alpha-Crystallin Promotes Assembly of a Trimeric Form of Mycobacterium tuberculosis Hsp16.3 in a Cell Free System
A. Abulimiti and Z. Chang*
Department of Biological Science and Biotechnology, School of Life Science, Tsinghua University, The Protein Science Laboratory of the Education Ministry, Beijing, 100084 P. R. China; fax: 86-10-6279-2995; E-mail: changzy@tsinghua.edu.cn* To whom correspondence should be addressed.
Received September 6, 2002
Hsp16.3, a small heat shock protein from Mycobacterium tuberculosis proposed to form specific trimer-of-trimers structures, acts as a molecular chaperone in vitro. The assembly mechanisms of this oligomeric protein were studied using in vitro transcription/translation systems. Analysis using a combination of non-denaturing pore gradient polyacrylamide gel electrophoresis and size-exclusion chromatography demonstrates that the predominant form of Hsp16.3 produced in the in vitro transcription/translation system is the trimer. Our result indicated that alpha-crystallin (molecular chaperone) remarkably promotes the trimer assembly of Hsp16.3, but does not convert the trimeric form to nonameric form. An “inert” Hsp16.3 dimer, which does not seem to participate in trimer assembly but may be involved in forming other forms of Hsp16.3, was also detected in the in vitro expression system. A latent phase of ~10 min in the appearance of the first detectable species indicated that Hsp16.3 assembly did not occur co-translationally.
KEY WORDS: small heat shock protein 16.3, alpha-crystallin, in vitro transcription/translation, protein assembly
Abbreviations: sHsps) small heat shock proteins; FPLC) fast protein liquid chromatography; BSA) bovine serum albumin.
Small heat shock proteins (sHsps) are a family of proteins that form
characteristic oligomeric structures having various numbers of subunits
[1]. Hsp16.3, originally identified as an
immunodominant antigen and later found to be a major membrane protein
[2, 3], is the small heat shock
protein from Mycobacterium tuberculosis. It is mainly
synthesized at stationary phase and strongly associated with the cell
wall thickening under low oxygen condition [4]. The
recombinant Hsp16.3 protein was found to exist as a nonamer, forming a
specific trimer-of-trimers structure [5]. This
oligomeric protein was revealed to be able to suppress the aggregation
of denatured proteins, thus having chaperone-like activity in vitro
[6-8]. Our previous
studies demonstrated that Hsp16.3 protein possess dynamic dissociation
and reassociation ability [9]. A stepwise model has
been proposed to account for the reassembly process of this protein
in vitro [6]. Whether such stepwise
reassembly mechanism can be extended to the assembly process of Hsp16.3
newly synthesized from the ribosomes is examined in the studies
reported here, using an in vitro transcription/translation
system. The advantage of using such in vitro expression system
to study protein folding and assembly mechanism of polypeptide chains,
although often more difficult to perform, is highly regarded [10, 11].
alpha-Crystallin functions as a molecular chaperone, a member of the sHsp family [12]. To judge how the chaperone-mediated protein assembly will be carried out in vivo, the effect of alpha-crystallin on Hsp16.3 assembly was examined. Our result demonstrated that the trimer is a predominant species in the in vitro transcription/translation system, alpha-crystallin likely converts “competent” dimer to trimer. No higher oligomers were observed. In addition to the trimers, a relatively “inert” dimer form was also repeatedly revealed when the Hsp16.3 protein was expressed using the in vitro transcription/translation system. Trypsin resistant reaction indicated the Hsp16.3 polypeptide assembly did not occur co-translationally.
MATERIALS AND METHODS
Materials. alpha-Crystallin and trypsin were purchased from Sigma (USA); 35S-labeled methionine (1000 Ci/mmol) was purchased from Amersham Corp. (USA). The GroEL/GroES proteins were kindly provided by Dr. Hai-Meng Zhou at the Department of Biological Science and Biotechnology, Tsinghua University (China). All other reagents were of analytical grade.
Plasmid construction. The construction of the expression plasmid vector for Hsp16.3 (pET-Hsp16.3) was described previously [5].
In vitro transcription and translation of Hsp16.3. Transcription and translation were performed using the in vitro E. coli T7 S30 extract system for circular DNA system (Promega, USA) according to the provider's instruction. Template DNA (being pET-Hsp16.3) of 1.8 µg was added per 50 µl reaction volume. Each protein expression reaction, having 10 µCi of 35S-labeled methionine, was performed at 37°C and was stopped by cooling on ice. The mixture was either immediately examined or stored at -20°C before use. alpha-Crystallin and other proteins were added to the reaction mixture before the in vitro transcription/translation reaction was initiated.
Electrophoresis and autoradiography. Cell-free translation products were analyzed using SDS-PAGE and non-denaturing pore gradient polyacrylamide gel electrophoresis. SDS-PAGE (15%) was performed as described with the protein components in the reaction mixture precipitated via acetone treatment, thus removing the free radioactive methionine and other wastes [13, 14]. Non-denaturing pore gradient polyacrylamide gel electrophoresis (5-20%) in a 125 × 100 × 1 mm gel was performed according to methods previously described (4°C, 150 V for 10 h) [9], with the following proteins used as molecular mass standards: soybean trypsin inhibitor (20 kD), chicken egg ovalbumin (43 kD), BSA monomer (67 kD), BSA dimer (135 kD), and BSA trimer (200 kD). Protein bands were visualized either by Coomassie Blue R-250 staining or by autoradiography (after the gel was dried). The radioactive protein bands were quantified by Phosphorimager analysis (Molecular Dynamics, USA).
Trichloroacetic acid (TCA) protein precipitation assay for amino acid incorporation. TCA precipitable radioactivity, representing the part that has been incorporated into newly synthesized proteins, was determined by spotting the sample aliquots onto Whatman GF/A glass fiber filters (USA) after NaOH hydrolysis. Radioactivity was measured using a 1209 RackBeta liquid scintillation counter (LKB, Sweden).
Detection of the compactness of protein conformation by proteolysis. In vitro transcription/translation reaction mixture that expresses the Hsp16.3 protein was acetone precipitated and then the pellet was subjected to trypsin (final concentration of 50 µg/ml) digestion at 37°C for 1 h. The reaction mixture was then applied to SDS-PAGE and autoradiography analysis.
Size-exclusion FPLC. Size-exclusion FPLC was performed using an AKTA explorer with an HR 10/30 Superdex 200 column (Pharmacia, Sweden). The translation mixture containing the newly synthesized Hsp16.3 protein (50 µl) was applied onto the column equilibrated with 50 mM sodium phosphate buffer, pH 7.0, containing 200 mM NaCl. Fractions of 0.5 ml were collected at 0.5 ml/min, with the radioactivity of each fraction counted after being precipitated with trichloroacetic acid. The column was calibrated with the following high molecular weight standards (Bio-Rad, USA): thyroglobulin, 670 kD; bovine gamma-globulin, 158 kD; chicken ovalbumin, 44 kD; equine myoglobin, 17 kD; vitamin B-12, 1.35 kD.
RESULTS
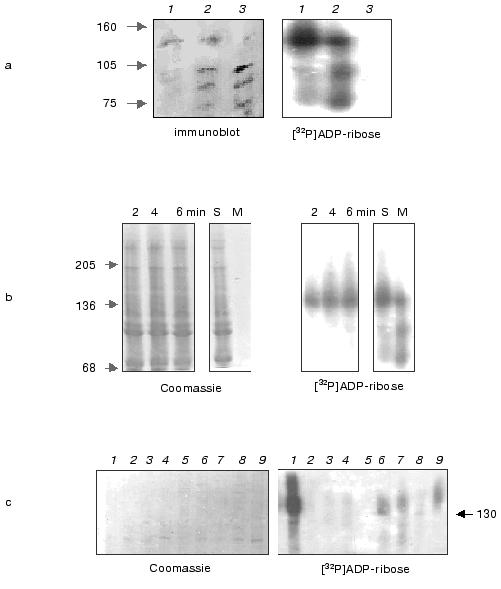
Hsp16.3 protein produced in the E. coli in vitro transcription/translation system exists mainly as trimers. The native size of Hsp16.3 protein produced in the E. coli cell-free system was first analyzed using non-denaturing pore gradient PAGE and size-exclusion chromatography. The protein, appearing after 1 min of the reaction (at 37°C) apparently existed mainly as trimers, with a small portion existing as dimers and monomers (Fig. 1a), as previously described [9]. The detection of only one single band of about 16 kD by SDS-PAGE (Fig. 1b) unequivocally demonstrated that all the protein bands (representing the Hsp16.3 forms having various sizes) detected by the non-denaturing pore gradient gel electrophoresis were indeed derived from Hsp16.3.
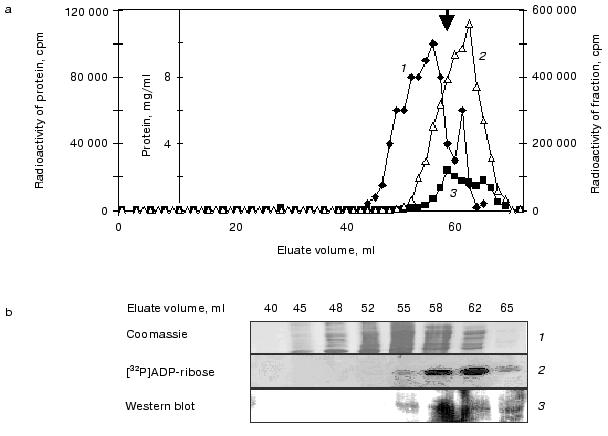
When the reaction mixture was analyzed by size-exclusion chromatography, three peaks of radioactivity, roughly corresponding to the sizes of trimers (~53 kD), dimers (~33 kD), and monomers (~17 kD) were detected (marked by the three bold arrows in Fig. 1c). In contrast to what was observed by the non-denaturing pore gradient gel electrophoresis, the trimer forms detected by size-exclusion FPLC are apparently present at a level lower than that of the dimers (Fig. 1c). It is likely that the dramatic dilution of the minute amount of Hsp16.3 via the size-exclusion chromatography have caused the dissociation of the trimer forms. Similarly, the nonameric form that was previously found to be the major form of purified Hsp16.3 [5], was not detected under these extremely diluting conditions.Fig. 1. Analysis of proteins produced from the pET-Hsp16.3 plasmid in the in vitro E. coli transcription/translation system. The nature of the proteins synthesized from pET-Hsp16.3 was analyzed by non-denaturing pore gradient polyacrylamide electrophoresis (5-20%) (a), SDS-PAGE (15%) (b), and size-exclusion chromatography (c). Panels (a) and (b) are the autoradiography results. Samples in lanes 1-5 in panels (a) and (b) correspond to the reaction time of 1, 10, 30, 60, and 120 min, respectively. Samples in lanes 6 in panels (a) and (b) represent the control reaction where no pET-Hsp16.3 construct was added. Marks on the right of panel (a) are the estimated oligomeric forms of Hsp16.3, and on the left are the molecular mass markers, respectively. Panel (c) was generated by plotting the radioactivity of each fraction versus its elution volume (2-h reaction mixture was applied for this size-exclusion chromatography analysis). The elution positions of the molecular weight standards are indicated on the top in panel (c).
The presence of alpha-crystallin significantly promotes trimer assembly of Hsp16.3. An intriguing question pertains to the mechanism by which the chaperone itself is folded and assembled into a functional oligomer. In order to understand the assembly mechanism of the Hsp16.3, as well as the fact that alpha-crystallin is able to act as a molecular chaperone, the following question was asked: would the purified alpha-crystallin be able to help the oligomeric forms (being predominantly trimers) of Hsp16.3 produced in the in vitro system to assemble into higher oligomeric forms? The data presented in Figs. 2a and 2b strongly suggest that alpha-crystallin had a significant effect on the assembly process of Hsp16.3 trimer. A careful examination of the autoradiographic bands revealed that the monomer forms disappeared almost completely and the dimeric forms decreased significantly in the presence of alpha-crystallin at high concentrations (curves 2 and 3 in Fig. 2b). Quantitative studies of the autoradiographic bands presented in Fig. 2 (via phosphorimager scanning) revealed that the density of the band corresponding to the trimeric form of Hsp16.3 increased significantly when alpha-crystallin was present at a high concentration (curve 1 in Fig. 2b). Furthermore, SDS-PAGE and quantitative measurement of the level of radioactivity in the precipitated proteins of the reaction mixture indicate that the addition of alpha-crystallin to the transcription/translation system seems to increase the total amount of Hsp16.3 protein produced (Fig. 2, c and d). The concentration of newly synthesized full-length Hsp16.3 in these experiments was estimated to be 2.0-2.5 ng/µl. Although alpha-crystallin was added in a large excess over newly synthesized polypeptide, the chaperone did not convert the trimer form to nonameric form. However, no such increased trimer was observed when alpha-crystallin was added after the transcription/translation reaction was stopped (data not shown). This suggested that the alpha-crystallin acted during (not after) the folding/assembly stage.
The effect of GroEL/GroES (a well-studied molecular chaperone) on the assembly of Hsp16.3 was also examined. The data (not presented here) indicate that trimers were still the predominant form present, no higher oligomers being observed. However, the total amount of Hsp16.3 protein synthesized was significantly decreased by the presence of GroEL/GroES. In addition, the production of Hsp16.3 protein in the transcription/translation system was almost completely inhibited when both GroEL/GroES and ATP were present (data not shown). Similar experiments on translation inhibition of luciferase by chaperones (GroEL/GroES) were carried out with E. coli S30 cell free translation system [15]. These results were somehow opposite to our expectation, considering that GroEL/GroES is supposed to aid the protein folding process. These results also ruled out the possibility that the absence of oligomers higher than trimer is due to the scarcity of GroEL/GroES in the in vitro transcription/translation system.Fig. 2. The Hsp16.3 trimer significantly increased in the in vitro transcription/translation system in the presence of alpha-crystallin. Shown in panels (a) and (c) are the autoradiographs of non-denaturing pore gradient polyacrylamide gel electrophoresis (5-20%) and SDS-PAGE (15%) analysis. Lanes 1-5 in panels (a) and (c) correspond to expression reactions in the presence of alpha-crystallin at the final concentrations of 0, 0.5, 2, 4, and 8 mg/ml, respectively. Marks on the right of panel (a) are the estimated oligomeric forms of Hsp16.3, and on the left are the molecular mass markers, respectively. Panel (b) is the kinetics of phosphorimager scan of the species detected after native-pore gradient PAGE in the presence of alpha-crystallin at the final concentrations of 0, 0.5, 2, 4, and 8 mg/ml, respectively (curves 1-3 correspond to trimer, dimer, and monomer, respectively). Panel (d) is the kinetics of amino acid incorporation of Hsp16.3 in cell free system in the presence of alpha-crystallin at the final concentrations of 0, 0.5, 2, 4, and 8 mg/ml, respectively.
Trypsin-resistant Hsp16.3 could be detected in significant amounts in the in vitro translation reaction after a latent phase of 10 min. It is well known that the native structure of a protein is much less protease labile than is the same protein when fully unfolded. This may be used to monitor the process of folding of polypeptides co- and post-translationally [10]. In vitro assembled Hsp16.3 was thus assayed by the resistance of 16.3 kD polypeptide band to trypsin digestion. The intensive bands in lanes 1-6 of Fig. 3 correspond to the intact radiolabeled Hsp16.3 expressed for 1, 10, 30, 60, 120, and 300 min, respectively. Conversely, no band of trypsin-resistant Hsp16.3 could be detected in significant amounts before 10 min (lane 1´ of Fig. 3). The radioactivity in the digested Hsp16.3 polypeptide band could only be determined after 10 min in vitro translation extract and trypsin-resistant Hsp16.3 band was found to represent 2% of the total Hsp16.3 polypeptides synthesized at 10 min (lane 2´ of Fig. 3, Fig. 4). This proportion increased gradually after 10 min from 10% at 30 min to a maximum of ~25% at 5 h (lane 6´ of Fig. 3, Fig. 4). Interestingly, a minor band, with size slightly smaller than that of the complete Hsp16.3, was consistently observed in all digested samples (lanes 1´ -6´, Fig. 3).
Fig. 3. Trypsin-resistance assay for Hsp16.3 synthesized in vitro translation system. Shown is the autoradiograph of the SDS-PAGE (15%) result of the reaction mixture after a 1-h trypsin digestion (with the final concentration of 50 µg/ml). Samples in lanes 1-6 correspond to the reaction time of 1, 10, 30, 60, 120, and 300 min, respectively. Lanes 1´-6´ represent the digested samples as examined in lanes 1-6.
Fig. 4. Kinetic analysis of in vitro translation Hsp16.3 assembly. Samples of in vitro translation reaction were analyzed by standard denaturing SDS-PAGE, and the radioactivity in the 16.3 kD band estimated before and after trypsin digestion. Radioactively labeled 16.3 kD protein was quantified by phosphorimager scan of autoradiograms such as presented in Fig. 3. The curve was generated by plotting the proportion of treated and untreated band intensity of each 16.3 kD band versus their time of in vitro translation reaction.
DISCUSSION
There is considerable interest in the study of Hsp16.3 polypeptide assembly in the in vitro transcription/translation system. Our principle discoveries include the following. First, non-denaturing pore gradient polyacrylamide gel electrophoresis and size-exclusion chromatography (data presented in Fig. 1) all suggest that trimer is the predominant form of Hsp16.3 for the Hsp16.3 proteins synthesized in the in vitro transcription/translation system. Second, the presence of alpha-crystallin has significant effect on trimer assembly of Hsp16.3, but does not convert the trimeric form to nonameric form. Third, a remarkably stable dimer form (Fig. 2) of Hsp16.3, which does not seem to participate in the formation of the trimer, was consistently detected from the in vitro transcription/translation products. Fourth, Hsp16.3 polypeptide assembly occurs post-translationally.
The nonameric form was found to be the major form of purified Hsp16.3 [5]. However, the nonameric form was not detected under our experiment conditions. We propose that the presence of trimeric form under our experiment conditions is due to the dynamic dissociation and reassociation property of Hsp16.3, as previously described [9]. It is likely that the minute amounts of Hsp16.3 facilitate the dissociation of nonameric forms to trimeric form rather than its reassociation.
Formation of an “inert” dimer is also proposed to account for the consistent appearance of the dimer band and its “inertness” towards Hsp16.3 trimer assembly under our experimental conditions (Fig. 2). The slight decrease in the band density of the dimeric form when higher concentration of alpha-crystallin was present (Fig. 2) may reflect co-existence of a “competent” dimer. The “competent” dimer may be further assembled to trimer, apparently via a certain conformation change before being converted to the trimer form. We propose that the conformational change of the “competent dimer” is due to the chaperone activity of alpha-crystallin, which might stabilize the folding intermediates and promote the Hsp16.3 assembly. However, as a result of dynamic dissociation and reassociation reaction [9], the increasing concentration of Hsp16.3 trimer in the presence of alpha-crystallin was not sufficient for further assembly of Hsp16.3 nonamer. Complete oligomerization of Hsp16.3 nonameric form in the cell free system was carried out in the presence of exogenous Hsp16.3 via trimer and hexamer intermediates and similar “inert” dimer was also observed by our recent study [9, 16]. A dimeric form of Hsp16.3 was also observed when the protein was examined by cryoelectron microscopy in the presence of certain type of lipid bilayers (Yong Chen et al., Tsinghua University, unpublished data). This “inert” form of dimer may participate in the formation of a layer of Hsp16.3 proteins that is formed on the outside surface of the Mycobacterium tuberculosis cells or the fiber structure formed in the cytoplasm under oxidative stress conditions [4].
The rate of assembly of the Hsp16.3 polypeptides was relatively slow, compared with the rate of synthesis, and the assembly process relatively inefficient in vitro; the amount of Hsp16.3 polypeptide assembled under our conditions never exceeded 25% of the total Hsp16.3 polypeptide chains synthesized (Fig. 4). A significant time lag of 10 min between the synthesis of Hsp16.3 polypeptides in vitro and the detectable assembled polypeptides suggested that Hsp16.3 polypeptide assembly did not occur co-translationally. In other words, homo-oligomeric proteins assemble after the synthesis of the whole polypeptide chain (i.e., post-translationally) and assemble spontaneously. Of course, one can argue that such assembly mechanism revealed by using the in vitro transcription/translation system still does not represent what actually happens inside the living cells.
To summarize, our studies of the assembly of Hsp16.3 protein using the in vitro transcription/translation system lead to the following conclusions: alpha-crystallin (molecular chaperone) remarkably promotes trimeric assembly of Hsp16.3, but does not convert the trimeric form to nonameric form. An “inert” Hsp16.3 dimer, which does not seem to participate in trimer assembly, was also detected in the in vitro expression system. The Hsp16.3 assembly did not occur co-translationally.
This work was supported by grants from the National Key Basic Research Foundation of China (No. G1999075607), National Science Foundation for Outstanding Young Scientists in China (No. G39725008) (to Dr. Zengyi Chang) and sponsored by Chinese Postdoctoral Foundation (to Dr. Abuduaini Abulimiti). We would like to thank Dr. Duanqing Pei at the Department of Pharmacology, School of Medicine, University of Minnesota, USA, and Dr. Wangwang Jiao at the Department of Biological Science and Biotechnology, Tsinghua University for helpful discussions.
REFERENCES
1.De Jong, W. W., Caspers, G., and Leunissen, J. A.
(1998) Int. J. Biol. Macromol., 22, 151-162.
2.Kingston, A. E., Salgame, P. R., Mitchison, N. A.,
and Colston, M. J. (1987) Infect. Immun., 55,
3149-3154.
3.Lee, B. Y., Hefta, S. A., and Brennan, P. J. (1992)
Infect. Immun., 60, 2066-2074.
4.Cunningham, A. F., and Spreadbury, C. L. (1998)
J. Bacteriol., 180, 801-808.
5.Chang, Z., Primm, T. P., Jakana, J., Lee, I. H.,
Chiu, W., Gilbert, H. F., and Quicho, F. A. (1996) J. Biol.
Chem., 271, 7218-7223.
6.Feng, X., Huang, S., Fu, X., Abulimiti, A., and
Chang, Z. (2002) Biochem. J., 363, 329-334.
7.Mao, Q., Ke, D., Feng, X., and Chang, Z. (2001)
Biochem. Biophys. Res. Commun., 284, 942-947.
8.Yang, H., Huang, S., Dai, H., Gong, Y., Zheng, C.,
and Chang, Z. (1999) Protein Sci., 8, 174-179.
9.Gu, L. C., Abulimiti, A., Li, W., and Chang, Z.
(2002) J. Mol. Biol., 319, 517-526.
10.Federov, A. N., and Baldwin, T. O. (1998)
Meth. Enzymol., 290, 1-17.
11.Jermutus, L. A., Ryabova, L., and Pluckthun, A.
(1998) Curr. Opin. Biotechnol., 9, 534-548.
12.Horwitz, J. (1992) Proc. Natl. Acad. Sci.
USA, 89, 10449-10453.
13.Ausubel, M. F., Brent, R., Kingston, E. R.,
Moore, D. D., Sedman, J. G., Smith, A. J., and Struhl, K. (1995)
Current Protocols in Molecular Biology, 3rd ed., John Wiley and
Sons, Inc., New York, pp. 10.2.18-10.2.21.
14.Lesley, S. A. (1995) Methods in Molecular
Biology, Vol. 37, Humana Press Inc., Totowa, pp. 265-278.
15.Vyacheslav, A. K., Eugeny, V. M., and Alexander,
S. S. (2000) J. Biol. Chem., 275, 16597-16601.
16.Abulimiti, A., Fu, X., Gu, L., Feng, X., and
Chang, Z. (2003) J. Mol. Biol., 326, 1013-1023.