Ordered Biochemical Program of Gene Expression in Cancer Cells
G. Weber
Laboratory for Experimental Oncology, Indiana University School of Medicine, 699 West Drive, Indianapolis, IN 46202-5119, USA; fax: 317-274-3939; E-mail: gweber1@iupui.edu
Received April 10, 2001
Our introduction of the molecular correlation concept and the key enzyme concept and the use of biologically meaningful tumor models and control systems resulted in the discovery of an ordered pattern of enzymic and metabolic imbalance and the elucidation of the linkage with transformation and progression. We showed that the biochemical and enzymic pattern of alterations was the result of a reprogramming of gene expression that was both quantitative and qualitative and was characteristic to neoplasia, since no similar pattern of imbalance was observed in any of the control normal, regenerating, or differentiating tissues. Important aspects of gene logic were identified. These include demonstration of operation of reciprocal control of activities of opposing key enzymes and antagonistic pathways of synthesis and catabolism in pyrimidine, purine, ornithine, and carbohydrate metabolism and recently in signal transduction. The extent of increase in the activities of key enzymes of pyrimidine and purine biosynthesis related to the absolute activity of the enzymes in resting liver. The qualitative alterations in gene expression included the isozyme shift of key regulatory enzymes. We identified a segment of gene expression that is essential for neoplasia. We pointed out the selective advantages that reprogramming of gene expression confers to cancer cells. Understanding these alterations in the enzymology and biochemistry of cancer cells made it possible to identify potentially sensitive targets for anticancer chemotherapy. In recent clinical studies we targeted the increased IMP dehydrogenase activity in leukemic blast cells by an inhibitor drug, tiazofurin, and achieved 77% responses, including complete remissions.
KEY WORDS: molecular correlation concept, key enzyme concept, biochemical strategy of the genome, spectrum of tumors of graded malignancy, isozyme pattern, specificity of neoplastic pattern, carbohydrate metabolism, key enzymes, pyrimidine metabolism--imbalance in cancer cells, reprogramming of gene expression in cancer cells--evidence, purine metabolism--imbalance in cancer cells, IMP dehydrogenase as target of tiazofurin treatment, leukemia treatment by an inhibitor of IMP dehydrogenase, signal transduction
I am contributing this paper in honor of Academician Prof. Dr. Sergei E. Severin whom I greatly admired. I met Dr. Severin in Leningrad at a meeting on carbohydrate metabolism where I was invited by Dr. V. S. Ilyin of Leningrad. During the course of the meeting Dr. Severin invited me to the subsequent meeting on similar topics which he was organizing for the next year in Moscow. We sat together several times at meals and Dr. Severin mentioned that there was interest in my “Advances in Enzyme Regulation” Symposia. As President of the All-Union Biochemical Society he hoped it might be possible for some scientists from the Soviet Union to participate. I assured him that I reciprocated his interest and that I would send invitations first to my hosts at the Leningrad Meeting, Drs. Ilyin and Shapot, and for the next meeting I would send invitations directly to him. The next year I participated in the Conference with Dr. Severin in Moscow. I also visited his apartment and his dacha on his invitation. There I met his son Eugene, his daughter Irene, and other family members. We discussed the arrangements to satisfy the bureaucracies and a cooperation was started which lasted over nineteen years.
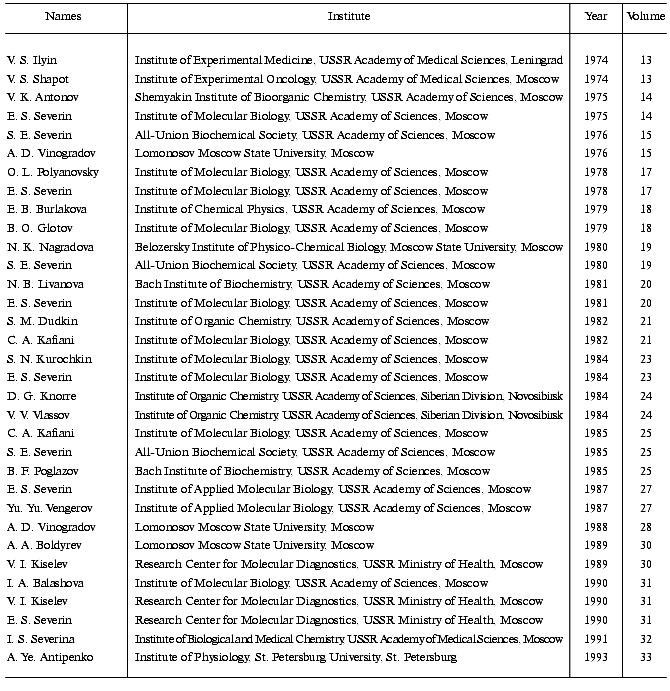
During the course of the collaboration 33 scientists from the Soviet Union/Russia participated in the “Advances” Symposia and presented excellent studies (table).
Thirty-three Soviet/Russian Participants
I was delighted that we were able to host three times, in Indianapolis, Dr. Severin [1-3] whose work and personality I respect very much. I was not surprised that the All-Union Biochemical Society kept him on as President far beyond his retirement age because he simply could not be replaced.
To honor Dr. Severin I have assembled an overview of part of my cancer biochemistry programs at Indiana University in a format that might be useful to those interested in the biochemistry and gene expression in cancer cells.
MATERIALS AND METHODS
The technical aspects of these studies are cited in the “References”.
RESULTS AND DISCUSSION
The purpose of the program of the Laboratory for Experimental Oncology is to discover strategic differences in the biochemistry of cancer cells in comparison with normal cells in order to design selective anticancer chemotherapy. We attempted to discover the pattern in the gene expression of cancer cells that should provide a rational design of chemotherapy directed against key enzymes.
In 1961 I introduced the ideas of the molecular correlation concept as a theoretical and experimental method for discovering the pattern of biochemical imbalance and its linking with neoplastic transformation and progression [4]. This approach provided precise testable predictions for anticipated alterations in the enzymic and metabolic pattern of cancer cells [5, 6]. Three main propositions were made.
1. The biochemical strategy of the genome in neoplasia can be identified by elucidating the pattern of gene expression as revealed in the activity, concentration, and isozyme program of key enzymes.
2. Activities of key enzymes and metabolic pathways and concentrations of strategic metabolites are stringently linked with neoplastic transformation and progression; those not stringently linked do not yield a pattern.
3. The linking of enzymic and metabolic imbalance with transformation and progression can be identified in a spectrum of tumors of graded malignancy, proliferative rate, and degree of differentiation.
Operational advantages of the molecular correlation concept: a unifying approach in cancer biochemistry. The operational advantage of the molecular correlation concept is its testability; it provided predictions that could be verified or disproved. That a theory must be testable to be useful, and thus susceptible to verification or refutation, is in line with views expressed in other research areas by Popper, Burnet and Medawar [7-9]. With the molecular correlation concept we determined that the enzymic and metabolic pattern of cancer cells has both ordered and random components [5, 6].
Gene logic in neoplasia. a) Reciprocal regulation of the activities of antagonistic key enzymes and overall metabolic pathways of synthesis and degradation amplifies the quantitative changes in the enzymic and metabolic capacity. b) The extent of rise in key enzymic activities and nucleotide concentrations is a function of their levels in resting normal liver. The lower the activities and concentrations in normal liver, the greater is the increase in the rapidly proliferating hepatomas. c) In the isozyme shift, the replacement of the liver-type isozymes that respond to nutritional and hormonal regulation by non-responsive, low-Km non-liver type isozymes is characteristic of cancer cells and the shift is linked with neoplastic progression [5, 6].
Thus, the reprogramming of gene expression in neoplasia is reflected in both quantitative and qualitative alterations. “Reprogramming” of gene expression is the appropriate term because the essential neoplasia-linked part of the program is displayed in an ordered, meaningful, and integrated enzymic and metabolic imbalance.
THE KEY ENZYME CONCEPT
The concept of key enzymes was developed in this laboratory along with methods for identifying such enzymes. Earlier studies by others on tumor metabolism failed to detect a meaningful pattern because, in part at least, of the ill-advised choice of enzymes and other biochemical variables.
Our observations and conceptual advances in studying carbohydrate metabolism led to the recognition that not all enzymes and biochemical parameters should be expected to be stringently linked with transformation and progression. The reason for this is that the integration of gene expression operates through the stringent control of a relatively small number of enzymes. I termed these the key enzymes because the regulation of the rate and direction of the flux of opposing and competing synthetic and catabolic pathways is achieved through their control [5, 6].
The term “pace maker” or “rate-limiting enzyme” is a narrower one, identifying an enzyme that has the lowest activity in a sequence of metabolic steps in one pathway. The key enzymes include the rate-limiting ones. The chief criteria for recognizing key enzymes include their biological role, place in the pathway, and regulatory properties. The key enzymes usually have relatively low activity, govern one-way metabolic reactions, and frequently are the first or the last in reaction sequences. They may be pathways in themselves or operate on both sides of reversible reactions with a one-way enzyme opposed by another one-way enzyme. Key enzymes are frequently the targets of feed-back or multiple regulation, may exhibit allosteric properties or are interconvertible enzymes and may have an isozyme pattern. In normal cells the activities and amounts of key enzymes are frequently subject to nutritional and hormonal regulation [5, 6].
In cancer cells the behavior of activities, amounts, and isozyme pattern of key enzymes is stringently linked with transformation and progression. Examples include: in carbohydrate metabolism: glucokinase-hexokinase, 6-phosphofructokine, pyruvate kinase, glucose-6-phosphatase, glucose-6-phosphate dehydrogenase; in pyrimidine metabolism: carbamoylphosphate synthetase II, CTP synthetase, ribonucleotide reductase, DNA polymerase, dihydrothymine dehydrogenase; in purine metabolism: amidophosphoribosyltransferase, xanthine oxidase, IMP dehydrogenase [5, 6].
The non-key enzymes. These are enzymes that apparently do not become limiting in the metabolic flux. Such enzymes are usually characterized by the following properties: they catalyze reversible equilibrium reactions and are shared by both synthetic and catabolic pathways; they exhibit high activities; and they are present in excess. The activities are usually not markedly altered by nutritional and endocrine regulation and do not correlate with transformation or progression in neoplasia. Examples include: in pyrimidine metabolism: dihydroorotate oxidase; in purine metabolism: GMP kinase; and in carbohydrate metabolism: phosphohexoseisomerase, lactate dehydrogenase, 6-phosphogluconate dehydrogenase [5, 6].
BIOLOGICAL MODEL SYSTEMS
We first reported evidence for the absence of an enzymic activity during hepatic carcinogenesis in observing that the activity of liver glucose-6-phosphatase (G6Pase) steadily decreased and was absent in the resulting primary hepatocellular carcinomas [4]. These results were rapidly confirmed in other laboratories. In 1957 we were the first to show the absence of an enzyme activity in transplantable cancer cells when we demonstrated that G6Pase activity was absent in the chemically-induced, transplantable, rapidly-growing Novikoff hepatoma [10]. There was an abstract by other workers in 1959 that in the transplantable Dunning hepatoma there was some G6Pase activity. Since the Dunning had a slower growth rate and was better differentiated than the Novikoff hepatoma it seemed to me that the different growth rates and the malignancy of tumors should be manifested also in gradations in the expression of clinical and biochemical properties. There was a need for hepatomas of different malignancy. Such tumors became available in collaboration from Dr. Harold P. Morris at the National Cancer Institute. From these I assembled a spectrum of hepatocellular carcinomas and tested the molecular correlation concept in these hepatomas [5, 6].
Quantitation of tumor malignancy. The rate of proliferation of these hepatomas was quantitated by several independent techniques: biological (tumor size and volume); cytological (mitotic count); histological (degree of differentiation); and biochemical (thymidine index). The malignancy of the tumor lines was ranked also by the time required in weeks for the neoplasms to reach a uniform diameter of 1.5 cm [5, 6]. All the methods yielded the same ranking order for the tumors. Most of these chemically induced transplantable solid tumor lines showed remarkable stability which indicated a stable gene pool and permitted good reproducibility of the studies in many laboratories. The mathematical relations between tumor proliferative rate and the extent of alterations in biochemistry are measured by correlation coefficients.
Specificity. Determination of specificity of the biochemical pattern to the neoplastic liver was made feasible by the ready availability of appropriate controls: normal, resting liver; regenerating liver; and differentiating, developing normal liver.
Generalization. To determine whether the biochemical pattern of enzymatic and metabolic imbalance originally discovered in the chemically-induced, transplantable hepatomas of the rat does or does not apply to other neoplasms in animals and in humans, a series of experimental neoplasms in animals and primary tumors in human was examined.
1. The kidney spectrum. On my request Dr. Morris produced a spectrum of kidney tumors and in a series of papers we elucidated the enzymology, nucleotide content and metabolism of these renal cell carcinomas.
2. Colon carcinoma and muscle sarcoma. The applicability of the biochemical imbalance discovered in hepatomas was also tested in chemically-induced, transplantable muscle sarcomas and in a series of chemically-induced, transplantable colon tumors in mouse and rat.
3. Human neoplasms. The presence of biochemical alterations observed in animal tumors was investigated with an emphasis on clinical samples of human primary hepatocellular carcinomas, renal cell carcinomas, colon carcinomas and lung neoplasia and in leukemic cells. Xenografts of colon carcinomas of different proliferative rates were also studied.
CARBOHYDRATE METABOLISM--IMBALANCE IN CANCER CELLS
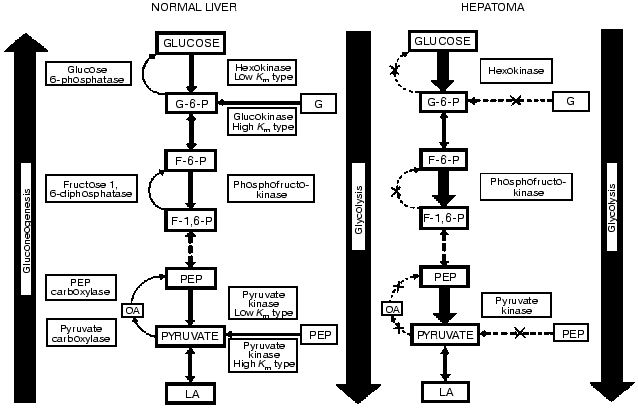
Liver and hepatomas are freely permeable to glucose; thus, the activity and the concentration of competing enzyme systems (in the presence of co-factors) determine the fate of the hexose. The degradative pathway of glucose (glycolysis) is opposed by the synthetic pathway of glucose production (gluconeogenesis) (Fig. 1). The three key enzymes of glycolysis (hexokinase-glucokinase, 6-phosphofructokinase, and pyruvate kinase) are opposed by the four key gluconeogenic enzymes (glucose-6-phosphatase, fructose-1,6-bisphosphatase, phosphoenolpyruvate carboxykinase, and pyruvate carboxylase). The rate and direction of the opposing pathways of glucogenesis and glycolysis are determined by control of the amount and activity of the triad of key glycolytic enzymes and the quartet of opposing key glycogenic enzymes.
Quantitative alterations. Early studies by Warburg demonstrated that in rapidly-growing, dedifferentiated tumors, which were the only ones available to him, aerobic glycolysis was high and respiration was low as compared to the various normal tissues he examined [11]. However, work carried out in the hepatoma spectrum where tumors were subcutaneously transplanted in inbred rats yielded different results. In these rats slow-growing and well-differentiated liver tumors could be transplanted and we were first to report that in these tumors aerobic glycolysis was not increased [4]. This was independently confirmed by others [12].Fig. 1. Biochemical strategy of cancer cells as revealed in the integrated reprogramming of gene expression manifested in the quantitative and qualitative imbalance in carbohydrate metabolism. The quantitative alterations entailed the decrease in the activities of the key gluconeogenic enzymes and the increase in those of the opposing key glycolytic ones. The qualitative change is the isozyme shift manifested in the loss of the high Km enzyme and the emergence of the low Km isozymes. The increase in enzyme concentration is denoted by thick arrows, and the decrease by interrupted arrows. The X in the arrows denotes the absence of those enzymes in the very rapidly growing hepatomas. Behavior of the non-key enzymes does not correlate with malignancy (e.g., phosphohexoseisomerase and lactate dehydrogenase). Further discussion is given in the text and references cited.
We determined that there are both ordered and random changes in the carbohydrate enzymology of the cancer cells. The activities of the key enzymes of the opposing pathways display a pattern of ordered alterations linked with the growth rates of the tumors. By contrast, activities of the non-key enzymes are present in excess (e.g., phosphohexoseisomerase, aldolase and lactate dehydrogenase) and exhibit random alterations, as expected. The following progression-linked alterations occur in the hepatoma spectrum. In parallel with the rise in tumor growth rates there is an increase in the activities of the key glycolytic enzymes and in aerobic glycolysis (conversion of glucose to lactate); a decrease in activities of the key glucogenic enzymes and in the pathway of gluconeogenesis (conversion of pyruvate to glucose).
Qualitative alterations. We discovered a pattern of isozyme shift which is expressed in the replacement of the liver type, high-Km isozymes (glucokinase, 6-phosphofructokinase, and pyruvate kinase) responsive to nutritional and hormonal regulation by non hepatic type, low-Km isozymes that are much less, or not at all, responsive to nutritional and endocrine regulation and feed-back controls.
As a result of the reprogramming of gene expression in cancer cells there is a biochemical imbalance with the predominance of glycolysis over glucogenesis. The imbalance in cancer cells entails both quantitative and qualitative changes. These alterations in gene readout are linked with the increased expression of malignant properties. This imbalance in carbohydrate metabolism should confer selective advantages to cancer cells [5, 6].
Further studies on the direct oxidative pathway showed a marked increase in the activity of the rate-limiting, one-way enzyme, G6P dehydrogenase, which routes G6P into this pathway. There is also an increase in the activity of transaldolase which channels fructose-6-phosphate to ribose-5-phosphate (R-5-P) production. R-5-P, this vital precursor for the biosynthesis of RNA and DNA, is generated by both direct oxidative and non-oxidative pathways. In tumor cells the channeling of the products of glucose degradation is particularly important. In addition, R-5-P is the precursor of phosphoribosylpyrophosphate (PRPP) which is an indispensable substrate for both de novo and salvage pathways of purine and pyrimidine biosynthesis [5].
The link between R-5-P and purine and pyrimidine metabolism is forged by PRPP synthetase, which uses R-5-P for production of PRPP. This enzymatic activity is increased in parallel with the growth rate and malignancy of the tumors [5].
PYRIMIDINE METABOLISM--IMBALANCE IN CANCER CELLS
The main diagnostic signs of the biological behavior of cancer cells are the commitment to proliferation and the altered control of cellular replication. Our results revealed that in cancer cells there is a clear-cut imbalance in pyrimidine and DNA metabolism which is stringently linked with transformation and progression. The biochemical strategy of gene expression here can also be best understood by an analysis of the balance of the activities of opposing pathways and key enzymes of synthesis and degradation.
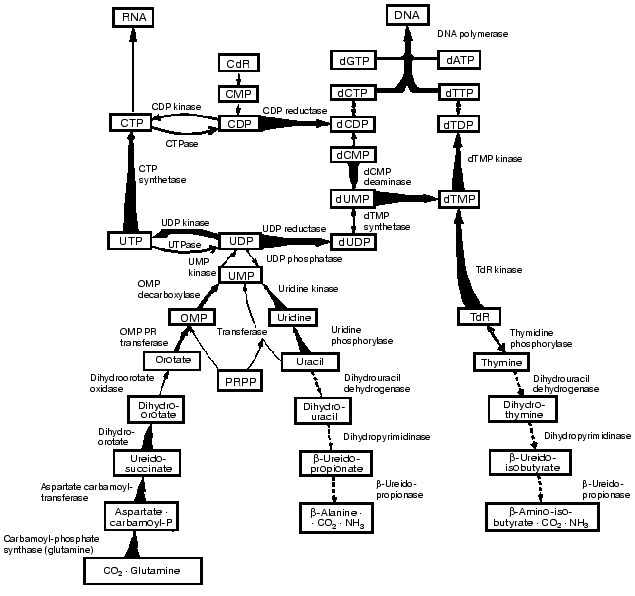
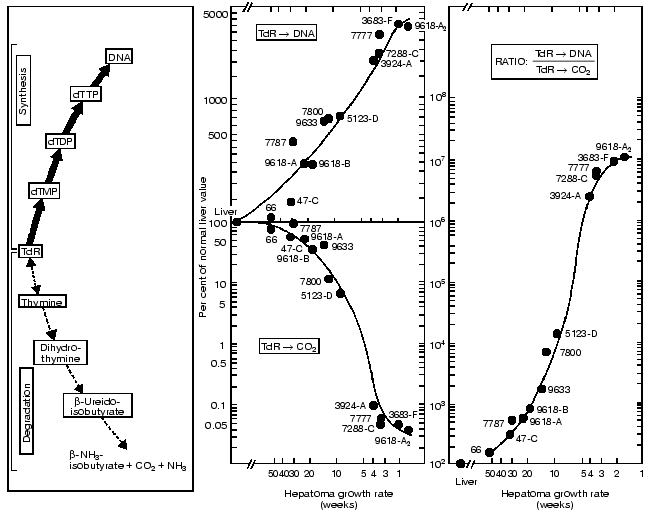
As shown in Fig. 2 the key enzymes in the de novo and salvage pathways of pyrimidine and DNA biosynthesis increase, and the opposing catabolic enzymes decrease, in parallel with tumor growth rates. In consequence, a progressively expressed imbalance emerges in the tumors between the capacities of the synthetic and catabolic enzymes and between the overall activities of the opposing pathways of anabolism and catabolism. As a result the ratios of the rate of thymidine incorporation into DNA and the rate of thymidine degradation to carbon dioxide yield the closest correlation with tumor growth rates among all the biochemical factors. The ratios can be used to measure the growth rates and malignancy of neoplasms. The studies carried out in enzyme systems were also confirmed in liver and hepatoma slices and in vivo in rats carrying subcutaneously transplanted, rapidly-growing hepatomas (Fig. 3) [6].
Fig. 2. Integrated reprogramming of gene expression manifested in the imbalance of key enzymes of pyrimidine biosynthesis and degradation and in the de novo and salvage biosynthetic pathways of DNA. Enzyme activities that are increased with growth rate are indicated by tapered thick arrows, and those that are decreased by interrupted arrows. The straight thick arrow for uridinediphosphate kinase indicates enzyme activity that is increased in all the tumors, and the dotted arrow for thymidine phosphorylase denotes an enzyme that is decreased in all the neoplasms. Arrows with normal thinness indicate that no relation was found with transformation or progression (dihydroorotate oxidase) or that the behavior has not yet been elucidated.
These observations indicate that the enzymes with the lowest activities in the liver of normal adult rats increased to the highest extent in the most rapidly growing hepatomas. By contrast, the synthetic enzymes with the highest activity in normal liver displayed the smallest extent of rise in the rapidly growing tumors. The catabolic enzymes (thymidine phosphorylase, dihydrouracil dehydrogenase and others) have much higher activities than the rate limiting synthetic enzyme of the de novo pathway (ribonucleotide reductase) and that of the final common pathway of the de novo and salvage pathways (DNA polymerase).Fig. 3. Covariance of the synthetic and catabolic utilization of thymidine (TdR) and the ratio of thymidine utilization and synthesis/thymidine degradation with malignancy (growth rate, measured in weeks required for tumor to reach a diameter of 1.5 cm) in the hepatoma spectrum.
The transformation- and progression-linked imbalance in pyrimidine and DNA metabolism provides an increased synthetic capacity for the tumor cells and the sharply curtailed catabolic potential, conferring selective reproductive advantages to the cancer cells. The increased activities of the key enzymes provide sensitive targets for chemotherapy which has been utilized in the clinical treatment of cancer patients.
PURINE METABOLISM--IMBALANCE IN CANCER CELLS
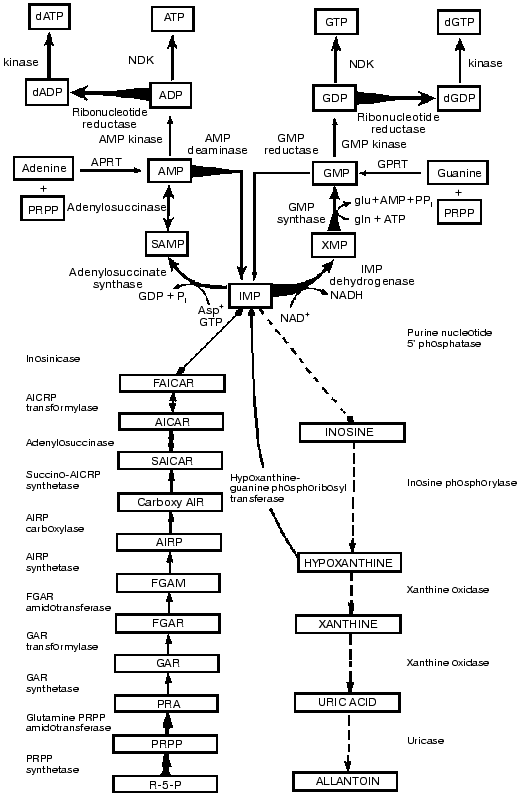
Our studies revealed that as in pyrimidine metabolism there was a clear-cut imbalance in purine metabolism in the activities of the antagonistic key enzymes and in the opposing synthetic and degradative pathways. As shown in Fig. 4 the activities of the key enzymes in the de novo and salvage pathways of purine metabolism increased and those of the opposing catabolic ones decreased in parallel with tumor growth rates. In purine metabolism as in pyrimidine metabolism, the activities of the catabolic enzymes (nucleotidase, inosine phosphorylase, xanthine oxidase, uricase) have much higher activities than those of the rate-limiting and subsequent synthetic enzymes of the de novo pathway (glutamine PRPP amidotransferase, amidophosphoribosyltransferase) [5, 6].
In the utilization of IMP, the central purine metabolite for ATP biosynthesis, the activities of the synthetic enzymes (adenylosuccinase, adenylosuccinate synthetase) are markedly elevated. In the de novo biosynthesis from IMP to GTP, the rate-limiting enzyme IMP dehydrogenase has the lowest activity among all the enzymes of purine biosynthesis. The activities of this enzyme (Fig. 4) and the subsequent GMP synthetase are markedly increased. The highest activities observed are those of ADP and GDP reductase which synthesize the dADP and dGDP leading to the production of dATP and dGTP. Since the activities of the powerful enzymes of degradation markedly declined, the imbalance favors the purine biosynthetic activities [6].Fig. 4. Imbalance in activities of key enzymes of biosynthesis, degradation and utilization of IMP in liver neoplasia. Biochemical strategy of cancer cells is revealed in the integrated reprogramming of gene expression manifested in imbalance in activities of key enzymes of biosynthesis, degradation and utilization of IMP in liver neoplasia. Arrows indicate linking of alterations in enzyme activity with transformation and growth rate as in legends to Figs. 1 to 3.
Isotope studies confirmed these enzymatic indications and studies involving freeze-clamping of tumors and HPLC assays of key metabolites revealed a progression-linked increase in the concentrations of dNTPs in hepatomas of different growth rates.
Reprogramming of gene expression in cancer cells. We provided evidence that the altered biochemical phenotype of cancer cells is based on the changed concentrations in the end products of gene expression, the amounts of specific catalytic proteins, the enzymes. These conclusions were supported by kinetic evidence which indicated that the activity of the enzyme measured under optimum substrate, co-factor and pH conditions yielding linear kinetics, was proportionate with the amount of the enzyme. A second approach showed by immunotitration that the enzyme protein amounts were altered in parallel with enzyme activities. These studies verified the changes in gene expression for 12 enzymes from various metabolic pathways [6].
Specificity to cancer cells of the enzymic and metabolic pattern. We determined that the pattern of imbalance in rat hepatoma cells is specific to neoplasia because no similar program was expressed in the developing and differentiating liver and in the regenerating and normal liver in adult rats [6].
Generalization. Our systematic studies revealed the presence of a strongly conserved program in gene expression in animal and human neoplasias: in chemically-induced, primary and transplantable rodent tumors, including carcinomas and sarcomas in rats and in mice and in myeloma, lymphosarcoma and leukemia in mice. The enzymic imbalance was also observed in a virus-induced, transplantable hepatoma in chickens. We determined the presence of the specific pattern in spontaneous human hepatocellular and renal cell carcinomas, in colon tumors of different degrees of differentiation and growth rates and in human primary lung and colon adenocarcinomas. The pattern was also detected in human leukemias and lymphomas [6].
Applicability of the molecular correlation concept and the key enzyme concept to other metabolic pathways. Recent and current studies in my laboratory showed that the two main concepts discussed for carbohydrate, purine and pyrimidine metabolism were also applicable to serine and ornithine metabolism, membrane cAMP metabolism and to the membrane signal transduction pathway of the conversion of phosphatidylinositol to inositol-1,4,5-trisphosphate (IP3) [13-19].
Application to chemotherapy. The molecular correlation concept suggested that the markedly increased activities of key enzymes should be sensitive targets for the design of chemotherapy. This approach was successfully tested in our clinical studies by targeting IMP dehydrogenase with the inhibitor drug, tiazofurin. In a study of patients with chronic myelogenous leukemia in blast crisis daily intravenous infusions of tiazofurin and administration of allopurinol by mouth resulted in 77% responses including some complete returns to the chronic phase [20]. These studies were carried out at Indiana University School of Medicine and similar trials are currently also ongoing at Houston, Boston and Little Rock., USA, and Ottawa, Canada.
Conclusions. Our introduction of the molecular correlation concept and the key enzyme concept resulted in the discovery in cancer cells of an ordered pattern of enzymic and metabolic imbalance and the elucidation of the linkage with transformation and progression. We showed that the biochemical pattern of alterations was the result of a reprogramming of gene expression that was both quantitative and qualitative. This pattern was characteristic to neoplasia, since no similar display of gene expression has been observed in any of the control normal tissues. Important stringently conserved aspects of gene logic were identified in various pathways in cancer cells. These include demonstration of operation of reciprocal control of activities of opposing key enzymes and antagonistic pathways of synthesis and catabolism. It was established that the extent of increase in activities of key enzymes related to the absolute activity of the enzymes in resting liver. The qualitative alterations in gene expression entailed the isozyme shift, particularly marked in the rapidly-growing hepatomas.
The pattern of gene expression was present in 16 different types of neoplasms including in human. With the concepts outlined above, insight was gained into strategic aspects of the biochemical commitment of cancer cells for replication. It has become possible to pinpoint the selective advantages that reprogramming of gene expression conferred to cancer cells. Understanding these biochemical alterations in cancer cells made it possible to identify sensitive targets for anticancer chemotherapy and to translate the results to the clinic.
This segment of my investigations was carried out with the assistance and collaboration of 120 scientists, medical and graduate students and technicians.
The project was supported by grant funds, primarily from the National Cancer Institute, NIH, the American Cancer Society and a Milan Panic Professorship.
REFERENCES
1.Severin, S. E., and Feigina, M. M. (1977) Advan.
Enzyme Regul., 15, 1-21.
2.Severin, S. E., and Stepanova, N. G. (1981)
Advan. Enzyme Regul., 19, 235-255.
3.Severin, S. E., Khailova, L. S., and Gomazkova, V.
S. (1986) Advan. Enzyme Regul., 25, 347-375.
4.Weber, G. (1961) Advan. Cancer Res.,
6, 403-494.
5.Weber, G. (1977) New Engl. J. Med.,
296, 486-493, 541-551.
6.Weber, G. (1983) Cancer Res., 43,
3466-3492.
7.Popper, K. R. (1968) in The Logic of Scientific
Discovery, Harper and Row, New York, p. 40.
8.Burnett, F. M. (1970) in Immunological
Surveillance, Pergamon Press, New York, p. 127.
9.Medawar, P. B. (1969) in Induction and Intuition
in Scientific Thought, Methuen and Co., London, p. 51.
10.Weber, G., and Cantero, A. (1955) Cancer
Res., 15, 105-108.
11.Warburg, O. (1956) Science, 123,
309-314.
12.Weinhouse, S. (1972) Cancer Res.,
32, 2007-2016.
13.Weber, G. (1980) in Renal Adenocarcinoma
(Sufrin, G., and Beckley, S. A., eds.) Geneva, pp. 44-50.
14.Weber, G., Burt, M. E., Jackson, R. C., Prajda,
N., Lui, M. S., and Takeda, E. (1983) Cancer Res.,
43, 1019-1023.
15.Weber, G., Hager, J. C., Lui, M. S., Prajda, N.,
Tzeng, D. Y., Jackson, R. C., Takeda, E., and Eble, J. N. (1981)
Cancer Res., 41, 854-859.
16.Weber, G., Lui, M. S., Takeda, E., and Denton, J.
E. (1980) Life Sci., 27, 793-799.
17.Williams-Ashman, H. G., Coppoc, G. L., and Weber,
G. (1972) Cancer Res., 32, 1924-1932.
18.Weber, G., Trevisani, A., and Heinrich, P. C.
(1974) Advan. Enzyme Regul., 12, 11-41.
19.Weber, G., Shen, F., Yang, H., Prajda, N., and
Li, W. (1999) Advan. Enzyme Regul., 39, 51-66.
20.Tricot, G., Jayaram, H. N., Weber, G., and
Hoffman, R. (1990) Intl. J. Cell Cloning, 8, 161-170.