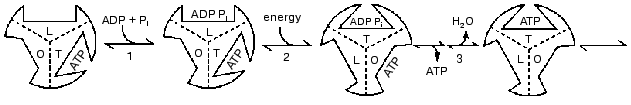Toward an Adequate Scheme for the ATP Synthase Catalysis
P. D. Boyer
Molecular Biology Institute, Boyer Hall, University of California, Los Angeles, CA 90095-1570, USA; fax: 310-206-7286; E-mail: pdboyer@ucla.edu
Received May 5, 2001
The suggestions from the author's group over the past 25 years for how steps in catalysis by ATP synthase occur are reviewed. Whether rapid ATP hydrolysis requires the binding of ATP to a second site (bi-site activation) or to a second and third site (tri-site activation) is considered. Present evidence is regarded as strongly favoring bi-site activation. Presence of nucleotides at three sites during rapid ATP hydrolysis can be largely accounted for by the retention of ADP formed and/or by the rebinding of ADP formed. Menz, Leslie and Walker ((2001) FEBS Lett., 494, 11-14) recently attained an X-ray structure of a partially closed enzyme form that binds ADP better than ATP. This accomplishment and other considerations form the base for a revised reaction sequence. Three types of catalytic sites are suggested, similar to those proposed before the X-ray data became available. During net ATP synthesis a partially closed site readily binds ADP and Pi but not ATP. At a closed site, tightly bound ADP and Pi are reversibly converted to tightly bound ATP. ATP is released from a partially closed site that can readily bind ATP or ADP. ATP hydrolysis when protonmotive force is low or lacking occurs simply by reversal of all steps with the opposite rotation of the gamma subunit. Each type of site can exist in various conformations or forms as they are interconverted during a 120° rotation. The conformational changes with the ATP synthase, including the vital change when bound ADP and Pi are converted to bound ATP, are correlated with those that occur in enzyme catalysis in general, as illustrated by recent studies of Rose with fumarase. The betaE structure of Walker's group is regarded as an unlikely, or only quite transient, intermediate. Other X-ray structures are regarded as closely resembling but not identical with certain forms participating in catalysis. Correlation of the suggested reaction scheme with other present information is considered.
KEY WORDS: adenosine triphosphate, ATP, ATP synthase, catalysis, binding change
[Footnote: It is a particular pleasure for me to contribute a paper to a volume commemorating the 100th birthday of the prominent Russian Biochemist Sergei E. Severin because three scientists trained in Russia, Vladimir Kasho, Marat Murataliev, and Yanick Milgrom, made fine contributions to the study of ATP synthase in my laboratory.]
In response to the opportunity to contribute to this memorial volume, I felt that a retrospective account of the development of some aspects of the ATP synthase mechanism might be appropriate. In particular, this brief presentation will focus on the how substrate binding, interconversion, and release steps are coordinated with conformational changes of the multiple catalytic sites. I will attempt to provide an account of how increasing information and evaluations over the past 25 years have led to changes in concepts of how the catalytic steps may occur. The contributions of many workers have led to the present understanding of the unusual ATP synthase catalysis, and it has been a privilege to participate in the accomplishments of which the field of bioenergetics can be proud. However, considerable uncertainty remains about the conformational forms that participate. The changing views of one investigator, as presented herein, including an important recommended revision of a previous scheme, may help to attain a better understanding of this remarkable enzyme.
THE ORIGINAL SCHEMES
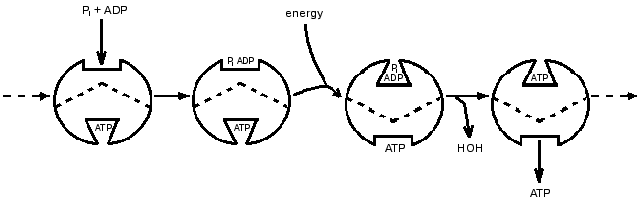
That the ATP synthase catalysis might have features not shown by most enzymes seemed likely in the 1970s when its unusual subunit composition began to be revealed. The remarkable versatility of proteins expressed through conformational changes was becoming more apparent. Accompanying these developments, probes by my group led to a proposal that a principal function of energy input was to bring about release of a tightly bound ATP by energy-driven conformational changes. Characteristics of isotope exchanges accompanying the synthesis of ATP could be explained if one catalytic site on the enzyme could not proceed with ATP release until substrate was available to bind at another catalytic site. At that time presence of only two catalytic sites on the enzyme seemed probable, and a 1977 presentation in the Annual Review of Biochemistry included a scheme (Fig. 1) in which two sites were depicted as proceeding in sequence through catalytic steps of binding, interconversion and release [1]. Soon thereafter the presence of three copies each of major subunits in the separated F1-ATPase became established, and the participation of three sites in sequence seemed logical. A three-site scheme presented by Cross in the 1981 Annual Review of Biochemistry [2] and shown in Fig. 2 became widely recognized.
Fig. 1. A 1977 depiction of alternating participation of two catalytic sites [1].
Fig. 2. A 1981 3-site model for the binding change mechanism [2].
ROTATIONAL CATALYSIS
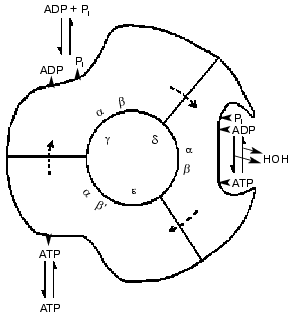
Advances in knowledge of the structure of the enzyme revealed by electron microscopy and chemical probes led to suggestions that the six large alpha and beta subunits were arranged alternately and circularly around a core of single-copy subunits. The three beta subunits had different chemical properties, as if they were in different conformations. Catalytic steps were modified by changes in the single copy gamma subunit. Such results, and the revelation from 18O exchange studies that all three sites conducted catalysis identically led to a 1981 model for a rotational catalysis (Fig. 3) in which rotation of an inner core is regarded as causing requisite binding changes in catalytic sites [3]. The model of Fig. 3 suggests that the asymmetry of the inner core arises from the presence of the three minor gamma, delta, and epsilon subunits; as now known, the gamma subunit alone serves this function. The presentation of the model was accompanied by the statement, “The figure suggests an additional stage that we have not considered before, namely that the conformation following the release of loosely bound ATP undergoes further change to better accommodate binding of ADP and Pi”. Although this seemed to be a desirable capacity, a number of years elapsed before experimental evidence for a conformation with preferential binding of ADP was attained.
Fig. 3. A 1981 model for rotational catalysis [3].
DEVELOPMENTS BY 1994
Although the occurrence of rotational catalysis remained controversial, to me the evidence as presented in my 1993 review [4] made such catalysis highly likely. By then other puzzling aspects of the catalysis had been clarified. The major characteristics of the important MgADP inhibition that has interfered in the interpretation of many studies had been revealed, mostly from contributions from the laboratories of Vinogradov, Boyer, and Allison. This included influences of the noncatalytic nucleotide binding sites on the inhibition, first detected by Milgrom and associates [5]. An important consequence was that a better evaluation could now be made of the concentration-velocity relationships during ATP hydrolysis. However, as discussed in a later section, ATP-velocity relationships still remain controversial in regard to whether binding of ATP to a second or to a second and third site is necessary to achieve near maximal hydrolysis rates.
A related concern is how many sites need to be filled with ADP for rapid net synthesis to occur. In our studies of photophosphorylation we found that a rapid rate of synthesis results when ADP binds to a second catalytic site [6]. As given later, my evaluation of these and related studies is that rapid net ATP photophosphorylation results when a second catalytic site fills with ADP and Pi. Similar bi-site activation would be expected for the hydrolysis of ATP, and the results with ADP have underlain my skepticism about suggestions that, in addition to nucleotides at a tight binding site, a second site and a third site must bind ATP for rapid ATPase activity.
Another result important for model considerations also came from our photophosphorylation studies. We made use of a hexokinase-accessibility procedure to determine the number of catalytic sites filled with ATP during net photophosphorylation. This gave evidence that adequate protonmotive force could shift the quasi-equilibrium at the tight catalytic site toward ATP. An attractive possibility was that after ADP bound, the next binding change step could give rise to both a catalytic capacity for conversion to ATP and an increased preference for ATP formation. Then the following binding change step could release ATP and not ADP from the tight site. A reaction scheme quite compatible with that of Fig. 3 was presented [6]. At this time exploration of the characteristics of the hydrolysis of trinitrophenyl phosphate-ATP (TNP-ATP) by Murataliev in my laboratory gave unexpected support for the presence of a form of the F1-ATPase that more readily bound ADP than ATP [7]. Evidence was obtained that TNP-ATP could readily enter the catalytic cycle for hydrolysis after two sites were already filled. ATP bound poorly to this third site, but ADP was a very effective inhibitor of the TNP-ATP binding. The possibilities of the participation of a form that preferentially binds ADP, and of a shift toward ATP at the tight sight, are explored further in the preferred scheme given later in this paper.
THE IMPACT OF X-RAY STRUCTURE AND ROTATIONAL DATA
In 1994 the three-dimensional structure of the major portion of the mitochondrial F1-ATPase was presented by the Walker group [8]. In 1995 Cross and associates demonstrated that catalysis caused rotational shifts between radio-labeled gamma and beta subunits [9]. In 1996 Junge and associates used polarized absorption relaxation to show intersubunit rotation [10]. In 1997 Yoshida and colleagues reported a striking visual demonstration of rotation by the ATPase [11]. These findings strongly supported the main features of the binding change mechanism and rotational catalysis. The reports and subsequent contributions have brought a new level of understanding and experimental design to the field. But the results have left considerable uncertainty about the patterns of catalytic site filling and the forms participating in the catalytic sequence.
In the Walker structure [8] one site, likely in the MgADP inhibited form, is regarded as representing the tight binding site where covalent catalysis occurs, and is designated as betaDP. Another form with imido-ATP present without covalent catalytic capacity is designated as betaTP. A third form without nucleotide present is designated as betaE. A reasonable assumption appeared to be that these forms were the equivalent of the tight (T), loose (L) and open (O) forms, respectfully, as suggested by Cross [2] for a binding change model in Fig. 2. In many subsequent reaction schemes the betaE form has been regarded as the initial site of binding of ADP for synthesis and ATP for hydrolysis. This has always seemed unlikely to me because ATP could then readily prevent ADP from entering a synthesis cycle.
Importantly, the direction of rotation observed by the Japanese group [11] indicated that if ADP and Pi entered the cycle by binding at the betaE form, then in the next binding change the site could be converted to a betaDP form where conversion to ATP could occur. This seemed reasonable to me, particularly if the betaE form would bind ADP more readily than ATP. If for ATP hydrolysis the ATP bound initially to a betaE form, then two binding changes would seem to be needed for the site to reach the betaDP form where hydrolysis could occur. Because of this and other reasons, attainment of satisfactory steps for ATP binding and subsequent events has been elusive. The number of sites that need to be filled for rapid ATP hydrolysis to occur remains controversial.
THE BI-SITE OR TRI-SITE ACTIVATION UNCERTAINTY
An unsettled and important aspect of the catalysis has centered on whether, after the initial very tight binding site is filled, rapid catalysis requires the binding of ATP at only one additional site (bi-site activation) or at two additional sites (tri-site activation). A valuable way to make direct measurements of nucleotide binding at catalytic sites was introduced by Senior and colleagues [12, 13]. By mutagenesis of the E. coli enzyme they put a tryptophan in the place of the catalytic-site tyrosine that would be adjacent to the adenine ring when nucleotide binds. The fluorescence of this tryptophan is quenched when nucleotide binds, providing a measure of site occupancy. Replacement of tryptophans at other locations in the enzyme enhanced specificity of the assay. From application of this approach they concluded that an enzyme with bound nucleotide at a tight site attained rapid hydrolysis only after a second and third site bound ATP [12, 13]. In contrast, my group felt that adequate kinetic evaluations of the dependency of velocity on ATP concentration show only one apparent Km value from below micromolar to millimolar concentrations [13], consistent with bi-site activation. Reasons why the fluorescence measurements might not give a satisfactory measure of site filling as related to catalytic rate were thus considered. The possibility that the presence of considerable MgADP inhibited enzyme or epsilon subunit inhibited enzyme resulted in nucleotide binding without accompanying activity [14, 15] was made unlikely by additional studies in the laboratories of Allison [16] and of Senior [17]. But problems still remained.
Adequate measurement of ATP binding by the fluorescence technique when catalysis is occurring is inherently difficult. Relatively high concentrations of enzyme are required and this results in hydrolysis of much of the added ATP during the time period that measurements are made. Binding of the ADP formed may readily occur. Indeed, the fluorescence technique appears to provide good measurements of ADP binding when catalysis is not occurring. Results show that in the presence of Mg2+ three ADPs are readily bound, one very tightly and the last two with Kd values in the range of 20-30 µM [12, 13]. This relatively high affinity for ADP means that the binding of ADP could cause considerable interference during the attempted assay of ATP binding.
Not only the rebinding of ADP may occur, but the departure of ADP may be a slow step in the catalysis. Such “sticky ADP” means that as ATP concentration is increased, soon ATP will be binding to enzyme that still retains an ADP. Thus as ATP concentration is increased and bi-site activation gives rapid hydrolysis, three sites on the enzyme will tend to be filled. The interference in the fluorescence assay by the slow release or rebinding of ADP, rather than tri-site activation, can account for the observation that during rapid ATP hydrolysis the enzyme has about two ADP and only one ATP bound [18]. How catalytic sites can tend to retain ADP that is formed during net ATP hydrolysis is considered later in a section on “ATP Synthesis and Hydrolysis” by a revised reaction pathway.
The probability of bi-site activation during ATP hydrolysis has gained unexpected support from another type of observation--the visual measurements of rotation accompanying ATP binding. At low ATP concentrations, where at most the one very tight binding site on the enzyme is filled, binding of an ATP results in a one-step rotational movement that is as rapid as the rotational steps observed at high ATP concentrations [11, 19].
Reaction schemes for ATP hydrolysis based on a tri-site activation have been suggested by Weber and Senior [20], by Allison [21], and by Leslie and Walker [22]. In these schemes an ATP molecule that binds does not undergo hydrolysis until another ATP binds and another rotation step has occurred or is occurring. Also, ATP hydrolysis rather than ATP binding is invoked as a principal driving force for rotation. It is recognized [20] that, at high ATP concentrations, ADP departure may by rate limiting. What is not recognized is that the essential features of bi-site activation, ATP binding and immediate rotational change, can occur whether or not a third site is occupied by ADP. An important feature they adequately justify is the suggestion that a betaE form is only a transitory intermediate. Suggestions of different pathways for synthesis and hydrolysis [20, 21] detract from the schemes. Acceptance of bi-site activation whether or not a third site has ADP present would make the schemes [20-22] unnecessary and would provide simpler schemes without objections such as mentioned above.
A REACTION PATHWAY WITH O, T, AND L FORMS
In the year 2000 I presented a scheme shown in Fig. 4 in which 120° rotational changes are correlated with changes in catalytic site forms [23]. The scheme accounts for bi-site activation and for the participation of a form that binds ADP better than ATP. But it retains the designation of O, T, and L forms (equivalent to the betaE, betaDP, and betaTP forms) as major forms that are interchanged by each 120° binding change. As noted in the preceding sections, recent data and further considerations point to the need for changes in the scheme of Fig. 4. It seems likely that the betaE form may not participate or at most is a quite transient intermediate. Similarly, the structures found by the X-ray analyses may not arise as such during actual catalysis, but are related to the forms that occur. The forms that are suggested in a revised scheme presented later are of three different types; tight binding forms where covalent changes occur, forms related to a site that binds ADP for synthesis, and forms related to a site that releases ATP during synthesis and binds ATP during hydrolysis. Before consideration of the properties of these forms and their possible relations to X-ray structures, I would like to call attention to some under-appreciated studies with fumarase that illustrate the types of forms that must be considered even in simple, single catalytic site enzymes.
Fig. 4. A reaction pathway that includes betaE, betaDP, and betaTP as major forms [22].
SUBSTRATES, PRODUCTS, AND THEIR INTERCONVERSION IN ENZYMIC CATALYSIS
Consideration of some important general properties of enzymes may contribute to a better understanding of the ATP synthase. Irwin Rose recently published papers on interconversion of malate and fumarate that are relevant to the mechanism of enzyme catalysis [24, 25] [Footnote: The yeast fumarase used by Rose was provided by Dr. J. Keruchenko of the Institute of Biochemistry, Russian Academy of Sciences, Moscow]. Rose considers that in catalysis by enzymes a conformational change in the enzyme accompanies the chemical transition from substrate to product, and the substrate and product binding and release steps. He demonstrates that such transitions occur with the enzyme fumarase. For example, he shows that malate combines with a conformation of the enzyme that accommodates malate better than fumarate and as the chemical transition occurs a form that accommodates fumarate better arises. The fumarase must now make a transition to the malate-preferring form for the net catalysis to proceed. The message emphasized by the fumarase results is that the minimum scheme that needs to be considered for catalysis by an enzyme is as follows:
Eo + S <--> ESS <--> EPP <--> E´ + P, (1)
where Eo is a form that has a better fit to S than P, and changes its conformation to Es when it combines with S. The conversion to product P is accompanied by a conformational change of the enzyme to EP that has a better fit to P than S. A further conformational change occurs when P dissociates to give a form E´. For catalysis to continue E´ must convert back to Eo. Such interconversion takes place in the absence of bound S or P. The four minimum required conformational changes may be accompanied by protonation or deprotonation of enzyme groups, or with shifts of water or solute molecules that influence catalytic steps. For many enzymes, conformational and other changes may not appear as detectable stages in the overall catalysis. For example, conversion of the E´ to the Eo form may take place as, or very rapidly after, P is released. Conformational changes following substrate binding may be small, or (as for ATP binding to ATP synthase) quite large. A key component is that the chemical interconversion step is accompanied by a conformational change--the substrate is taken on a guided tour through the transition state. This conformational adaptability is made possible by the wondrous properties of enzymes. Hail to proteins.
For the more complicated ATP synthase more than four conformational forms must participate in catalysis. Suggested forms are given in the following sections.
A REVISED REACTION SCHEME FOR THE ATP SYNTHASE CATALYSIS
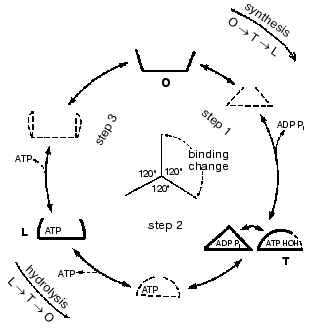
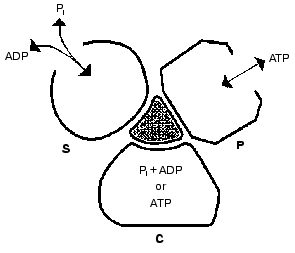
Important support for a revised reaction pathway was provided by the recent report of Menz, Leslie, and Walker [26] giving the X-ray structure for the F1-ATPase with two sites filled with nucleotide-aluminum fluoride complexes and a third site filled with ADP. They consider that this third “half-closed” ADP site and a related “half-closed” site for ATP participate in the catalytic sequence. Importantly, the structure of the ADP binding site was such that it would be expected to have low affinity for ATP. Their accomplishment provides proof for the existence of, and the structure for, a type of site that had been postulated as advantageous for the synthesis of ATP [3, 6] and for which we had obtained some experimental evidence [7]. The proposed reaction pathway involves three types of sites that each beta subunit must pass through as net catalysis proceeds. These are a site at which ADP and Pi must be present for net ATP synthesis to occur, a tight-binding site where interconversion of ADP and Pi to ATP is catalyzed, and a site at which ATP must be present for net ATP hydrolysis to occur. The suggested participating types of sites and some of their properties are given in Fig. 5 and the accompanying legend.
Present evidence suggests additional important properties for the three types of sites. The S site readily binds ADP but not ATP, the C site can tightly bind either ADP and Pi or ATP, and the P site has good affinity for ADP as well as ATP. The three sites can assume intermediate conformations as catalysis proceeds, including different forms depending on whether or not reactants are bound and the relative positions of the beta and gamma subunits. There is thus an abundance of forms, some more transitory than others, that must be considered as possible intermediate states during catalysis. The X-ray structures provide an important reference as to the nature of the conformations that may be attained but must of necessity be stable structures. It is thus not justified to propose the betaE, betaDP, and betaTP structures as intermediate catalytic forms but they may be closely related to participating forms. Relations that might exist between X-ray structures and participating forms are mentioned in the following sections.Fig. 5. A depiction of the three major site types that all catalytic sites must pass through sequentially. The type S site readily binds the substrates ADP and Pi. The type C site (for catalysis) tightly binds either ADP and Pi or ATP and catalyzes their interconversion. The type P site readily dissociates the product ATP, or binds ATP for hydrolysis. Each type of site may attain two or more different conformations (forms) and must pass through intermediate conformations as sites undergo binding changes and rotation of the gamma subunit (shaded) occurs.
The minimal forms that arise in enzyme catalysis as shown by the studies with fumarase mentioned earlier were indicated in Eq. (1). For the ATP synthase more than four forms must be considered but they will include the four forms represented by Eq. (1). Thus the empty S-type site (Fig. 5) would be form Eo, and form ESS would arise when ADP and Pi bind and the transition to a C-type site occurred. The form ESS would be converted to EPP. ATP departure would give rise to the E´ form like the empty P-type site. The E´ to Eo transition would represent the P to S-type site transition. Each of the three catalytic sites would go through these and any other intermediate forms sequentially.
THE BINDING OF ADP AND Pi
For simplicity, the binding of ADP and Pi are indicated as single steps in most reaction schemes and in Fig. 5. The ADP-binding form observed in the X-ray structure has a sulfate ion bound [26] and this indicates a capacity for binding of Pi as well as ADP. Small or extensive conformational changes undoubtedly occur when ADP and Pi bind. The form after ADP and Pi bind is likely closely related to the ADP-binding form reported recently by Menz et al. [26]. Conformational signals accompanying the binding of ADP and Pi must be necessary to allow rotational catalysis to proceed. The binding of ADP appears to be particularly important in this regard as it allows a shift of reactants at the tight C site toward ATP formation [6]. Conformational changes also increase the affinity of Pi. That Pi binding is promoted by an energy input was noted with the mitochondrial enzyme many years ago [27] and has been substantiated by later studies with the chloroplast [28] and E. coli enzymes [29]. As mentioned earlier, direct ADP binding measurements show that the onset of rapid photophosphorylation with excess Pi present occurs when ADP binds to a second catalytic site [6], as expected for bi-site activation [Footnote: Although these studies were limited, it is an oversight for a recent review [20] to state “Direct MgADP binding measurements in the presence of a proton gradient have not been reported.” Also, the direct measurement of catalytic site ATP by a hexokinase accessibility assay [6, 32] is overlooked]. These studies also show the promotion of ADP binding by protonmotive force during photophosphorylation. Kinetic studies of Kayalar et al. [30] with the mitochondrial enzyme and of Perez and Ferguson [31] with the Paracoccus denitrificans enzyme give evidence that the binding of ADP and Pi is random. The study by Perez and Ferguson was particularly well designed and extensive. It includes convincing demonstration that two Km values for ADP can be observed, one well below micromolar for filling the very tight site, and only one at micromolar concentrations consistent with bi-site activation for ADP binding. A similar behavior was observed earlier in my laboratory for the chloroplast enzyme [33] but deviations from strict Michaelis-Menten behavior were noted in later studies that may have arisen from changes in protonmotive force as ADP concentrations were increased (unpublished observations). Richard and Gräber used special techniques to keep the protonmotive force constant during photophosphorylation. They found only a single Km in the micromolar range [34]. Such results give added support to bi-site activation.
ATP SYNTHESIS AND HYDROLYSIS
The conformation after ADP and Pi bind must undergo further changes as rotation of the gamma subunit occurs and a C-type site arises in a conformation with tightly bound ADP and Pi present. During net ATP synthesis this form, as reactants pass through the transition state, is now converted to a form with preferential ATP binding. These two C-site type forms are likely closely related to the betaDP form of the Walker structure. As rotational catalysis continues, the C site with tightly bound ATP present then undergoes the vital conformational change that allows the P-type site to arise. This site is akin to a half-closed ATP site postulated by Menz et al. [26] as an intermediate stage but for which a related X-ray structure is not available. The betaTP form may be like an intermediate in the C- to P-type transition or may have features that the half-closed ATP form (a P type form) assumes when ATP is bound. During net ATP synthesis the ATP may be released at this stage or, as noted in the following paragraph, during subsequent conformational changes accompanying the P to S transition. In each 120° rotational change all three sites must of course undergo coordinated conformational changes and at any one time are in different conformations. The nature of the forms involved in the P to S transition remains to be clarified. It would seem that only minor changes in the half-closed ADP-binding form of Menz et al. [26] would be required for the site to gain an ATP binding capacity. Thus a form like a betaE may not be even a transitory intermediate. It is possible though, that in net ATP synthesis the appearance of a betaE-related form could help by promoting ATP dissociation. Alternatively, such assistance might also result because as the half-closed ADP-binding form is arising, its lack of capacity for ATP binding could hasten departure of any ATP that remained. The essential requirements for rotationally-driven net synthesis of ATP are adequate protonmotive force and the presence of ADP and Pi at an S site. Rapid net ATP formation can likely proceed whether or not the P site has bound ATP or ADP present, although nucleotide occupancy of the P site might have some effect on reaction rate. Also, it seems possible that the initial ADP binding could sometimes be at a P-type site, with the ADP remaining as S-type site arises (if a form like a betaE is not an intermediate in the P to S transition). Clever experimentation will hopefully help supply more details about such matters.
In net ATP hydrolysis in the absence of a protonmotive force, reaction steps and rotational changes are reversed. At lower ATP concentrations ADP and Pi formed in a previous binding change will have dissociated from the S site and both the S and P sites will be empty. As ATP concentration and hydrolysis rates increase, the ADP dissociation may be from an S-type site or later following the S to P conversion if a form like betaE is not a required intermediate. The ATP binding to the P site causes conformational changes that drive rotation and conformational changes that favor the presence of ADP and Pi at the C site. Thus when the binding change is completed ADP and Pi, and not ATP, are released. At lower concentrations of ATP there is time for ADP to dissociate from the S site and hydrolysis would continue when another ATP binds to a P site. Only one or two sites would be filled during steady state catalysis at lower ATP concentrations. Even at higher ATP concentrations that suffice for near maximal velocity, the ATP would not bind to an S site even if it is empty. But it would bind to an empty P site as soon as it became available. ADP release is a likely slow step in the catalysis, and if any ADP remained at the S or the P site, nearly three catalytic sites would readily become filled. Even if the release of ADP is not a slow step, the data from Senior's group shows that ADP very readily binds to all three sites on the enzyme. Thus the binding of medium ADP at both the S and P site could tend to keep nearly three sites filled as maximal ATP hydrolysis rates are approached. About one bound ATP and two bound ADP are present as maximal hydrolysis rates are attained [18]. The total of about one bound ATP may reflect that present both at a P and a C-type site.
The important point, as mentioned earlier, is that at higher ATP concentrations nearly three catalytic sites will be filled, although the essential step for rapid catalysis is the binding of ATP to a P-type site.
COMPATIBILITY WITH OTHER DATA
The reaction sequences proposed above appear to be compatible with a wide range of present data. For example, they can readily account for the fluorescence measurements of ADP binding in presence of Mg2+ and of site filling during rapid catalysis. When ADP binding in presence of Mg2+ is measured, the MgADP-inhibited betaDP form of the enzyme is probably present. And, as mentioned above, the other two catalytic sites readily bind ADP. As noted by Weber and Senior [20] this gives evidence that no betaE site is present in the enzyme as tested and that, besides the betaDP site, there are two additional sites present that can readily and reversibly bind ADP. These could be the S and P forms of Fig. 5. The betaE form likely arises as a stable form in the enzyme as used for the X-ray studies from the conformational changes produced when a first site is blocked by inhibitory MgADP and a second site binds imido-ATP. As noted recently by Menz, Leslie, and Walker, with the MgADP-inhibited enzyme the betaE site remains empty even in the presence of 5 mM imido-ATP [35]. The catalytic sites of the uninhibited enzyme used in the studies by Senior and associates seem to be more closely related to those during actual catalysis and have little or no betaE conformation present.
The ADP binding data support another suggested property. This is that form P of Fig. 5 that can bind ATP must also be able to bind ADP. Otherwise three sites would not fill at relatively low ADP concentrations. A desirable aspect of the new scheme is that the participation of a site that binds ADP better than ATP nicely satisfies a metabolic need of being able to synthesize ATP with relatively low cellular ADP and high ATP concentrations. Nature has developed a clever way so that the ADP binding for oxidative and photophosphorylation can readily occur even in the presence of otherwise overwhelming ATP. ADP and ATP do not compete for the same binding form and the energy-linked rotation assures that the form preferentially binding ADP will arise. In addition, as noted previously, the transition from the P to the S form during net ATP synthesis could promote release of any remaining ATP from the changing P site. The ability of ADP to readily bind to the P form could explain why ADP is a potent inhibitor of ATP hydrolysis even though ATP is a poor inhibitor of synthesis.
Reaction patterns based on Fig. 5 have an attractive symmetry of ADP or ATP addition. Both participate in bi-site activation for synthesis or hydrolysis by combining just one binding change step away from being brought into the chemical interconversion stage. The steps in the scheme are readily reversible, and with only the change in rotational direction determining whether synthesis or hydrolysis occurs. This is what would be expected for a reversible catalysis by a single enzyme. That separate pathways do not exist seems likely from the observation that even under conditions of rapid ATP synthesis in mitochondria the 18O and 32P exchange data show that rapid reversal of synthesis steps is occurring [36]. The situation is akin to the rapid glycolysis that can occur with the participating enzyme reactions not far from equilibrium. The net forward reaction is accompanied by a considerable flux in the reverse direction readily detected by isotopic labels.
CORRELATION WITH OTHER OBSERVATIONS
In a continuation of their innovative experimentation, Yoshida and Kinosita and colleagues have measured rotational steps with enzyme preparations not slowed or damped by a long actin filament. They measured rotation by oblique attachment of a small gold bead, and use of a microscope equipped with a laser beam and a camera device capable of recording up to 8,000 frames/sec. They demonstrated that the 120° rotation step consists of about 90° and 30° substeps, each taking only a fraction of a millisecond. The ~90° substep is driven by ATP binding, followed by two ~1-msec steps, then an ~30° rotational step [19]. They regard their observations as quite consistent with bi-site activation and present a reaction scheme largely compatible with that suggested herein. Uncertainty remains as to what changes are occurring in the about 2 msec dwell time between rotational movements. Yasuda et al. [19] indicate that the one step could logically be the chemical change, with product release steps also involved. In terms of the reaction pattern suggested here the 90° step might occur after the initial ATP binding and a subsequent conversion of the P-type site to a C-type site. The 30°step could result from the C-type to the S-type site transition. Further studies will hopefully give needed clarification.
In their valuable modeling of the ATPase reaction Oster and Wang [37] suggest that ATP binding energy triggers elastic bending in the catalytic beta subunit, which then torques the gamma subunit. Although they visualize ATP binding to a subunit in the betaE conformation, their treatment likely can adapt to a binding to the half-closed P-type site. Similarly, the catalytic pathway suggested herein would seem to be adaptable to the theoretical considerations of Junge and coworkers [38] and of others. There is obviously much yet to learn, and what is suggested here needs more appraisal and experimental evidence. It is likely now apparent to the reader why the phrase “Toward an Adequate Concept.” was included in the title. Hopefully, the suggestions made over the years are moving in the right direction. The reaction pattern presented here has to me the type of “feeling right” that accompanied my initial suggestion that energy served primarily for release of a tightly bound ATP. Time will tell.
REFERENCES
1.Boyer, P. D. (1977) Annu. Rev. Biochem.,
46, 957-966.
2.Cross, R. L. (1981) Annu. Rev. Biochem.,
50, 681-714.
3.Boyer, P. D., and Kohlbrenner, W. E. (1981) in
Energy Coupling in Photosynthesis (Selman, B., and
Selman-Reiner, S., eds.) Elsevier/North Holland, NY, pp. 231-240.
4.Boyer, P. D. (1993) Biochim. Biophys. Acta,
1140, 215-250.
5.Milgrom, M. M., Ehler, L. L., and Boyer, P. D.
(1990) J. Biol. Chem., 265, 18725-18728.
6.Zhou, J.-M., and Boyer, P. D. (1993) J. Biol.
Chem., 268, 1531-1538.
7.Murataliev, M. B., and Boyer, P. D. (1994) J.
Biol. Chem., 269, 15431-15439.
8.Abrahams, J. P., Leslie, A. G., Lutter, R., and
Walker, J. E. (1994) Nature, 370, 621-628.
9.Duncan, T. M., Bulygin, V. V., Hutcheon, M. S., and
Cross, R. L. (1995) Proc. Natl. Acad. Sci. USA, 92,
10964-10968.
10.Sabbert, D., Engelbrecht, S., and Junge, W.
(1996) Nature, 381, 623-625.
11.Noji, H., Yasuda, R., Yoshida, M., and Kinosita,
K., Jr. (1997) Nature, 386, 299-302.
12.Weber, J., Wilke-Mounts, S., Lee, R. S.-F.,
Grell, E., and Senior, A. E. (1993) J. Biol. Chem., 268,
20126-20133.
13.Weber, J., and Senior, A. E. (1997) Biochim.
Biophys. Acta, 1319, 19-58.
14.Milgrom, Y. M., Murataliev, M. B., and Boyer, P.
D. (1998) Biochem. J., 330, 1037-1043.
15.Boyer, P. D. (1997) Annu. Rev. Biochem.,
66, 717-749.
16.Dou, C., Fortes, P. A., and Allison, W. S. (1998)
Biochemistry, 37, 16757-16764.
17.Weber, J., Dunn, S. D., and Senior, A. E. (1999)
J. Biol. Chem., 274, 19124-19128.
18.Weber, J., Bowman, C., and Senior, A. E. (1996)
J. Biol. Chem., 271, 18711-18718.
19.Yasuda, R., Noji, H., Yoshida, M., Kinosita, K.,
and Hiroyasu, I. (2001) Nature, in press.
20.Weber, J., and Senior, A. E. (2000) Biochim.
Biophys. Acta, 1458, 300-309.
21.Allison, W. S. (1998) Accts. Chem. Res.,
31, 819-826.
22.Leslie, A. G. W., and Walker, J. E. (2000)
Phil. Trans. R. Soc. Lond. B, 355, 465-472.
23.Boyer, P. D. (2000) Biochim. Biophys.
Acta, 1458, 252-262.
24.Rose, I. A. (1997) Biochemistry,
36, 12346-12354.
25.Rose, I. A. (1998) Biochemistry,
37, 17651-17658.
26.Menz, R. I., Leslie, A. G. W., and Walker, J. E.
(2001) Cell, in press.
27.Rosing, J., Kayalar, C., and Boyer, P. D. (1977)
J. Biol. Chem., 252, 2478-2485.
28.Feldman, R. I., and Sigman, D. S. (1983) J.
Biol. Chem., 252, 12178-12183.
29.Al-Shawi, M. K., Parsonage, D., and Senior, A. E.
(1990) J. Biol. Chem., 265, 4402-4410.
30.Kayalar, C., Rosing, J., and Boyer, P. D. (1977)
J. Biol. Chem., 252, 2486-2491.
31.Perez, J. A., and Ferguson, S. J. (1990)
Biochemistry, 29, 10503-10518.
32.Rosen, G., Gresser, M., Vinkler, C., and Boyer,
P. D. (1979) J. Biol. Chem., 254, 10654-10661.
33.Stroop, S. D., and Boyer, P. D. (1985)
Biochemistry, 24, 2304-2310.
34.Richard, P., and Gräber, P. (1992) Eur.
J. Biochem., 210, 287-291.
35.Menz, R. I., Leslie, A. G. W., and Walker, J. E.
(2001) FEBS Letts., 494, 11-14.
36.Berkich, D. A., Williams, G. D., Masiakos, P. T.,
Smith, M. B., Boyer, P. D., and LaNoue, K. F. (1991) J. Biol.
Chem., 266, 123-129.
37.Wang, H., and Oster, G. (1998) Nature,
396, 279-282.
38.Cherepanov, D. A., Mulkidjanian, A. Y., and
Junge, W. (1999) FEBS Letts., 449, 1-6.