DNA-methyltransferases were observed in bacteria and then in eukaryotes; these enzymes transferred methyl groups from S-adenosyl-L-methionine (AdoMet) selectively onto definite cytosine or adenine residues in DNA chains [5]. It became clear that minor bases (m5C and m6A) do not incorporate into DNA in a ready-made form but originate there as a result of enzymatic modification (methylation) of common bases (C or A, respectively) in DNA chains that are forming or already formed. Nevertheless, the specificity and functional role of DNA methylations were still unknown for a long time. Moreover, the concept that these minor bases do not have any essential significance both in the structure of DNA itself and in its functioning was quite widely disseminated.
The classical object of traditional genetics, Drosophila, served mistakenly very often as an “irresistible” evidence for this postulate. In fact, m5C in the genome of this insect escaped detection for a very long time. Therefore, it was concluded by many investigators that this DNA modification does not play a significant role in the life of eukaryotic organisms because Drosophila lives comfortably without any DNA methylation. This situation did not bring very much enthusiasm to DNA methylation research in many world-renowned molecular biology laboratories and allowed us to study this matter without any race for many years.
Actually, we have been involved in this research for about 50 years. Similarly to the great Russian physiologist (Nobel Prize winner) I. P. Pavlov, who erected a memorial to dog (his beloved experimental animal), we have to erect a memorial to Drosophila not because it like a dog is a very useful object in biological experiments, but because it allowed us to peacefully investigate DNA methylation starting from the early beginnings without being tired out by enormous competition and agiotage. Besides, a long time ago we already noted that the Drosophila genome is very much deficient in CpG sequences that usually serve as the main substrates for in vivo DNA methylation in eukaryotes; according to our opinion this strong CpG suppression in Drosophila genome could be due to only methylation of cytosine residues [6]. As we could not detect the proper DNA-methyltransferase activity in Drosophila at that time [7], we designated this putative DNA modification as a “fossil” DNA methylation [6]. Recently it was clearly shown that DNA in Drosophila is methylated (it contains m5C), this DNA modification being important for normal insect development, and specific cytosine DNA-methyltransferase has been detected at early insect developmental stages [8, 9].
We were always sure that minor methylated bases in DNA as well as the enzymatic genome modification should not be traceless in the genome organization and they must obligatorily effect cell functions.
DNA methylation and its influence on DNA structure and interaction with proteins. Now a few words on the effect of methylation on DNA structure. We have been lucky to find unusual natural double-stranded DNA (DNA of AR9 bacteriophage for Bacillus brevis) in which thymine is completely substituted by a typical RNA base, uracil. Roughly speaking, uracil is thymine lacking a methyl group. This bacteriophage DNA containing uracil melted at significantly lower temperature compared with normal thymine containing DNA of the equivalent base composition [10]. It became clear that methylation of cytosine residues is not indifferent to DNA structure: it stabilizes double helix. But it was more attractive to find that methylation strongly affects DNA binding with various proteins. In particular, we isolated from plant nuclei proteins that bind specifically to regulatory elements of ribosomal RNA genes (135 bp subrepeat element) and showed that binding of these proteins is modulated (inhibited) by in vitro methylation of cytosine residues in CCGG sites [11].
In many cases cytosine DNA methylation prohibits binding of specific nuclear proteins (factors) involved in realization of various genetic processes including transcription, replication, and DNA repair. On the other hand, there are m5C-DNA-binding proteins that arrange specifically on DNA an entire ensemble of protein complexes controlling gene expression [12, 13].
Non-enzymatic DNA methylation. It is curious that if DNA was incubated in the presence of methyl-labeled S-adenosyl-L-methionine without any proteins added, the radioactivity was detected in DNA and, particularly, in 5-methylcytosine and thymine residues newly formed there. In this way we discovered non-enzymatic DNA methylation [14]. Interestingly, the amount of labeled thymine formed was much more than that of labeled m5C. Thus, it was shown that non-enzymatic DNA methylation in water solution is conjugated with rapid oxidative deamination of the m5C residues formed with their transformation into thymine residues (see Scheme). This was real evidence that methylation of cytosine residues in DNA may often result in C → T transition, and 5-methylcytosine residues in DNA are “hot” mutation points. This event is a reason for disappearance (suppression) of some CpG sequences from genes and genomes of various organisms, and it is a main route of natural mutagenesis and evolution.
Specificity of DNA methylation. The existence of potential non-enzymatic DNA methylation in nature was effectively used by specific enzymatic proteins that appeared with evolution; these proteins, DNA-methyltransferases, unlike random non-enzymatic methylation, modify cytosine and adenine residues specifically in definite DNA nucleotide sequences. We deciphered one of the first such sequences methylated in bacterial DNA [15]. It appeared that in Bacillus brevis cells the cytosine DNA-methyltransferase methylates a cytosine residue (underlined) in the symmetrical nucleotide sequence (5´) N´ G C T G C N (3´). The specific endonuclease splitting this sequence when unmethylated was later isolated from similar bacilli by Dr. R. Roberts. It turned out that DNA methylation in bacteria is a part of host restriction-modification phenomenon. This, in particular, means that bacteriophage grown in cells of a given bacterial strain acquires host specificity due to DNA methylation and afterwards is able to infect only the cells of this particular host (the set of future hosts is extremely restricted). Bacteriophage DNA methylated with host DNA-methyltransferases is protected against hydrolysis with methyl-sensitive endonucleases in these particular host cells but not in others. We have established, in particular, that specificity of the adenine methylation sites in T2 bacteriophage DNA and its host E. coli DNA is the same [16]. To some extent, these early data along with others served as a sort of explanation of the chemical nature of host restriction-modification in bacteria. Before that, we had been assured that in various bacteria DNA is methylated in a different fashion; thus, in fact, strain and species specificity of DNA methylation in microorganisms was described [17]. Moreover, it was shown that DNA methylation pattern changes on dissociation (R- and S-form) [17], spore formation [18], and piocyne induction [19] in bacteria. It seems that these data were the first indication that DNA methylation is associated with cellular differentiation in microorganisms.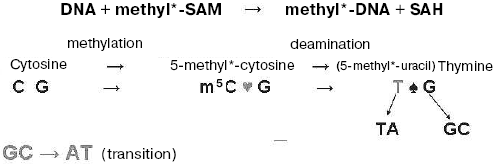
It was unknown but important to determine what is the chemical and biological specificity of DNA methylation in eukaryotes including plants and animals. Even before the methods of DNA sequencing appeared we analyzed pyrimidine sequences (clusters) isolated from DNA by chemical hydrolysis and showed that in plant genomes 5-methylcytosine is located in sequences such as Pu-m5C-Pu, Pu-m5C-T-Pu, Pu-m5C-C-Pu, and Pu-m5C-m5C-Pu [20]. This corresponded to data of Razin's group that appeared simultaneously on methylation of cytosine residues in plant and animal DNAs [21]. According to our data for plant DNA, up to about 30% of 5-methylcytosine is localized in m5CNG sequences [20]. The presence of m5C in CNG sites, particularly, in animal DNA was distrusted and even neglected for a long time. But, in fact, this DNA methylation was shown to be real in animal cells also [22-26] and it has an essential biological significance. Methylation of cytosine residues in these and asymmetrical DNA sequences is mainly observed on gene methylation directed by small double-stranded RNA molecules associated with gene silencing [27]. In plants an unusual enzyme was recently found that methylates cytosine residues in any context except for CpG sequences [28]. It should be noted that DNA methylation in plants is much richer and more diverse compared with animals; it is carried out by a larger set of specific DNA-methyltransferases, some of which are controlled and utilized via ubiquitin [29, 30].
Thus, in principle the nature of the chemical specificity of DNA methylation in plants and animals is established. There is no doubt that along with specific action of DNA-methyltransferases DNA methylation depends also on the structural organization of chromatin and accessibility in it of respective DNA sites to be modified. This certainly means that a study of DNA methylation itself requires serious research on chromatin; further progress in this area is impossible without deciphering of fine fluctuating chromatin structures in the nucleus.
As for biological specificity of DNA methylation, we learned a long time ago that it is species-specific. In many invertebrates methylation degree of the genome is very low; as we have already mentioned, m5C amount in Drosophila DNA is very small and it could not even be detected for a very long time, whereas in vertebrate DNAs it is always present in noticeable amounts [31]; in plant DNA this base cannot even be called a minor one because it is quite comparable with cytosine [32, 33].
We have established that along with species-specificity there are also tissue- (cellular), subcellular- (organelle), and age-specificity of DNA methylation in animals and plants. It seems to be appropriate to cite here one of our American colleagues, Dr. Craig Cooney ([34], Preface XV, pp. 124, 182): “Dr. Boris Vanyushin and co-workers in Moscow first showed in the 1960s and published in 1967 that salmon lost DNA methylation with age. The same group of scientists later showed that what was true in salmon was also true for most of the organs they studied in cattle and rats. Later studies by several groups of scientists in the United States and Japan indicated that many organs in mice also showed loss of DNA methylation with aging”. Original data on decrease in m5C content in DNA of different animals on aging are presented in [35-42] (see Table 1). Now the age-dependent demethylation of DNA is quite obvious, and some investigators are inclined even to consider DNA methylation degree as a sort of biological clock that measures the age and forecasts lifespan. Distortions in DNA methylation may lead to premature aging. We have to keep it always in mind and to avoid a deficiency, at least, in folic acid and B12 vitamin that take part in formation of methionine, a donor of methyl groups in the cell.
Table 1. Content of 5-methylcytosine in DNA
of Pacific salmon [35]
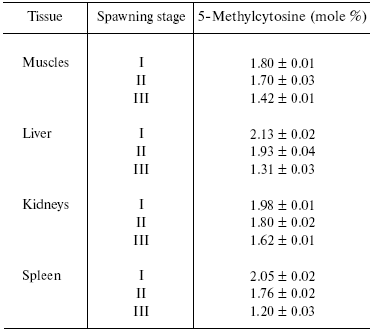
Note: I, see life period; II, beginning of migration into river;
III, soon after spawning.
We have observed that DNAs in the cells of one and the same organism are methylated in a different fashion. Detection of tissue specificity of DNA methylation allowed us first to declare that DNA methylation should be a mechanism of regulation of gene expression and cell differentiation [31, 43]. These data have drawn the attention of many investigators in this country and abroad and pushed ahead the intense study of DNA methylation throughout the world.
We also found that DNA methylation in mitochondria and the nucleus of one and the same cell is different. 5-Methylcytosine was detected in a bovine heart mitochondrial DNA [44]. The distribution pattern of m5C among pyrimidine isopliths in mitochondrial and nuclear DNAs occurred to be different [45]. Besides, we isolated cytosine DNA-methyltransferase from bovine heart mitochondria and found that this enzyme has different site specificity action compared with a nuclear one [46]. Thus, the subcellular (organelle) specificity of DNA methylation was also discovered.
Functional role of DNA methylation. As fate decrees, we were the ones who first, in a very skeptical general atmosphere, ask the question: What biological significance may DNA methylation have? Therefore, we were obliged to find different illustrative and adequate biological models that could be used for getting evidence of the exclusive significance of this genome modification. Initially we thought that if DNA methylation is really associated with some biological functions, it should be changed itself when these functions are induced. Thus, we started to investigate what could happen with DNA methylation in models induced such as hydrocortisone-liver and training (memory)-neurons. Indeed, DNA methylation pattern in rat liver changed significantly after hydrocortisone introduction and these changes were accompanied by induction of various genes [39, 42, 47-49]. DNA methylation pattern in neurons but not other brain cells was changed during rat training [50-52]. Changes in the neuron DNA methylation pattern induced by training were among the first evidences that the genome takes part in memory formation [50]. In plants DNA methylation strongly changes on seed germination [53], transition to flowering [54], and after infection with various fungi, parasitic plants, and viruses [55, 56]. It became clear that infective agents can more or less delicately act on plants in their own favor by modulation of DNA methylation in host cells.
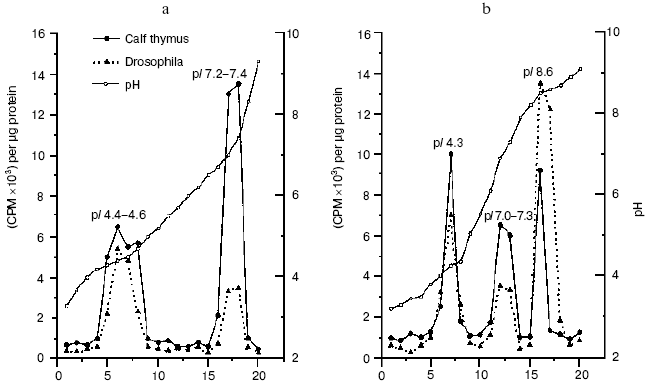
“As early as 1977 a Russian team looked at normal cells from cows, then compared those with cells from cows who had a type of cancer known as lympholeukemia. In general, the overall DNA methylation was lower in cells from animals with the cancer. This was one of the early clues that methylation, DNA methylation, at least, might be involved in cancer, either as a cause or as a result” [34]. In fact, we have shown that in lymphocytes of cows suffering from chronic lympholeucosis the DNA methylation pattern is drastically changed compared with normal cells [57-60]. In malignant cells with very high DNA-methyltransferase activity the level of total DNA methylation was lower but in palindrome highly-reiterated sequences it was higher than that in normal lymphocytes [61]. Unlike in normal cells, two DNA-methyltransferase activities, at least, were found in the nuclei from cows suffering from lympholeucosis. One of these enzymes differed strongly in site specificity from DNA-methyltransferase activity of lymphocytes from healthy animals [62]. All these findings allowed us to first firmly conclude that serious distortions in DNA methylation are a way to cancer. Now this idea is an axiom; it was confirmed lately and strongly developed now in brilliant works done by Drs. S. Baylin, M. Ehrlich, R. Jaenisch, P. Jones, M. Szyf, G. Pfeifer, W. Doerfler, P. Volpe, and many others; gene methylation pattern now is a safe diagnostic index of many cancers even at the very early stages of malignancy. We have observed recently an increased level of DMT1 gene expression, changes in the set and site specificity of DNA-methyltransferases, and an appearance of new proteins with DNA-methyltransferase activities in tumor (hepatoma) cells of rats fed with methyl-deficient diet (Fig. 1) [63]. Thus, our original idea [57] of the triggering of malignancy by distortion of DNA methylation has been confirmed again.
We have established that DNA methylation in animals and plants is under hormonal control [39, 42, 47-49] and is modulated by antioxidants [42]. Thus, ionol (BHT) modulates DNA methylation in rats; in liver it induces de novo DNA methylating enzymes and stimulates transcription of genes for AdoMet-synthetase and p53 [64]. It was shown that highly purified dexamethasone-receptor complexes from rat liver bind specifically to GC-rich DNA regions [65-67]. This was the first evidence that specific sites for binding of hormone-receptor complexes (HRC) exist in DNA; this was a sort of a surprise for leaders of the “empire” of molecular endocrinology of corticosteroid hormones. HRC bind predominantly with a fraction of homologous DNA, transcription of which is stimulated by cortisol [67]. In rat liver chromatin, cortisol induces transposition of specific sites binding HRC from potentially active DNA fraction to DNA fraction transcribed [67]. A very important fact was described at that time--DNA methylation effectively controls hormonal signal: in vitro methylation of rat liver nuclear DNA with homologous DNA-methyltransferase prevents HRC binding [68]. Thus, there is a mutual control in cells; on one hand, hormones control genome methylation and, on the other hand, DNA methylation is not indifferent to realization of the hormonal signals. This is true for both animals and plants.Fig. 1. DNA-methyltransferase activities from rat liver (a) and hepatoma (b) cells [63].
In plants, hormones (phytohormones of various classes) and other specific plant growth regulators effectively control DNA methylation [69-71]. For example, they can diminish global DNA methylation in a cell cycle. Besides, phytohormones inhibit methylation of newly formed DNA chains but not the Okazaki fragments (Table 2) [70, 72]. This was the first evidence that phytohormones can influence genome by modulation of its methylation status. In general, we are inclined to think that modulation of DNA methylation is one of the principal mechanisms of hormone action in plants and animals. It cannot be ruled out that HRC can recognize and compete well with respective DNA-methyltransferases for specific DNA binding sites.
Table 2. Methylation degree of newly formed
DNA in L-cells and suspension culture of tobacco cells [71]
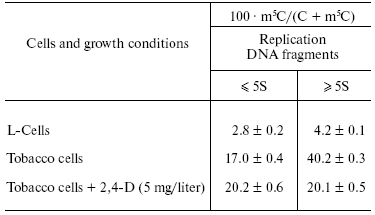
Replicative DNA methylation and inheritance of DNA methylation pattern. We have been interested to learn when and to what extent DNAs are methylated in the cell cycle in plants and animals. It is well known that synthesis of one of the DNA chains during replication of double-stranded DNA is continuous but other chain is synthesized discontinuously with formation of relatively short fragments that then are to be ligated. We decided to isolate these fragments and to learn whether they are methylated or not. Fortunately, we were glad to observe that DNA synthesis in plant and animal cells grown at their high concentration in a medium is mainly limited to formation and accumulation of these short DNA fragments as their ligation is strongly inhibited [69-74]. Therefore, it was a great opportunity to obtain these DNA fragments in sufficient amounts and investigate their methylation. In fact, they represented the Okazaki fragments that in chase-experiments were ligated with formation of normal long DNA chains. It turned out that Okazaki fragments in plants and animals are already methylated (Table 2).
Thus, the proper replicative DNA methylation in eukaryotes was discovered and well documented; besides, it was suggested that some DNA-methyltransferases can be constituents of the replicative complex [69-76]. Okazaki fragments differed in degree and site specificity of methylation from ligated replication intermediates and mature DNA (Table 2). Unlike methylation of ligated DNA products, methylation of Okazaki fragments was relatively resistant to various inhibitors of the methylation reaction (S-isobutyladenosine and others) and it was not inhibited by hormones (auxins in plants). We concluded that the nucleus should contain a few DNA-methyltransferases, each of them serving at a particular replication stage and differing from others in site specificity and sensitivity to respective inhibitors [69, 74]. This is now in a full agreement with modern data on the multiplicity of DNA-methyltransferases in plant and animal cell.
Then we were able to discriminate between replicative and postreplicative DNA methylations in plants [76-79]. These types of DNA methylation are different in site specificity and regulation by phytohormones and inhibitors [71-79].
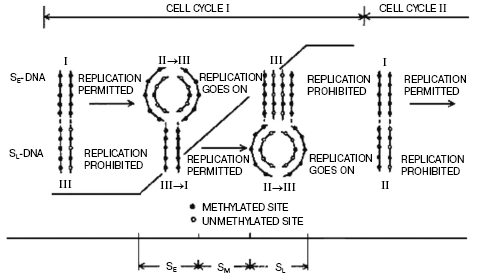
We have suggested a mechanism of regulation of DNA replication by methylation (Fig. 2). It has been established that during replication hemimethylated sites in plant DNA are formed, and the methylation pattern of newly synthesized DNA duplexes is asymmetric as daughter strand is essentially undermethylated compared with the parental one. We observed that this asymmetry of chain DNA methylation decreases up to the end of the cell cycle, and before a new replication round the hemimethylated sites become again fully methylated due mainly to maintenance DNA-methylation. This was proved by analysis of methylation degree of DNA isolated at different cell cycle stages with separation of individual chains by CsCl density gradient centrifugation. Replication of hemimethylated DNA in the eukaryotic cell seems to be prohibited as it will result in a loss of the epigenetic signal [71, 79]. Much later, it was clearly experimentally demonstrated that in bacteria this type of regulation of replication is realized by dam-methylation. Now it is more or less evident how this signal (methylation pattern) can be inherited. As already mentioned, replication produces hemimethylated sites in DNA formed, and most genes seem to be functional when DNA in an interphase nucleus is in this particular state. Before the next round of replication and cell division the maintenance DNA-methyltransferases, particularly dmt1 responsible for maintenance of genome methylation status, methylate hemimethylated sites with formation of fully methylated ones. m5C in one DNA chain serves as a signal for methylation of the opposite cytosine residue in a complementary chain. This stops gene transcription but permits replication when the cell cycle is completed (Fig. 2).
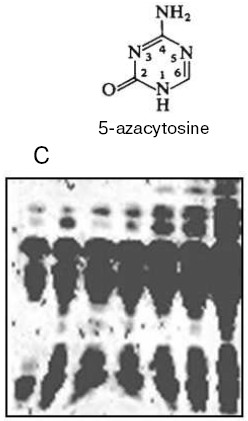
Thus, DNA methylation pattern is inheritable and this has been demonstrated in many experiments. For instance, treatment of wheat plants which are ready to flower with 5-azacytidine (a DNA methylation inhibitor) results in an increase in the seed storage protein content by more than 30% (Fig. 3) [80]. This is a fantastic event, which plant geneticists could only dream about. Unfortunately, this extraordinary seed quality induced is inherited for a few generations only. Usually the seed storage protein genes seem to be quite significantly repressed. As a result of plant treatment with 5-azacytidine, DNA becomes strongly demethylated and elevated expression of these genes in a forming seed takes place. Usually the amount of storage proteins in mature seed formed is enough for future plant development, and excess of these proteins is not needed. The excessive formation of storage proteins in seeds induced by 5-azacytidine can be very useful in biotechnology. DNA demethylation induced by 5-azacytidine also activates apoptosis strongly [81]. Sometimes distorted DNA methylation pattern seems to be inherited, then in one case it will be a good but in another case it will be a disaster associated with inheritable diseases and death. Nevertheless, the cell aspires always to recover the initial DNA methylation pattern, and there are many special delicate mechanisms that correct effectively the distorting attacks of fate and medium. Unfortunately, we still know very little about this wisdom of living cell.Fig. 2. Regulation of DNA replication in a cell cycle by DNA methylation [71].
Anyway, the skeptics mentioned above are prostrated, and today it is well known that DNA methylation in a cell is not a trifle but it controls all (!) genetic processes including transcription, replication, recombination, gene transposition, DNA repair, and X-chromosome inactivation. Therefore, this small DNA modification strongly attracts the interests of very many scientists in the modern world.Fig. 3. Increased storage protein content in wheat seeds induced by DNA-demethylating agent 5-azacytidine (C, control) [80].
Methylation of adenine residues in plant DNA. N6-Methyladenine was directly detected in DNA of eukaryotes a long time ago. It occurs in DNA of algae [82] and their viruses [83], fungi [84, 85], protists [86-91], and plants [92, 93]. Certainly, this could mean that in eukaryotes the genomes can be methylated on both cytosine and adenine residues. We have found N6-methyladenine in total DNA isolated from many higher plants [92], in mitochondrial DNA of wheat seedlings [94], and shown that, unlike in animals, the mitochondria in plants (wheat) possess adenine DNA-methyltransferases but not cytosine DNA-methyltransferases [95]. In plants a new class of heterogeneous in contour length minicircular heavy mitochondrial DNAs containing N6-methyladenine but not 5-methylcytosine was observed [96]. These DNA were found in all archegoniate and flowering plants investigated [97]; synthesis of these DNAs is most strongly pronounced in aging plant organs (for instance, aging coleoptiles in cereals) [96]. Intense replication of these mtDNAs is one of the specific apoptotic features in plants [98, 99].
By computer analysis of available databases we have established that the open reading frames (ORF) coding for proteins that possess a high degree of homology with prokaryotic DNA-(amino)methyltransferases are present in the genomes of Leishmania major, Saccharomyces cerevisiae, Schizosaccharomyces pombe, Arabidopsis thaliana, Drosophila melanogaster, Caenorhabditis elegans, and Homo sapiens [100]. Conservative motifs typical for bacterial DNA-(amino)methyltransferases have been detected in the amino acid sequences of these putative proteins. The ORFs of all putative DNA-(amino)methyltransferases found are encoded in nuclear DNA. In mitochondrial genomes including a few fully sequenced higher plant mtDNAs the nucleotide sequences significantly homologous to genes of prokaryotic DNA-(amino)methyltransferases were not found. Thus, ORFs homologous to bacterial adenine DNA-methyltransferases are present in nuclei of protozoa, yeasts, insects, nematodes, higher plants, vertebrates, and other eukaryotes [100]. A special search for corresponding proteins and, in particular, adenine DNA-methyltransferases in these evolutionarily distant organisms and a study of their functions are quite perspective.
We now have in our hands only one of these eukaryotic enzymes that modifies DNA on adenine residues. Being engaged in the study of apoptosis in plants, we first detected, isolated, and characterized the subcellular structures (vesicles) specific for apoptotic plant cells [99]. We were fortunate to isolate this enzyme exactly from these vesicles. The enzyme (wadmtase) methylates an internal adenine residue in the TGATCA sequence predominantly in a single-stranded DNA and seems to take part in regulation of mitochondrial DNA replication [101]. Unfortunately, up to now we do not know whether this enzyme modifies adenine residues in nuclear DNA as well or nuclear DNA is methylated by another adenine DNA-methyltransferase.
Anyhow, there is the impression that DNA methylation in plants is richer and more complex than in animals. Plants possess a larger set of DNA-methyltransferases, some of which are unique for these organisms and have a ubiquitin-binding domain [29, 30]. Unlike in animals, knockout of enzyme homologous to animal dmt1 is not lethal for plants and it seems to be compensated by other DNA-methyltransferases in plant cells. As one and the same plant gene can be methylated on both adenine and cytosine residues [102], a mutual control system for both of these genome modifications should exist in plants. It seems that adenine DNA methylation can influence cytosine DNA methylation and vice versa. For example, in Arabidopsis plants the methylation of adenine residues in GATC sites of cytosine DNA-methyltransferase gene DRM2 well pronounced in wild-type plants was strongly decreased in transgenic plants with expressed antisense construct of cytosine DNA-methyltransferase gene MET1 [102].
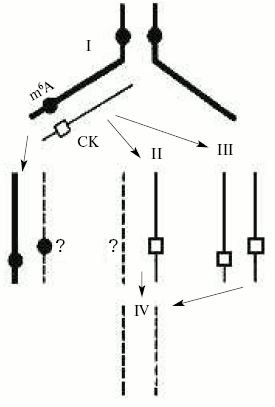
Unfortunately, the functional role of adenine DNA methylation in higher eukaryotes is still practically unknown. We suggest that, like cytosine methylation, it may control DNA replication and gene expression in eukaryotes. Besides, the control for adenine DNA methylation by cytokinins (N6-derivatives of adenine) as a result of their incorporation into DNA may be a mechanism of regulation of gene expression and cellular differentiation in plants (Fig. 4) [72]. In fact, it was recently established that the cytokinin 6-benzylaminopurine inhibits DNA methylation in sycamore plastid culture and induces there an expression of enzymes servicing photosynthesis [103]. We have evidence that phytohormone cytokinins (natural adenine derivatives) can be directly incorporated in DNA in plants [104] and the protist Tetrahymena pyriformis [105], organisms that have been shown to have N6-methyladenine in DNA or adenine DNA methylation enzymes. Natural incorporation of cytokinins substituting for adenine residues into sites recognized by adenine DNA-methyltransferases should prohibit adenine methylation in these particular sites. Thus, adenine undermethylated DNA may occur and this may result in a significant change in gene transcription and DNA replication.
DNA methylation and other epigenetic signals. In fact, the origin and materialization of a new science, epigenetics, are mainly due to observation and description of the exclusive role of DNA methylation in the life of various organisms. It is usually understood that epigenetics is the knowledge of total organism properties that are not encoded directly in the genome but they can and by definition should be hereditarily transmitted. Often epigenetics is even considered as an entire program of organism development. It should not be forgotten that organisms have many powerful regulatory elements in genomes or even entire cell regulatory systems that control gene expression depending on various abiotic factors and biosphere signals. These signals collide with genetics and they very often answer the crucial question--to be or not to be? In fact, even excellent genetics cannot be realized if epigenetics is wrong. According to Medawar's known expression, the “genetics proposes but epigenetics disposes”. For a long period epigenetics was not recognized, and very often it was modestly or even advisedly passed over in silence. Now it has become clear that one of the real epigenetic signals is an enzymatic modification (methylation) of the genetic template.Fig. 4. Possible mechanism of formation of unmethylated adenine sites due to cytokinin (CK) incorporation in DNA [72].
The way to decipher the nature and specificity and to understand the biological role of this enzymatic DNA modification (methylation) was very uneasy and we are glad to realize that our small landmarks on this hard way are also visible. I would like to stress that this research was started under the late Academician A. N. Belozersky and it is always associated with the name and memories of this open-hearted person and outstanding scientist who discovered DNA in plants and founded the nucleic acid school in Russia.
There are many other different epigenetic signals in a cell. Chromatin proteins rather than DNA are most significant in the realization of some of these particular signals. Today we can already speak of “histone code” [106]. This code enlarges the potential of the DNA genetic code. The chromatin structure changes due to various histone modifications, and this may lead to inheritable changes in gene transcription. The proper histone modifications ((de)methylation, (de)acetylation, (de)phosphorylation, and others) often predetermine gene activity. There are also very many special nonhistone regulatory proteins forming intricate DNA-protein complexes that trigger transcription or make genes silent. We are now on the doorstep to understanding these peculiarly complicated events.
There are many data showing that DNA methylation and histone modifications are closely connected. In Neurospora, the methylation of lysine 9 in H3 histone is critical for cytosine DNA methylation and normal development of the fungus [107]. In other words, histones can be the signal transmitters for genome methylation. On the other hand, in Arabidopsis plants CpG DNA methylation precedes and directs lysine 9 methylation in H3 histone [108]. In plants and animals, DNA methylation is associated with histone deacetylation [109-112]. For instance, the histone deacetylase gene is essential for small RNA (dsRNA)-directed DNA methylation [112, 113]. Indeed, “methylation meets acetylation” [114].
RNA-directed DNA methylation. Investigation of mechanisms and of biological role of RNA-directed DNA methylation (involved in specific gene silencing) is of special interest. In the presence of small signaling RNA the site-specific cytosine DNA-methyltransferases methylate de novo CG and other sites in DNA sequences recognized by small RNA; this methylation mobilizes respective enzymes that modify histones [115]. Histone modification in its own turn leads to induction and intensification of methylation in CNG sites that is maintained then without any RNA-trigger. In this chain of events the DNA methylation can be a cause or a consequence of gene silencing [115]. It is not surprising that significant progress in the study of the RNA-directed gene methylation was achieved due to investigation of DNA methylation in plants rather than animals. Perhaps this is due to the fact that particularly in plants the methylation of CNG and nonsymmetrical sites in DNA is well expressed [20], and these methylation types are mainly involved in RNA-directed genome methylation.
Thus, enzymatic DNA methylation is a constituent element of complex epigenetic control for practically all genetic functions of the cell. Speaking of this genome modification we have to realize that, in fact, we are dealing with at least three components of this complicated reaction or even system: they are the reaction substrate or DNA itself, the enzyme (DNA-methyltransferase), and the donor of methyl groups (S-adenosyl-L-methionine). It is clear that control for DNA modification and its effectiveness in the cell is performed at the level of each and all of these components and in addition other various representatives of cell metabolism may be also involved in control. Very often, even when the enzyme is active and the amount of S-adenosyl-L-methionine is sufficient, the reaction in a nucleus is impossible due to inaccessibility of DNA in a peculiar flexible chromatin structure. In this particular case, the structure of chromatin is most important for a reaction. In addition to many histone modifications modulating chromatin organization and DNA availability to enzymes, many other proteins can effectively compete with DNA-methyltransferases for DNA binding. In particular, various hormone-receptor complexes seem to perform this business. This could explain, at least in part, the mechanism of regulation of DNA methylation by hormones in plants and animals observed and the action of hormones in the cell. Anyway, further progress in the study of genome methylation in eukaryotes depends now mainly on research in the detail of fine chromatin structure and its various functional modulations in the nucleus.
In conclusion, I would like to thank cordially all my coworkers, numerous graduate and postgraduate students, who performed with me fruitful research in one of the most fascinating areas of modern molecular biology, as well as all colleagues and friends who shared with us here their knowledge and views on DNA methylation on the pages of this specialized issue of Biochemistry (Moscow). I am very sorry that the list of contributors to the field is quite far from complete, and I make my apologies to all of them whose work was not described or mentioned in this short review.
This work was partially supported by grants of the Russian Foundation for Basic Research (02-04-48005) and the Ministry of Industry, Science, and Technologies (NSH-1019.2003.4).
REFERENCES
1.Hotchkiss, R. D. (1948) J. Biol. Chem.,
175, 315-332.
2.Wyatt, G. R. (1950) Nature, 166,
237.
3.Dunn, D. B., and Smith, J. D. (1955) Nature,
175, 336-337.
4.Dunn, D. B., and Smith, J. D. (1958) Biochem.
J., 68, 627-636.
5.Gold, M., and Hurwitz, J. (1964) J. Biol.
Chem., 239, 3858-3865.
6.Mazin, A. L., and Vanyushin, B. F. (1988) Mol.
Biol. (Moscow), 22, 1399-1404.
7.Mazin, A. L., Mukhovatova, L. M., Shuppe, N. G.,
and Vanyushin, B. F. (1984) Dokl. Akad. Nauk SSSR, 276,
760-762.
8.Gowher, H., Leismann, O., and Jeltsch, A. (2000)
EMBO J., 19, 6918-6923.
9.Lyko, F., Ramsahoye, B. H., and Jaenisch, R. (2000)
Nature, 408, 538-540.
10.Vanyushin, B. F., Belyaeva, N. N., Kokurina, N.
A., Stelmashchyuk, V. Ya., and Tikhonenko, A. S. (1970) Mol.
Biol. (Moscow), 4, 724-729.
11.Ashapkin, V. V., Antoniv, T. T., and Vanyushin,
B. F. (1995) Gene, 157, 273-277.
12.Jaenisch, R., and Bird, A. (2003) Nature
Genetics, 33, 245-252.
13.Bird, A., and Wolffe, A. P. (1999) Cell,
99, 451-454.
14.Mazin, A. L., Gimadutdinov, O. A., Turkin, S. I.,
Burtseva, N. N., and Vanyushin, B. F. (1985) Mol. Biol.
(Moscow), 19, 903-914.
15.Vanyushin, B. F., and Dobritsa, A. P. (1975)
Biochim. Biophys. Acta, 407, 61-72.
16.Vanyushin, B. F., Buryanov, Ya. I., and
Belozersky, A. N. (1971) Nature, New Biology, 230,
25-27.
17.Vanyushin, B. F., Belozersky, A. N., Kokurina, N.
A., and Kadirova, D. X. (1968) Nature, 218,
1066-1067.
18.Dobritsa, S. V., Dobritza, A. P., and Vanyushin,
B. F. (1976) Dokl. Akad. Nauk SSSR, 227, 995-998.
19.Vanyushin, B. F., Kokurina, N. A., and
Belozersky, A. N. (1970) Dokl. Akad. Nauk SSSR, 193,
215-218.
20.Kirnos, M. D., Aleksandrushkina, N. I., and
Vanyushin, B. F. (1981) Biokhimiya, 46, 1458-1474.
21.Gruenbaum, Y., Naveh-Many, T., Cedar, H., and
Razin, A. (1981) Nature, 292, 860-862.
22.Salomon, R., and Kaye, A. M. (1970) Biochim.
Biophys. Acta, 204, 340-351.
23.Sneider, T. W. (1972) J. Biol. Chem.,
247, 2872-2875.
24.Woodcock, D. M., Crowther, P. J., and Diver,
W. P. (1987) Biochem. Biophys. Res. Commun., 145,
888-894.
25.Toth, M., Müller, U., and Doerfler, W.
(1990) J. Mol. Biol., 214, 673-683.
26.Marinitch, D. V., Vorobyev, I. A., Holmes, J. A.,
Zakharchenko, N. S., Dyachenko, O. V., Buryanov, Ya. I., and Shevchuk,
T. V. (2004) Biochemistry (Moscow), 69, 340-349.
27.Wassenegger, M., Heimes, S., Riedel, L., and
Sanger, H. L. (1994) Cell, 76, 567-576.
28.Wada, Y., Ohya, H., Yamaguchi, Y., Koizumi, N.,
and Sano, H. (2003) J. Biol. Chem., 278, 42386-42393.
29.Finnegan, E. J., and Kovac, K. A. (2000) Plant
Mol. Biol., 43, 189-210.
30.Finnegan, E. J., Peacock, W. J., and Dennis, E.
S. (2000) Curr. Opin. Genet. Devel., 10, 217-223.
31.Vanyushin, B. F., Tkacheva, S. G., and
Belozersky, A. N. (1970) Nature, 225, 948-949.
32.Vanyushin, B. F., and Belozersky, A. N. (1959)
Dokl. Akad. Nauk SSSR, 129, 944-946.
33.Drozhdenyuk, A. P., Sulimova, G. E., and
Vanyushin, B. F. (1977) Biokhimiya, 42, 1439-1444.
34.Cooney, C., and Lawren, B. (1999) Methyl
Magic, Andrews McMeel Publishing, Kansas City, USA.
35.Berdyshev, G. D., Korotaev, G. K., Boyarskikh, G.
V., and Vanyushin, B. F. (1967) Biokhimiya, 32,
988-993.
36.Zinkovskaya, G. G., Berdyshev, G. D., and
Vanyushin, B. F. (1978) Biokhimiya, 43, 1883-1892.
37.Romanov, G. A., Zinkovskaya, G. G., Berdyshev, G.
D., and Vanyushin, B. F. (1979) Biokhimiya, 44,
1576-1581.
38.Vanyushin, B. F., Zinkovskaya, G. G., and
Berdyshev, G. D. (1980) Mol. Biol. (Moscow), 14,
857-866.
39.Romanov, G. A., and Vanyushin, B. F. (1981)
Biochim. Biophys. Acta, 653, 204-218.
40.Vanyushin, B. F., Nemirovsky, L. E., Klimenko, V.
V., Vasiliev, V. K., and Belozersky, A. N. (1973) Gerontologia
(Basel), 19, 138-152.
41.Kudryashova, I. B., and Vanyushin, B. F. (1976)
Biokhimiya, 41, 1106-1115.
42.Vanyushin, B. F., and Romanenko, E. B. (1979)
Biokhimiya, 44, 78-85.
43.Vanyushin, B. F., Mazin, A. L., Vasiliev, V. K.,
and Belozersky, A. N. (1973) Biochim. Biophys. Acta, 299,
397-403.
44.Vanyushin, B. F., and Kirnos, M. D. (1974)
FEBS Lett., 39, 195-199.
45.Vanyushin, B. F., and Kirnos, M. D. (1977)
Biochim. Biophys. Acta, 475, 323-336.
46.Kudryashova, I. B., Kirnos, M. D., and Vanyushin,
B. F. (1976) Biokhimiya, 41, 1968-1977.
47.Kudryashova, I. B., and Vanyushin, B. F. (1976)
Biokhimiya, 41, 215-222.
48.Romanov, G. A., Kiryanov, G. I., Dvorkin, V. M.,
and Vanyushin, B. F. (1976) Biokhimiya, 41,
1038-1043.
49.Smirnov, V. G., Romanov, G. A., and Vanyushin, B.
F. (1977) Dokl. Akad. Nauk SSSR, 232, 961-963.
50.Vanyushin, B. F., Tushmalova, N. A., and Guskova,
L. V. (1974) Dokl. Akad. Nauk SSSR, 219, 742-744.
51.Vanyushin, B. F., Tushmalova, N. A., Guskova, L.
V., Demidkina, N. P., and Nikandrova, L. R. (1977) Mol. Biol.
(Moscow), 11, 181-187.
52.Guskova, L. V., Burtseva, N. N., Tushmalova, N.
A., and Vanyushin, B. F. (1977) Dokl. Akad. Nauk SSSR,
233, 993-996.
53.Sulimova, G. E., Drozhdenyuk, A. P., and
Vanyushin, B. F. (1978) Mol. Biol. (Moscow), 12,
496-504.
54.Khvojka, L., Sulimova, G. E., Bulgakov, R.,
Bashkite, E. A., and Vanyushin, B. F. (1978) Biokhimiya,
43, 996-1000.
55.Guseinov, V. A., and Vanyushin, B. F. (1975)
Biochim. Biophys. Acta, 395, 229-238.
56.Guseinov, V. A., Kiryanov, G. I., and Vanyushin,
B. F. (1975) Mol. Biol. Repts., 2, 59-63.
57.Burtseva, N. N., Azizov, Yu. M., Itkin, B. Z.,
and Vanyushin, B. F. (1977) Biokhimiya, 42,
1690-1696.
58.Burtseva, N. N., Demidkina, N. P., Azizov, Yu.
M., and Vanyushin, B. F. (1978) Biokhimiya, 43,
2082-2091.
59.Burtseva, N. N., Azizov, Yu. M., and Vanyushin,
B. F. (1979) Biokhimiya, 44, 1296-1302.
60.Burtseva, N. N., Romanov, G. A., Azizov,
Yu. M., and Vanyushin, B. F. (1979) Biokhimiya, 44,
2006-2072.
61.Burtseva, N. N., Romanov, G. A., Gimadutdinov, O.
A., and Vanyushin, B. F. (1983) Dokl. Akad. Nauk SSSR,
268, 1251-1255.
62.Burtseva, N. N., Gimadutdinov, O. A., and
Vanyushin, B. F. (1987) Biokhimiya, 52, 290-301.
63.Lopatina, N. G., Vanyushin, B. F., Cronin, G. M.,
and Poirier, L. A. (1998) Carcinogenesis, 19,
1777-1781.
64.Vanyushin, B. F., Lopatina, N. G., Wise, C. K.,
Fullerton, F. R., and Poirier, L. A. (1998) Eur. J. Biochem.,
256, 518-527.
65.Romanova, N. A., Romanov, G. A., Rozen, V. B.,
and Vanyushin, B. F. (1979) Biokhimiya, 44, 529-542.
66.Romanov, G. A., Romanova, N. A., Rozen, V. B.,
and Vanyushin, B. F. (1983) Biochem. Int., 6,
339-348.
67.Romanov, G. A., Kuzmenko, A. P., Dashkevich, V.
S., Salganik, R. I., and Vanyushin, B. F. (1984) FEBS Lett.,
165, 35-38.
68.Zhavoronkova, E. N., and Vanyushin, B. F. (1987)
Biokhimiya, 52, 870-877.
69.Bashkite, E. A., Kirnos, M. D., Kiryanov, G. I.,
Aleksandrushkina, N. I., and Vanyushin, B. F. (1980) Biokhimiya,
45, 1448-1456.
70.Vanyushin, B. F., Bashkite, E. A., and Khvojka,
L. A. (1981) Biokhimiya, 46, 47-54.
71.Vanyushin, B. F., and Kirnos, M. D. (1988)
Gene, 74, 117-121.
72.Vanyushin, B. F. (1984) Curr. Top. Microbiol.
Immunol., 108, 99-114.
73.Kiryanov, G. I., Kirnos, M. D., Demidkina, N. P.,
Alexandrushkina, N. I., and Vanyushin, B. F. (1980) FEBS Lett.,
112, 225-228.
74.Kiryanov, G. I., Isaeva, L. V., Kirnos, M. D.,
Ganicheva, N. I., and Vanyushin, B. F. (1982) Biokhimiya,
47, 153-161.
75.Kirnos, M. D., Aleksandrushkina, N. I., Kutueva,
L. I., and Vanyushin, B. F. (1988) Biokhimiya, 53,
1397-1406.
76.Kirnos, M. D., Aleksandrushkina, N. I., and
Vanyushin, B. F. (1988) Biokhimiya, 53, 1667-1678.
77.Kirnos, M. D., Artyukhovskaya, N. A.,
Aleksandrushkina, N. I., Ashapkin, V. V., and Vanyushin, B. F. (1986)
Biokhimiya, 51, 1875-1885.
78.Kirnos, M. D., Aleksandrushkina, N. I., Kutueva,
L. I., Artyukhovskaya, N. A., and Vanyushin, B. F. (1987)
Biokhimiya, 52, 625-637.
79.Kirnos, M. D., Aleksandrushkina, N. I., Kutueva,
L. I., and Vanyushin, B. F. (1988) Biokhimiya, 53,
355-367.
80.Vanyushin, B. F., Sevostyanova, S. S., Kirnos, M.
D., and Saidova, N. S. (1990) Izv. Akad. Nauk SSSR, Ser. Biol.,
1, 75-83.
81.Vanyushin, B. F., Shorning, B. Yu., Seredina, A.
V., and Aleksandrushkina, N. I. (2002) Fiziol. Rast., 49,
558-564.
82.Pakhomova, M. V., Zaitseva, G. N., and
Belozersky, A. N. (1968) Dokl. Akad. Nauk SSSR, 182,
712-714.
83.Nelson, M., Burbank, D. E., and van Etten, J. L.
(1998) Biol. Chem., 379, 423-428.
84.Buryanov, Ya. I., Il'in, A. V., and Skryabin, G.
K. (1970) Dokl. Akad. Nauk SSSR, 195, 728-730.
85.Rogers, S. D., Rogers, M. E., Saunders, G., and
Holt, G. (1986) Curr. Genet., 10, 557-560.
86.Gorovsky, M. A., Hattman, D., and Pleger, G. L.
(1973) J. Cell Biol., 56, 697-701.
87.Zaitseva, G. N., Kolesnikov, A. A., Yatsenko, I.
Ya., Kirnos, M. D., and Vanyushin, B. F. (1974) Dokl. Akad. Nauk
SSSR, 219, 243-245.
88.Kirnos, M. D., Merkulova, N. A., Borkhsenius, S.
N., and Vanyushin, B. F. (1980) Dokl. Akad. Nauk SSSR,
255, 225-227.
89.Pratt, K., and Hattman, S. (1981) Mol. Cell.
Biol., 1, 600-608.
90.Cummings, D. J., Tait, A., and Godard, J. M.
(1974) Biochim. Biophys. Acta, 374, 1-11.
91.Rae, P. M., and Spear, B. B. (1978) Proc.
Natl. Acad. Sci. USA, 75, 4992-4996.
92.Vanyushin, B. F., Kadyrova, D. Kh., Karimov, Kh.
Kh., and Belozersky, A. N. (1971) Biokhimiya, 36,
1251-1258.
93.Buryanov, Ya. I., Eroshina, N. V., Vagabova, L.
M., and Il'in, A. V. (1972) Dokl. Akad. Nauk SSSR, 206,
992-994.
94.Vanyushin, B. F., Alexandrushkina, N. I., and
Kirnos, M. D. (1988) FEBS Lett., 233, 397-399.
95.Kirnos, M. D., Aleksandrushkina, N. I., Shorning,
B. Yu., Bubenshchikova, S. N., and Vanyushin, B. F. (1997)
Biochemistry (Moscow), 62, 1348-1357.
96.Kirnos, M. D., Alexandrushkina, N. I.,
Zagorskaya, G. Ya., Kireev, I. I., and Vanyushin, B. F. (1992) FEBS
Lett., 298, 109-112.
97.Kirnos, M. D., Aleksandrushkina, N. I.,
Goremykin, V. V., Kudryashova, I. B., and Vanyushin, B. F. (1992)
Biokhimiya, 57, 1566-1573.
98.Kirnos, M. D., Bakeeva, L. E., Volkova, S. A.,
Ganicheva, N. I., and Vanyushin, B. F. (1983) Biokhimiya,
48, 1505-1512.
99.Bakeeva, L. E., Kirnos, M. D., Aleksandrushkina,
N. I., Kazimirchyuk, S. B., Shorning, B. Yu., Zamyatnina, V. A.,
Yaguzhinsky, L. S., and Vanyushin, B. F. (1999) FEBS Lett.,
457, 122-125.
100.Shorning, B. Yu., and Vanyushin, B. F. (2001)
Biochemistry (Moscow), 66, 753-762.
101.Fedoreyeva, L. I., and Vanyushin, B. F. (2002)
FEBS Lett., 514, 305-308.
102.Ashapkin, V. V., Kutueva, L. I., and Vanyushin,
B. F. (2002) FEBS Lett., 532, 367-372.
103.Ngernprasirtsiri, J., Kobayashi. H., and
Akazawa, T. (1988) Proc. Natl. Acad. Sci. USA, 85,
4750-4754.
104.Kudryashova, I. B., and Vanyushin, B. F. (1986)
Biokhimiya, 51, 321-327.
105.Mazin, A. L., and Vanyushin, B. F. (1986)
Izv. Akad. Nauk SSSR, Ser. Biol., 1, 122-124.
106.Jenuwein, T., and Allis, C. D. (2001)
Science, 293, 1074-1080.
107.Tamaru, H., and Selker, E. U. (2001)
Nature, 414, 277-283.
108.Soppe, W. J. J., Jasencakova, Z., Houben, A.,
Kakutani, T., Meister, A., Huang, M. S., Jacobsen, S. E., Schubert, I.,
and Fransz, P. F. (2002) EMBO J., 21, 6549-6559.
109.Nan, X., Ng, H. H., Johnson, C. A., Laherty, C.
D., Turner, B. M., Eisenman, R. N., and Bird, A. (1998) Nature,
393, 386-389.
110.Jones, P. L., Veenstra, G. J., Wade, P. A.,
Vermaak, D., Kass, S. U., Landsberger, N., Strouboulis, J., and Wolffe,
A. P. (1998) Nature Genetics, 19, 187-191.
111.Deplus, R., Brenner, C., Burgers, W. A.,
Putmans, P., Kouzarides, T., de Launoit, Y., and Fuks, F. (2002)
Nucleic Acids Res., 30, 3831-3838.
112.Aufsatz, W., Mette, M. F., van der Winden, J.,
Matzke, M., and Matzke, A. J. (2002) EMBO J., 21,
6832-6841.
113.Aufsatz, W., Mette, M. F., van der Winden, J.,
Matzke, A. J. M., and Matzke, M. (2002) Proc. Natl. Acad. Sci.
USA, 94, 11721-11725.
114.Bestor, T. H. (1998) Nature, 393,
311-312.
115.Matzke, M., Aufsatz, W., Kanno, T., Daxinger,
L., Papp, I., Mette, M. F., and Matzke, A. J. M. (2004) Biochim.
Biophys. Acta, 1677, 129-141.